TL;DR#
Current unified vision-language models struggle with medical data due to limited data scale and quality and conflicts between comprehension and generation tasks. These models often excel at one task at the expense of the other. This necessitates a paradigm shift in the way we approach multi-modal learning in the medical domain.
The researchers introduce HealthGPT, which uses a novel Heterogeneous Low-Rank Adaptation (H-LoRA) technique. H-LORA efficiently manages the complexities of multiple tasks by separating the learning processes for comprehension and generation. Combined with a hierarchical visual perception approach and a three-stage training strategy, HealthGPT achieves state-of-the-art results on diverse medical visual comprehension and generation tasks. The researchers also contribute a new dataset (VL-Health) specifically designed for training such models.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical AI and vision-language modeling. It addresses the critical challenge of unifying comprehension and generation tasks in medical visual data, a significant limitation of current models. By introducing a novel approach and dataset, it paves the way for more capable and versatile medical AI systems with wider applications in diagnostics, treatment planning, and education. Its innovative H-LoRA technique could also inspire improvements in other multi-modal learning scenarios.
Visual Insights#

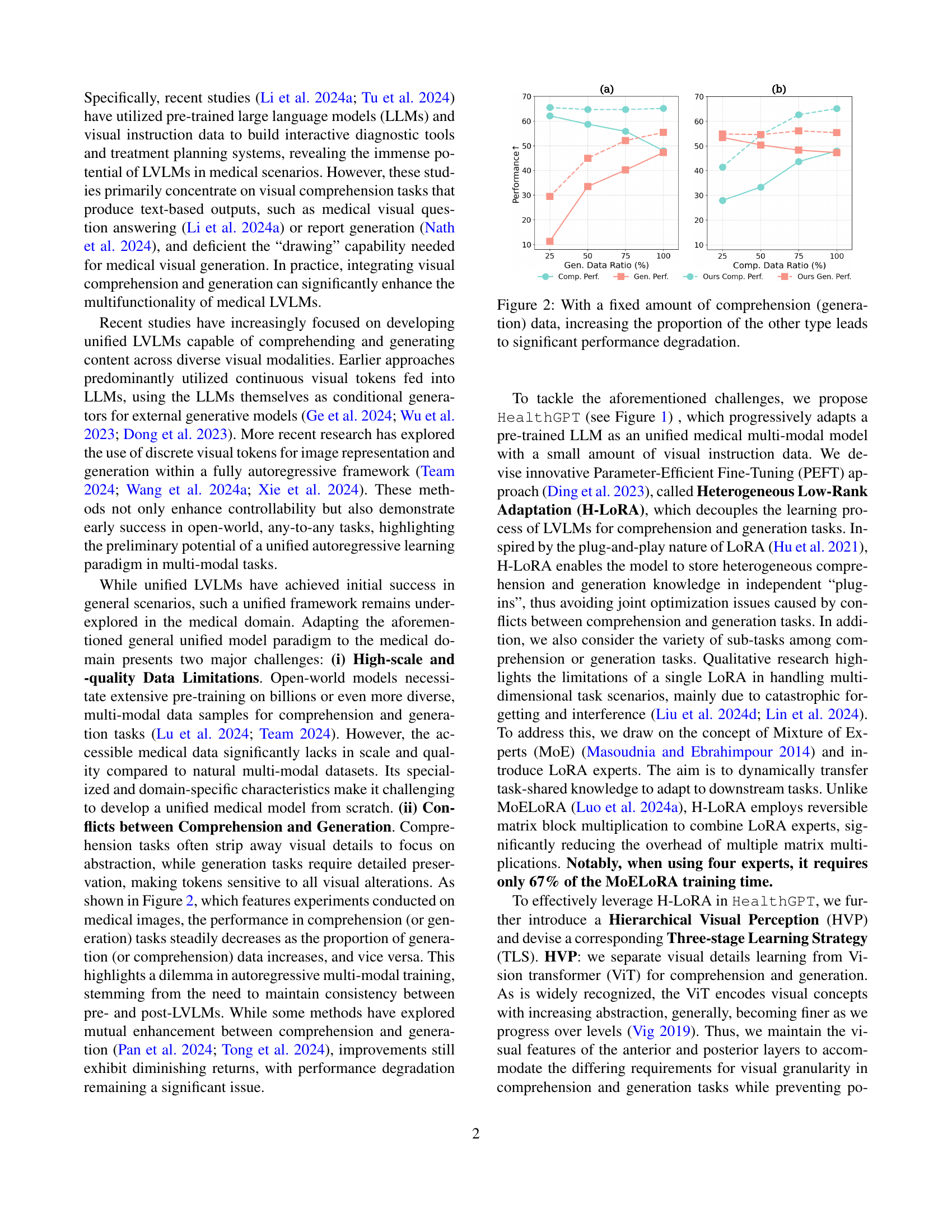
🔼 This figure showcases HealthGPT’s capabilities in handling medical multi-modal tasks, including both comprehension and generation. It demonstrates superior performance compared to other state-of-the-art models, both general-purpose unified visual models and those specifically designed for medical applications. The results displayed highlight HealthGPT’s ability to effectively address complex medical tasks. The metrics ‘Comp. Perf.’ and ‘Gen. Perf.’ represent the performance scores achieved in the comprehension and generation tasks, respectively. Various example tasks are shown, emphasizing the wide range of capabilities.
read the caption
Figure 1: HealthGPT enables medical multi-modal comprehension and generation, outperforming both state-of-the-art unified visual models and medical-specific models across various tasks. This highlights its superior capability in tackling complex tasks in healthcare applications. Comp.Perf. and Gen.Perf. denote the results of comprehension and generation.
| VQA-RAD ↑ | SLAKE ↑ | PathVQA ↑ | ||||||||||
| Type | Model | # Params | Medical LVLM | close | all | close | all | close | all | MMMU -Med ↑ | OMVQA↑ | Avg. ↑ |
| Comp. Only | Med-Flamingo | 8.3B | ✓ | 58.6 | 43.0 | 47.0 | 25.5 | 61.9 | 31.3 | 28.7 | 34.9 | 41.4 |
| LLaVA-Med | 7B | ✓ | 60.2 | 48.1 | 58.4 | 44.8 | 62.3 | 35.7 | 30.0 | 41.3 | 47.6 | |
| HuatuoGPT-Vision | 7B | ✓ | 66.9 | 53.0 | 59.8 | 49.1 | 52.9 | 32.0 | 42.0 | 50.0 | 50.7 | |
| BLIP-2 | 6.7B | ✗ | 43.4 | 36.8 | 41.6 | 35.3 | 48.5 | 28.8 | 27.3 | 26.9 | 36.1 | |
| LLaVA-v1.5 | 7B | ✗ | 51.8 | 42.8 | 37.1 | 37.7 | 53.5 | 31.4 | 32.7 | 44.7 | 41.5 | |
| InstructBLIP | 7B | ✗ | 61.0 | 44.8 | 66.8 | 43.3 | 56.0 | 32.3 | 25.3 | 29.0 | 44.8 | |
| Yi-VL | 6B | ✗ | 52.6 | 42.1 | 52.4 | 38.4 | 54.9 | 30.9 | 38.0 | 50.2 | 44.9 | |
| InternVL2 | 8B | ✗ | 64.9 | 49.0 | 66.6 | 50.1 | 60.0 | 31.9 | 43.3 | 54.5 | 52.5 | |
| Llama-3.2 | 11B | ✗ | 68.9 | 45.5 | 72.4 | 52.1 | 62.8 | 33.6 | 39.3 | 63.2 | 54.7 | |
| Comp. & Gen. | Show-o | 1.3B | ✗ | 50.6 | 33.9 | 31.5 | 17.9 | 52.9 | 28.2 | 22.7 | 45.7 | 42.6 |
| Unified-IO 2 | 7B | ✗ | 46.2 | 32.6 | 35.9 | 21.9 | 52.5 | 27.0 | 25.3 | 33.0 | 33.8 | |
| Janus | 1.3B | ✗ | 70.9 | 52.8 | 34.7 | 26.9 | 51.9 | 27.9 | 30.0 | 26.8 | 33.5 | |
| HealthGPT-M3 | 3.8B | ✓ | 73.7 | 55.9 | 74.6 | 56.4 | 78.7 | 39.7 | 43.3 | 68.5 | 61.3 | |
| HealthGPT-L14 | 14B | ✓ | 77.7 | 58.3 | 76.4 | 64.5 | 85.9 | 44.4 | 49.2 | 74.4 | 66.4 | |
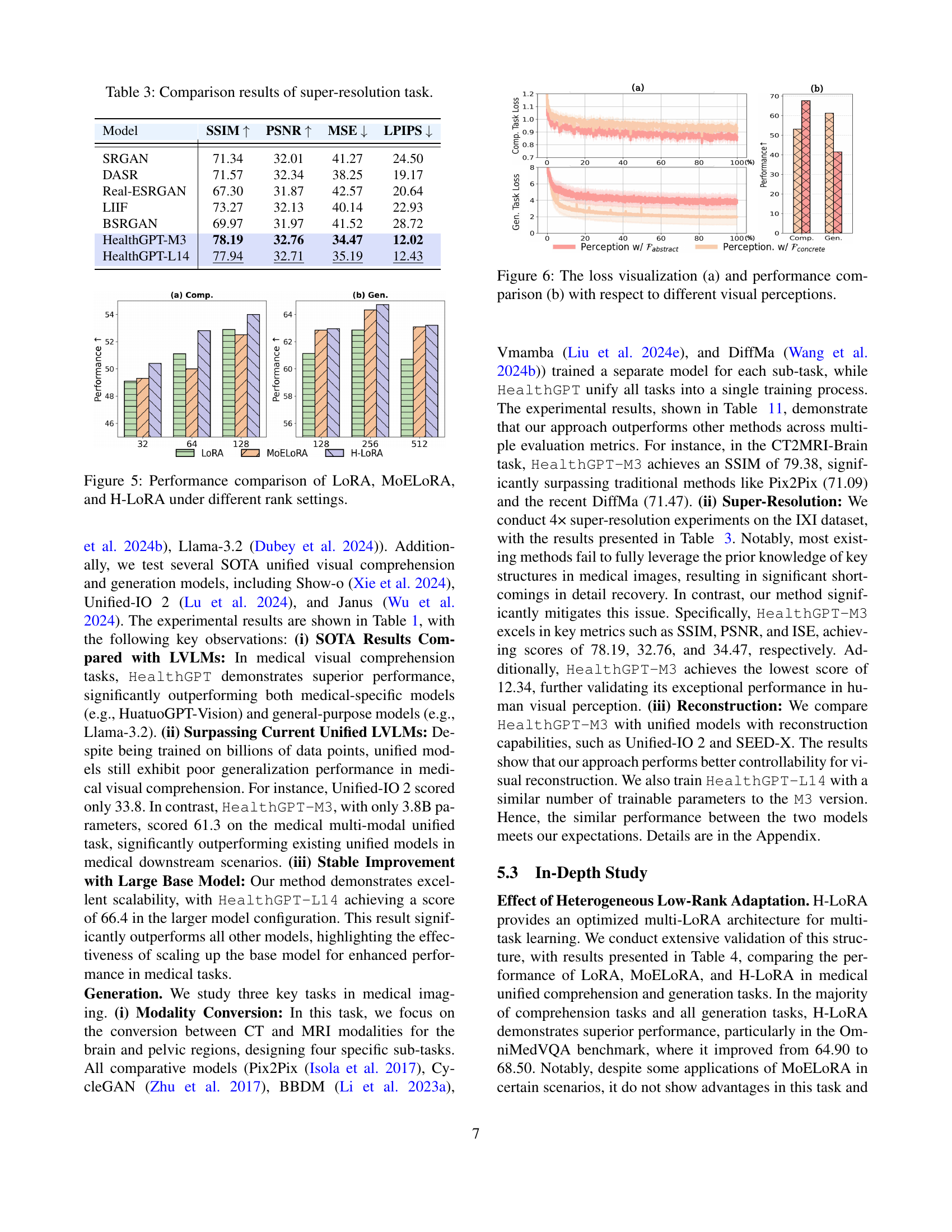
🔼 This table compares the performance of HealthGPT against other Large Vision-Language Models (LVLMs) and unified multi-modal models on several medical visual comprehension tasks. The tasks assess the models’ ability to understand and answer questions about medical images. The metrics used to evaluate performance include scores from various benchmarks (VQA-RAD, SLAKE, PathVQA, MMMU-Med, OMVQA). Higher scores indicate better performance. The table also lists the number of parameters for each model, providing a context for comparing performance relative to model size. Bold and underlined scores indicate the top and second-top performing models for each metric.
read the caption
Table 1: Comparison of HealthGPT with other LVLMs and unified multi-modal models on medical visual comprehension tasks. Bold and underlined text indicates the best performance and second-best performance, respectively.
In-depth insights#
Med-LVLM Unification#
The concept of ‘Med-LVLM Unification’ centers on the integration of visual comprehension and generation capabilities within a single, unified medical large vision-language model (Med-LVLM). This approach aims to overcome the limitations of current models which tend to excel in either comprehension or generation tasks, but not both effectively. Unified Med-LVLMs offer the advantage of enhanced multi-functionality, enabling a broader range of applications in healthcare, from diagnosis and treatment planning to image analysis and report generation. However, achieving true unification presents significant challenges, particularly concerning the inherent conflicts between comprehension (requiring abstraction and generalization) and generation (demanding preservation of fine-grained visual details). The paper explores techniques to address these challenges using innovative parameter-efficient fine-tuning methods and hierarchical visual perception to optimize the learning process and improve model performance across both task types. This unified approach not only aims to improve efficiency but also to enhance the overall consistency and quality of medical visual processing by avoiding the performance degradation often observed when independently training comprehension and generation models.
H-LORA Adaptation#
The proposed H-LORA (Heterogeneous Low-Rank Adaptation) is a novel parameter-efficient fine-tuning method designed to address the challenges of training unified vision-language models for both comprehension and generation tasks. Its core innovation lies in decoupling the learning processes for these often-conflicting tasks. This is achieved by using separate low-rank adapters for comprehension and generation, avoiding the negative interactions that can occur when training them jointly. The use of independent ‘plug-ins’ for each task allows the model to store and utilize different types of knowledge, enhancing its adaptability and performance. Further improving efficiency, H-LORA leverages a mixture-of-experts approach to dynamically route information to the relevant modules, reducing computational overhead compared to other methods. This strategy enables effective knowledge adaptation from limited medical datasets, a crucial benefit given the scarcity of high-quality, large-scale medical multi-modal data.
VL-Health Dataset#
The VL-Health dataset is a crucial component of the HealthGPT research, providing the foundation for training a unified medical vision-language model. Its construction involved careful consideration of data diversity and quality, encompassing various medical imaging modalities and tasks. The dataset’s design addresses the scarcity of high-quality medical data, a significant challenge in developing robust medical AI models. By including a diverse range of image types and associated annotations, VL-Health enables HealthGPT to learn rich representations of visual and textual information in a medical context. This multi-faceted approach, including both comprehension and generation tasks, allows for a more comprehensive understanding of medical images and their associated information. The careful curation of VL-Health, including data processing steps to ensure consistency and quality, is paramount to the success of HealthGPT’s unified capabilities. The scale and diversity of VL-Health are key strengths, allowing the model to generalize well to unseen medical data, a crucial aspect of effective healthcare applications. The detailed description of VL-Health’s creation highlights the importance of robust, representative data in advancing the field of medical AI.
Three-Stage Training#
The three-stage training strategy in HealthGPT is a crucial innovation for effectively leveraging heterogeneous low-rank adaptation (H-LORA). The first stage focuses on multi-modal alignment, separately training visual adapters and H-LORA submodules for comprehension and generation tasks to establish initial alignment and incorporate initial knowledge. This decoupled approach avoids early conflicts between the tasks. Stage two introduces heterogeneous H-LORA plugin adaptation, fine-tuning the shared components (word embedding and output heads) to enhance compatibility and avoid catastrophic forgetting. This ensures seamless integration of diverse knowledge. Finally, the third stage performs visual instruction fine-tuning, exclusively training H-LORA plugins with task-specific data for specific downstream tasks, allowing for rapid adaptation while preserving the established unified foundation. This three-stage approach is particularly important due to the limited size and quality of medical datasets, addressing the common conflicts between visual comprehension and generation tasks in unified models. The strategy’s effectiveness is validated by the superior performance of HealthGPT compared to other unified and specialized models, highlighting the efficacy of this carefully designed training regime.
Future Med-LLMs#
Future Med-LLMs hold immense potential for revolutionizing healthcare. Improved data quality and quantity are crucial; larger, higher-quality datasets will be essential for training more robust and accurate models. Enhanced multimodality is key; future Med-LLMs must seamlessly integrate various data types, including images, text, and sensor data, for a more holistic understanding of patient conditions. Explainability and trustworthiness are paramount; future Med-LLMs must be transparent and understandable, building trust among clinicians and patients. Addressing bias and fairness is critical; careful attention must be given to mitigate biases present in training data and ensure equitable access and outcomes. Integration with existing clinical workflows is necessary for successful implementation; Med-LLMs must be easily incorporated into existing healthcare systems. Finally, robustness and safety are vital; rigorous testing and validation are necessary to ensure accuracy and reliability, especially given the high stakes in medical applications.
More visual insights#
More on figures

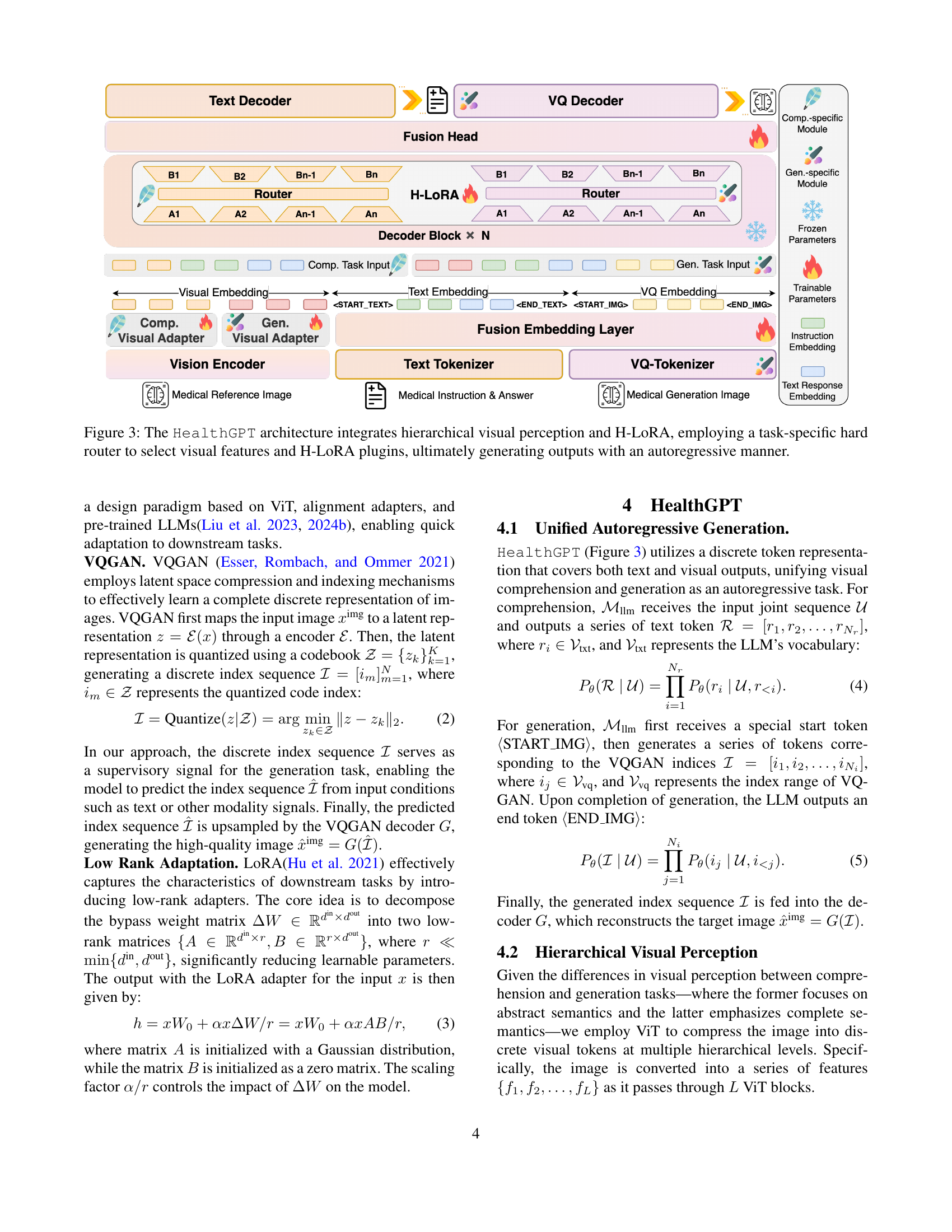
🔼 This figure shows the impact of data imbalance on the performance of a unified vision-language model. Two graphs are presented, one showing comprehension performance and the other showing generation performance. Both graphs show performance on the y-axis and the ratio of comprehension to generation data on the x-axis. As the ratio of one type of data increases (while the amount of the other type is held constant), the model’s performance on the task with less training data decreases significantly. This highlights a challenge in training unified models that perform both comprehension and generation tasks, where an optimal balance of data is needed for both tasks to achieve good performance on both.
read the caption
Figure 2: With a fixed amount of comprehension (generation) data, increasing the proportion of the other type leads to significant performance degradation.

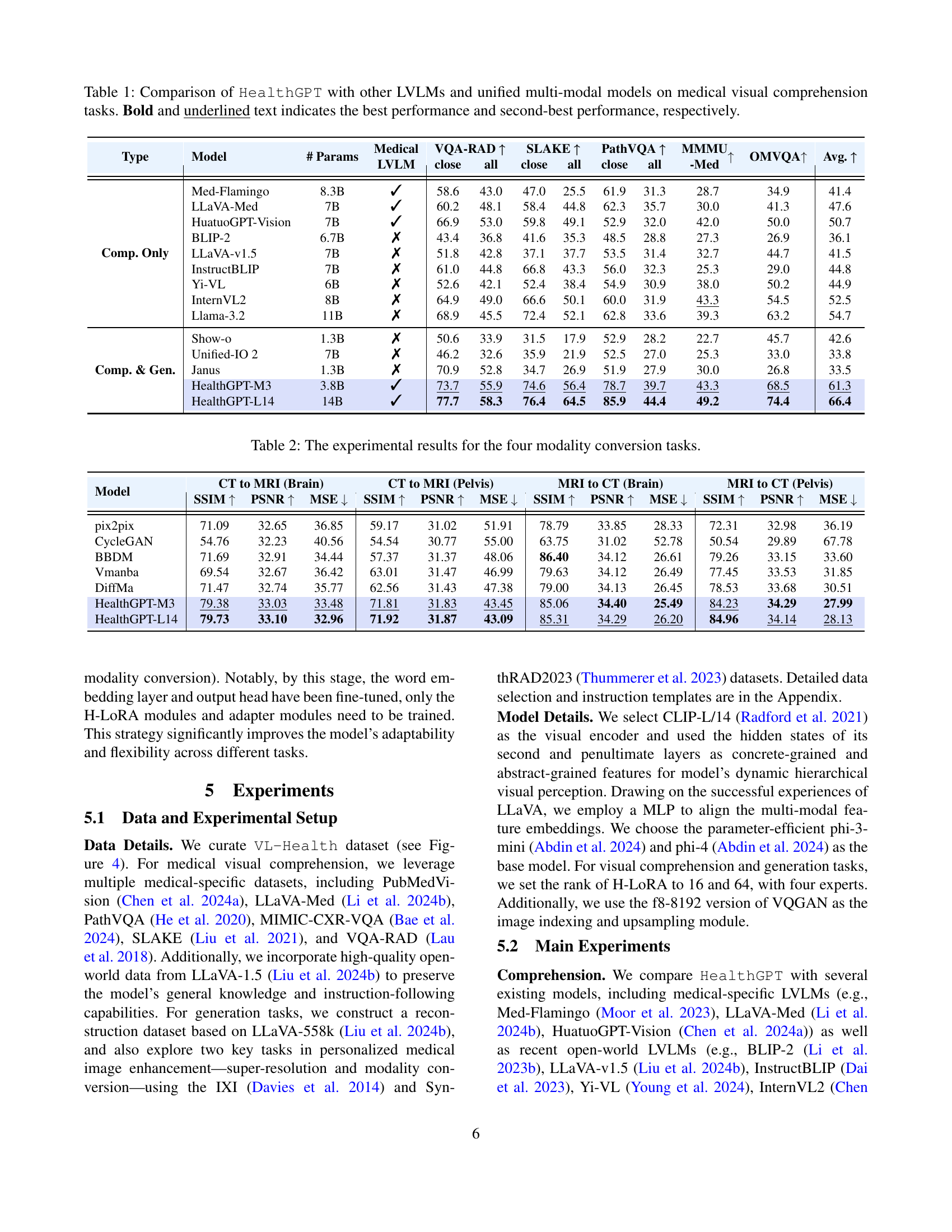
🔼 This figure illustrates the architecture of HealthGPT, a medical large vision-language model. HealthGPT uses a hierarchical visual perception approach, processing images through multiple levels of a Vision Transformer (ViT) to extract both concrete (detailed) and abstract (semantic) visual features. These features are then routed to task-specific H-LoRA (Heterogeneous Low-Rank Adaptation) plugins. The H-LoRA plugins, designed for efficient parameter-tuning, are responsible for adapting the pre-trained language model to both comprehension and generation tasks. A hard router dynamically selects the appropriate visual features (concrete or abstract) based on the task (comprehension or generation). The model then generates outputs (text or images) in an autoregressive manner.
read the caption
Figure 3: The HealthGPT architecture integrates hierarchical visual perception and H-LoRA, employing a task-specific hard router to select visual features and H-LoRA plugins, ultimately generating outputs with an autoregressive manner.

🔼 This figure shows the statistical distribution of data within the VL-Health dataset, a key component of the HealthGPT model. Panel (a) illustrates the number of images and text-image pairs used for different types of medical visual tasks including fundus images, OCT, MRI, ultrasound, X-ray, microscopy, digital photography, dermoscopy, and tomography. Different generation tasks such as image reconstruction and modality conversion are also represented. Panel (b) provides a breakdown of the number of data samples used for each specific dataset in the VL-Health dataset including IXI, MIMIC-CHEST-XRAY, and LLaVA-558k among others. This breakdown further categorizes the data into different task types such as comprehension and generation.
read the caption
Figure 4: Data statistics of VL-Health.

🔼 This figure compares the performance of three parameter-efficient fine-tuning (PEFT) methods: LoRA, MoELORA, and H-LoRA, across different rank settings. The methods were used to adapt large language models for medical visual comprehension and generation tasks. The x-axis represents the rank (dimensionality of the low-rank matrices), and the y-axis shows the performance. It demonstrates how the performance of each method changes as the rank increases, showing the trade-off between model size and performance.
read the caption
Figure 5: Performance comparison of LoRA, MoELoRA, and H-LoRA under different rank settings.

🔼 This figure shows the impact of different visual perception strategies on the performance of the HealthGPT model. Part (a) visualizes the training loss curves for comprehension and generation tasks, separately using either abstract or concrete visual features. The curves illustrate how the choice of features impacts the training process and potential conflicts between comprehension and generation. Part (b) presents a bar chart comparing the final performance (measured by some unspecified metric) for both comprehension and generation tasks using either abstract or concrete features. This comparison highlights the effectiveness of the chosen hierarchical visual perception approach, which utilizes both abstract and concrete features depending on the task.
read the caption
Figure 6: The loss visualization (a) and performance comparison (b) with respect to different visual perceptions.

🔼 This figure presents a case study demonstrating the capabilities of the HealthGPT model in generating chest X-ray (CXR) images based on textual descriptions. Panel (a) provides a baseline with a normal CXR image for comparison. Panels (b) and (c) show generated CXR images illustrating the model’s ability to produce images reflecting different levels of severity and different affected regions within the lungs, as indicated by varying levels of pulmonary edema and pleural effusion. The highlighted areas in (b) and (c) represent the regions described as having abnormal conditions in the text prompts.
read the caption
Figure 7: Case study of report-to-CXR under different instructions. (a) shows a normal CXR image for comparison. (b) and (c) illustrate generated cases with varying severity and affected regions. The graffiti areas indicate abnormal conditions.

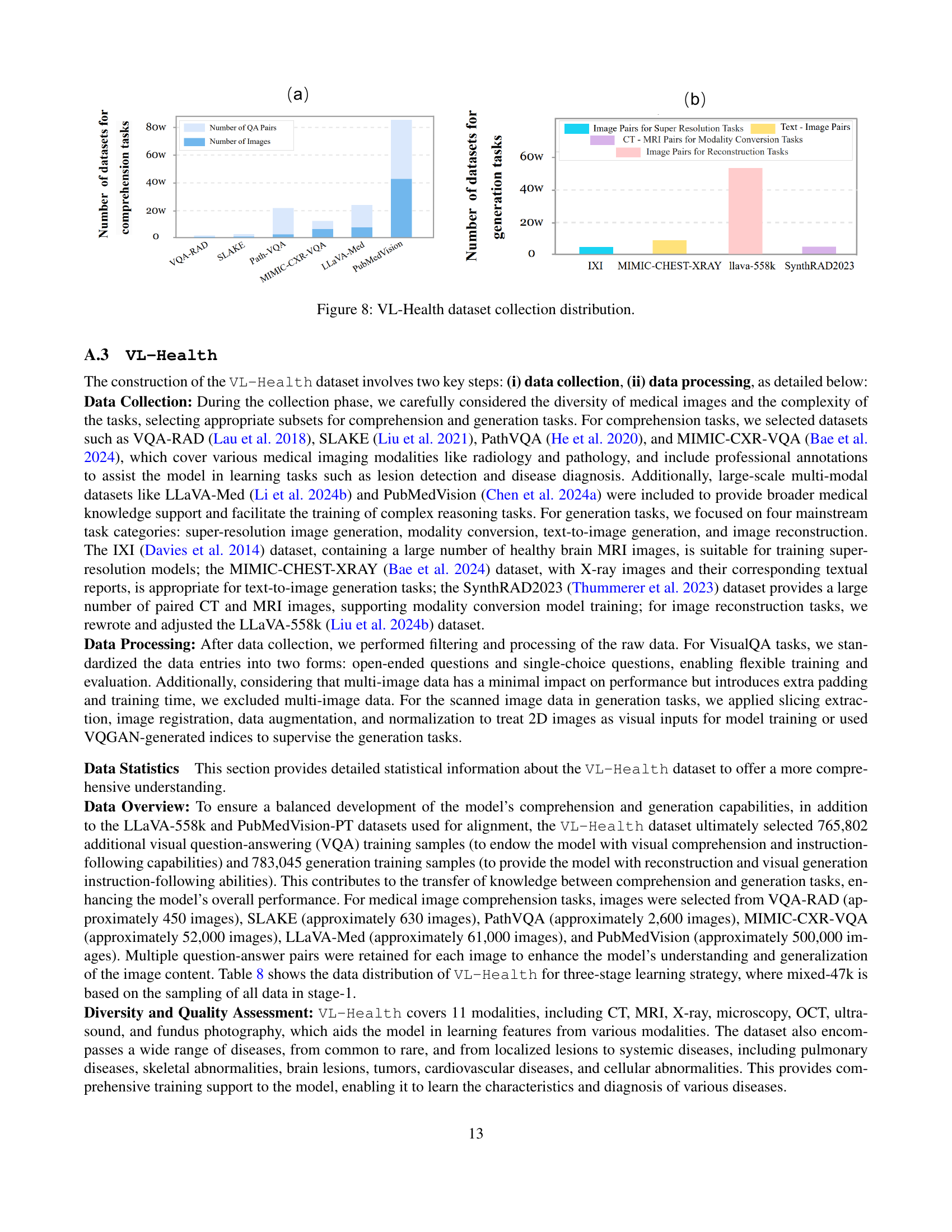
🔼 This figure shows the distribution of data used to create the VL-Health dataset. Panel (a) presents a bar chart illustrating the number of question-answer pairs and images for each of the datasets used for medical visual comprehension tasks (VQA-RAD, SLAKE, PathVQA, MIMIC-CXR-VQA, LLaVA-Med, PubMedVision). Panel (b) displays a bar chart showing the number of image pairs used for various medical image generation tasks. These tasks include super-resolution, modality conversion (CT to MRI, MRI to CT), and image reconstruction. The datasets used for generation tasks are IXI, MIMIC-CHEST-XRAY, LLaVA-558k, and SynthRAD2023.
read the caption
Figure 8: VL-Health dataset collection distribution.

🔼 This figure displays a bar chart visualizing the performance changes in medical visual comprehension and generation tasks before and after the second stage of training. The x-axis represents the task type (comprehension or generation), and the y-axis represents the performance metric. The bars show a comparison of performance before and after the introduction of the heterogeneous low-rank adaptation (H-LORA) in stage 2 of the training process, highlighting the impact of this stage on the model’s ability to handle both tasks effectively.
read the caption
Figure 9: Performance changes before and after the stage-2.

🔼 Figure 10 presents the results of a human evaluation comparing HealthGPT’s performance to other LLMs on medical visual question answering tasks. Part (a) is a pie chart showing the proportion of times each model’s response was ranked as the best among multiple responses to the same question by human evaluators (clinicians). Part (b) briefly describes the human evaluation dataset used, which includes questions from VQA-RAD, SLAKE, and PathVQA.
read the caption
Figure 10: (a) Proportion of model responses selected as the best in human evaluation. (b) Human Evaluation Dataset.

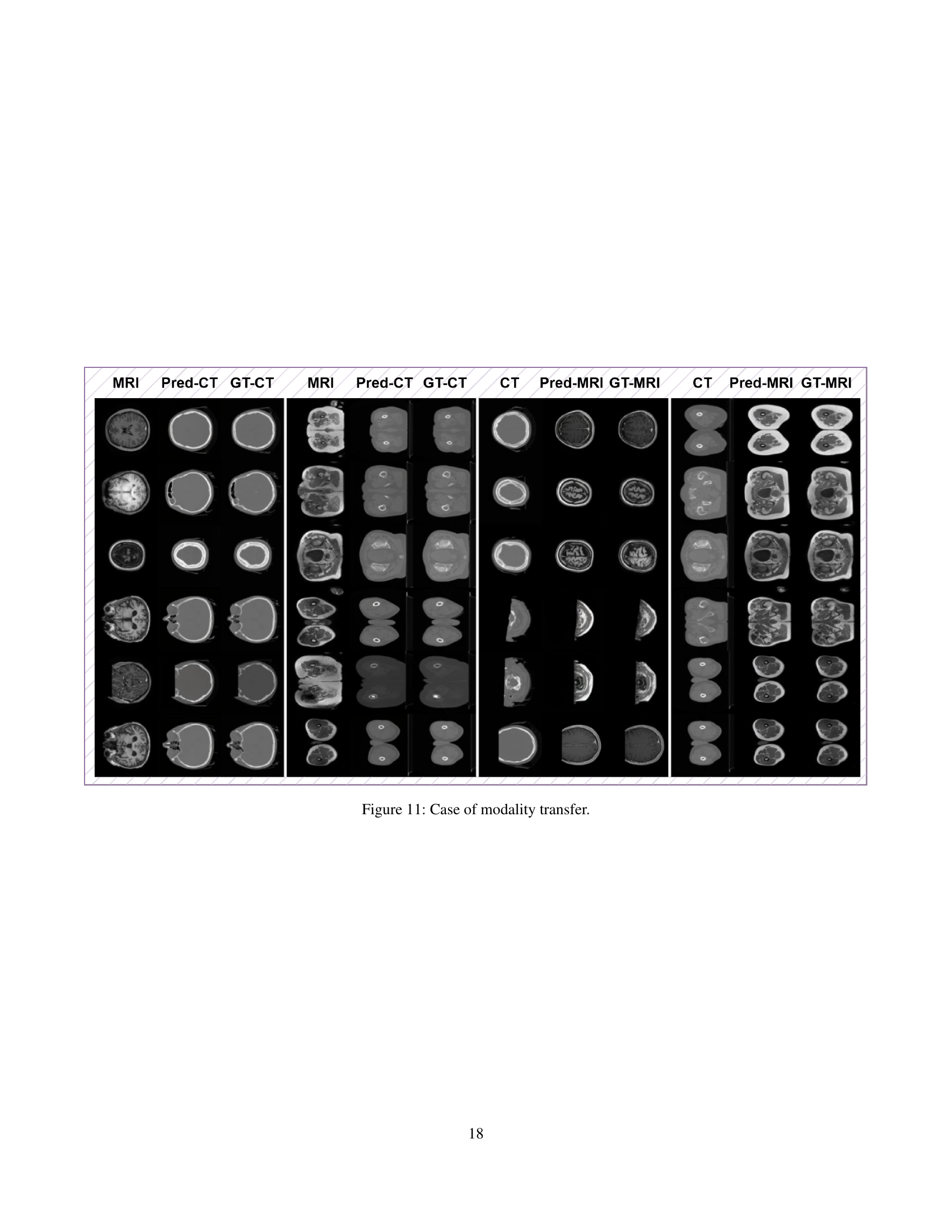
🔼 This figure showcases examples of modality transfer, specifically converting between CT and MRI scans. It presents pairs of images for four different scenarios: brain CT to brain MRI, pelvic CT to pelvic MRI, brain MRI to brain CT, and pelvic MRI to pelvic CT. Within each scenario, the original image, the prediction generated by the HealthGPT model, and the ground truth image are shown side-by-side for comparison. This visual comparison demonstrates the model’s ability to accurately transform medical images between modalities.
read the caption
Figure 11: Case of modality transfer.
More on tables
| CT to MRI (Brain) | CT to MRI (Pelvis) | MRI to CT (Brain) | MRI to CT (Pelvis) | |||||||||
| Model | SSIM | PSNR | MSE | SSIM | PSNR | MSE | SSIM | PSNR | MSE | SSIM | PSNR | MSE |
| pix2pix | 71.09 | 32.65 | 36.85 | 59.17 | 31.02 | 51.91 | 78.79 | 33.85 | 28.33 | 72.31 | 32.98 | 36.19 |
| CycleGAN | 54.76 | 32.23 | 40.56 | 54.54 | 30.77 | 55.00 | 63.75 | 31.02 | 52.78 | 50.54 | 29.89 | 67.78 |
| BBDM | 71.69 | 32.91 | 34.44 | 57.37 | 31.37 | 48.06 | 86.40 | 34.12 | 26.61 | 79.26 | 33.15 | 33.60 |
| Vmanba | 69.54 | 32.67 | 36.42 | 63.01 | 31.47 | 46.99 | 79.63 | 34.12 | 26.49 | 77.45 | 33.53 | 31.85 |
| DiffMa | 71.47 | 32.74 | 35.77 | 62.56 | 31.43 | 47.38 | 79.00 | 34.13 | 26.45 | 78.53 | 33.68 | 30.51 |
| HealthGPT-M3 | 79.38 | 33.03 | 33.48 | 71.81 | 31.83 | 43.45 | 85.06 | 34.40 | 25.49 | 84.23 | 34.29 | 27.99 |
| HealthGPT-L14 | 79.73 | 33.10 | 32.96 | 71.92 | 31.87 | 43.09 | 85.31 | 34.29 | 26.20 | 84.96 | 34.14 | 28.13 |
🔼 This table presents a quantitative evaluation of the model’s performance on four medical image modality conversion tasks: converting CT images to MRI images (for both brain and pelvis), and vice versa. For each task, the table displays the performance metrics achieved by the model, including Structural Similarity Index (SSIM), Peak Signal-to-Noise Ratio (PSNR), and Mean Squared Error (MSE). These metrics provide a comprehensive assessment of the model’s ability to accurately transform images between different modalities.
read the caption
Table 2: The experimental results for the four modality conversion tasks.
| Model | SSIM | PSNR | MSE | LPIPS |
| SRGAN | 71.34 | 32.01 | 41.27 | 24.50 |
| DASR | 71.57 | 32.34 | 38.25 | 19.17 |
| Real-ESRGAN | 67.30 | 31.87 | 42.57 | 20.64 |
| LIIF | 73.27 | 32.13 | 40.14 | 22.93 |
| BSRGAN | 69.97 | 31.97 | 41.52 | 28.72 |
| HealthGPT-M3 | 78.19 | 32.76 | 34.47 | 12.02 |
| HealthGPT-L14 | 77.94 | 32.71 | 35.19 | 12.43 |
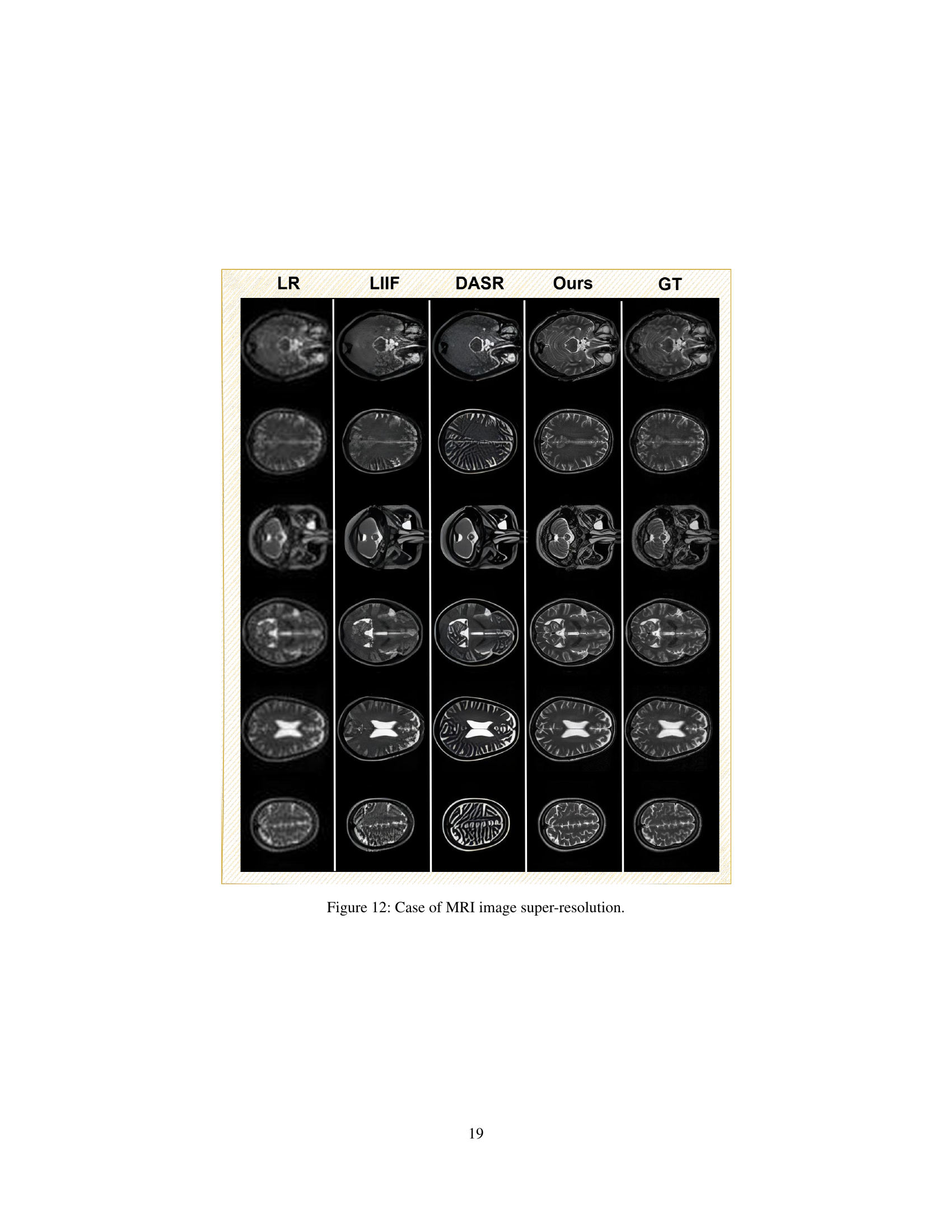
🔼 This table presents a comparison of different super-resolution models’ performance. The models compared include SRGAN, DASR, Real-ESRGAN, LIIF, BSRGAN, and two versions of the HealthGPT model (HealthGPT-M3 and HealthGPT-L14). The performance is evaluated based on four metrics: Structural SIMilarity index (SSIM), Peak Signal-to-Noise Ratio (PSNR), Mean Squared Error (MSE), and Learned Perceptual Image Patch Similarity (LPIPS). Lower MSE and LPIPS values indicate better performance, while higher SSIM and PSNR values indicate better performance. This table showcases the superior performance of the HealthGPT models, specifically in terms of LPIPS, compared to other existing super-resolution methods.
read the caption
Table 3: Comparison results of super-resolution task.
| Comp. | Gen. | ||||||||||||
| VQA-RAD | SLAKE | PathVQA | |||||||||||
| Model | close | all | close | all | close | all | MMMU -Med | OMVQA | RECOM | MTRANS | SR | Training Time | |
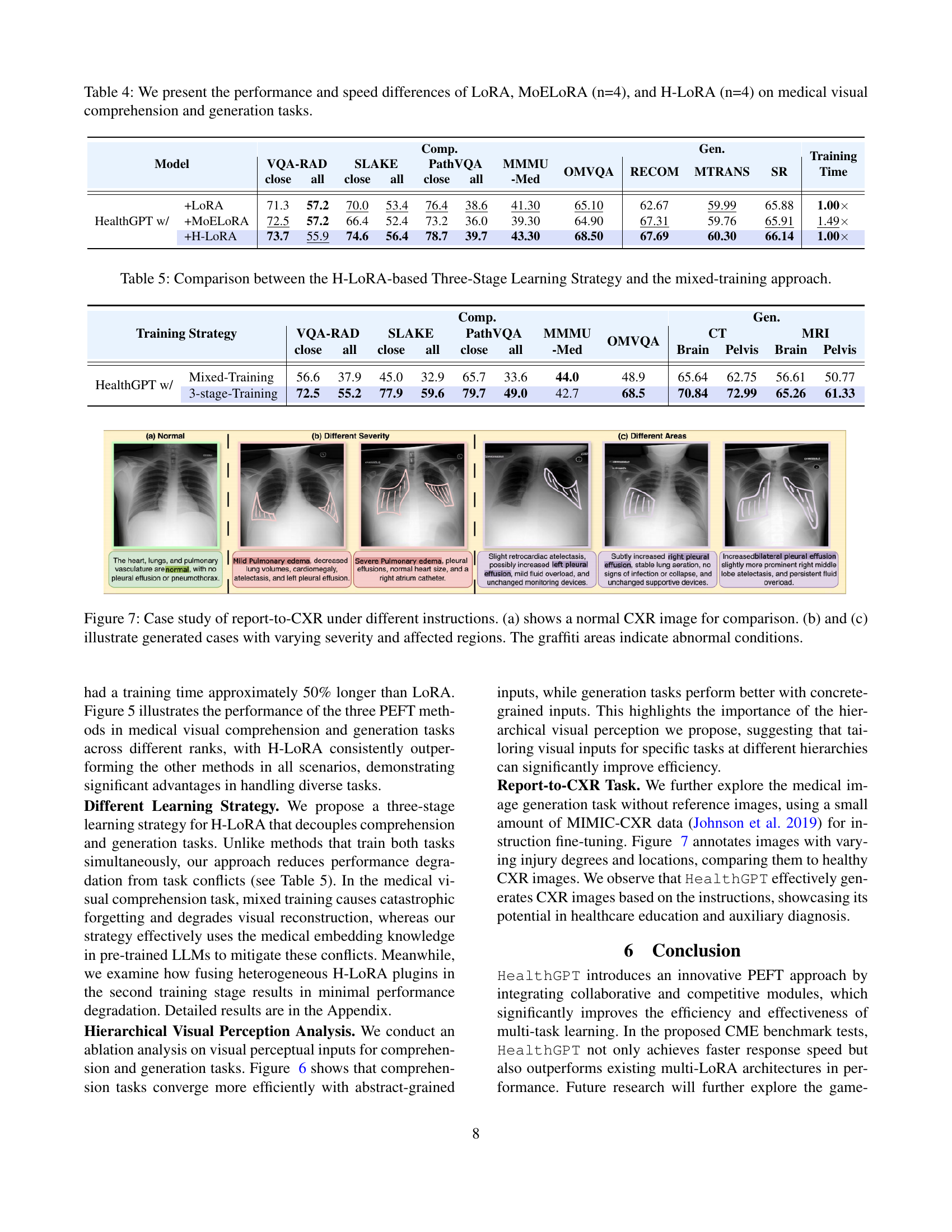
| HealthGPT w/ | +LoRA | 71.3 | 57.2 | 70.0 | 53.4 | 76.4 | 38.6 | 41.30 | 65.10 | 62.67 | 59.99 | 65.88 | 1.00 |
| +MoELoRA | 72.5 | 57.2 | 66.4 | 52.4 | 73.2 | 36.0 | 39.30 | 64.90 | 67.31 | 59.76 | 65.91 | 1.49 | |
| +H-LoRA | 73.7 | 55.9 | 74.6 | 56.4 | 78.7 | 39.7 | 43.30 | 68.50 | 67.69 | 60.30 | 66.14 | 1.00 | |
🔼 This table compares the performance and training speed of three different parameter-efficient fine-tuning (PEFT) methods: LoRA, MoELORA, and H-LORA. All three methods are applied to both medical visual comprehension and generation tasks. The comparison includes metrics for several specific tasks (VQA-RAD, SLAKE, PathVQA, MMMU, OMVQA, RECOM, MTRANS, SR) to assess the effectiveness of each PEFT method across a range of applications. The training time is normalized relative to LoRA to showcase the relative efficiency gains or losses.
read the caption
Table 4: We present the performance and speed differences of LoRA, MoELoRA (n=4), and H-LoRA (n=4) on medical visual comprehension and generation tasks.
| Comp. | Gen. | ||||||||||||
| VQA-RAD | SLAKE | PathVQA | CT | MRI | |||||||||
| Training Strategy | close | all | close | all | close | all | MMMU -Med | OMVQA | Brain | Pelvis | Brain | Pelvis | |
| Mixed-Training | 56.6 | 37.9 | 45.0 | 32.9 | 65.7 | 33.6 | 44.0 | 48.9 | 65.64 | 62.75 | 56.61 | 50.77 | |
| HealthGPT w/ | 3-stage-Training | 72.5 | 55.2 | 77.9 | 59.6 | 79.7 | 49.0 | 42.7 | 68.5 | 70.84 | 72.99 | 65.26 | 61.33 |
🔼 This table compares the performance of two training strategies for the HealthGPT model: a three-stage learning approach using the proposed heterogeneous low-rank adaptation (H-LORA) method and a mixed-training approach. It evaluates their effectiveness on various medical visual comprehension and generation tasks, showing metrics such as VQA-RAD, SLAKE, PathVQA, MMMU-Med, OmniMedVQA, and modality conversion tasks (CT, MRI) results. The purpose is to demonstrate the superiority of the three-stage H-LORA approach in avoiding conflicts and improving performance.
read the caption
Table 5: Comparison between the H-LoRA-based Three-Stage Learning Strategy and the mixed-training approach.
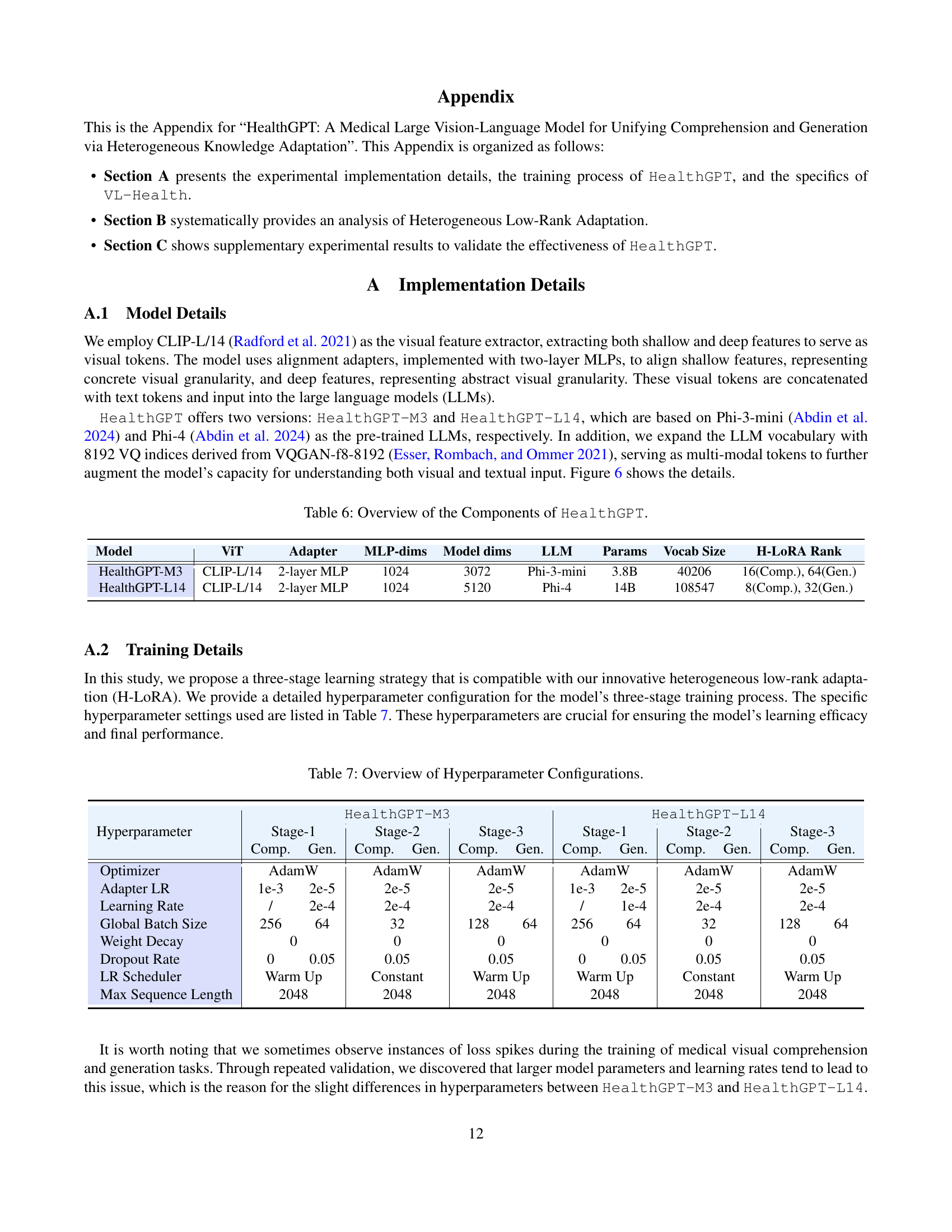
| Model | ViT | Adapter | MLP-dims | Model dims | LLM | Params | Vocab Size | H-LoRA Rank |
| HealthGPT-M3 | CLIP-L/14 | 2-layer MLP | 1024 | 3072 | Phi-3-mini | 3.8B | 40206 | 16(Comp.), 64(Gen.) |
| HealthGPT-L14 | CLIP-L/14 | 2-layer MLP | 1024 | 5120 | Phi-4 | 14B | 108547 | 8(Comp.), 32(Gen.) |
🔼 This table details the architecture of the HealthGPT model, showing the components used in both the HealthGPT-M3 and HealthGPT-L14 versions. It lists the specific Vision Transformer (ViT) used, the type of adapter layers, the dimensions of the Multilayer Perceptron (MLP), the model dimensions, the specific Large Language Model (LLM) used, the number of parameters in the LLM, the vocabulary size of the LLM, and the rank used for the Heterogeneous Low-Rank Adaptation (H-LoRA). This information provides a comprehensive overview of the model’s building blocks and their configurations.
read the caption
Table 6: Overview of the Components of HealthGPT.
| HealthGPT-M3 | HealthGPT-L14 | |||||||||||
| Stage-1 | Stage-2 | Stage-3 | Stage-1 | Stage-2 | Stage-3 | |||||||
| Hyperparameter | Comp. | Gen. | Comp. | Gen. | Comp. | Gen. | Comp. | Gen. | Comp. | Gen. | Comp. | Gen. |
| Optimizer | AdamW | AdamW | AdamW | AdamW | AdamW | AdamW | ||||||
| Adapter LR | 1e-3 | 2e-5 | 2e-5 | 2e-5 | 1e-3 | 2e-5 | 2e-5 | 2e-5 | ||||
| Learning Rate | / | 2e-4 | 2e-4 | 2e-4 | / | 1e-4 | 2e-4 | 2e-4 | ||||
| Global Batch Size | 256 | 64 | 32 | 128 | 64 | 256 | 64 | 32 | 128 | 64 | ||
| Weight Decay | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Dropout Rate | 0 | 0.05 | 0.05 | 0.05 | 0 | 0.05 | 0.05 | 0.05 | ||||
| LR Scheduler | Warm Up | Constant | Warm Up | Warm Up | Constant | Warm Up | ||||||
| Max Sequence Length | 2048 | 2048 | 2048 | 2048 | 2048 | 2048 | ||||||
🔼 This table details the hyperparameter settings used during the three-stage training process of the HealthGPT model. It shows the optimizer, learning rates for different model components (adapter and main model), batch size, weight decay, dropout rate, learning rate scheduler, and maximum sequence length used in each of the three training stages. Different configurations are provided for the two variants of the model, HealthGPT-M3 and HealthGPT-L14.
read the caption
Table 7: Overview of Hyperparameter Configurations.
| Medical Task | Stage-1 | Stage-2 |
| Comp. | LLaVA-558k, PubMedVision-PT | Mixed-47k |
| Gen. | LLaVA-558k | |
| Medical Task | Stage-3 | |
| Comp. | LLaVA_Med, MIMIC_CXR_VQA, PubMedVision-FT, LLaVA-665k, PathVQA, SLAKE, VQA-RAD | |
| Gen. | IXI, SynthRAD2023, MIMIC-CHEST-XRAY | |
🔼 This table details the data distribution used in the three stages of the HealthGPT model training. It breaks down the datasets used for comprehension and generation tasks in each of the three training stages: Stage 1 (Multi-modal Alignment), Stage 2 (Heterogeneous H-LoRA Plugin Adaptation), and Stage 3 (Visual Instruction Fine-Tuning). For each stage, it lists the specific datasets used for both comprehension and generation tasks, offering a clear view of the data composition throughout the model’s training process.
read the caption
Table 8: Data distribution of VL-Health in three-stage learning strategy.
| OmniMedVQA ↑ | |||||||||||
| Type | Model | # Params | Medical LVLM | CT | X-ray | FDM | MiS | OCT | MRI | USS | Avg. |
| Comp. Only | Med-Flamingo | 8.3B | ✓ | 30.1 | 33.9 | 25.5 | 37.0 | 60.0 | 27.6 | 30.4 | 34.9 |
| LLaVA-Med | 7B | ✓ | 28.4 | 32.8 | 42.7 | 31.6 | 55.3 | 45.0 | 53.6 | 41.3 | |
| HuatuoGPT-Vision | 7B | ✓ | 35.3 | 41.5 | 51.4 | 62.3 | 59.3 | 40.4 | 60.1 | 50.0 | |
| BLIP-2 | 6.7B | ✗ | 26.6 | 29.1 | 22.3 | 36.9 | 29.1 | 22.7 | 21.4 | 26.9 | |
| LLaVA-v1.5 | 7B | ✗ | 28.0 | 55.7 | 35.5 | 42.1 | 49.2 | 52.9 | 49.7 | 44.7 | |
| InstructBLIP | 7B | ✗ | 20.1 | 22.2 | 34.1 | 30.6 | 38.6 | 31.9 | 25.5 | 29.0 | |
| Yi-VL | 6B | ✗ | 51.2 | 47.1 | 27.7 | 62.6 | 67.6 | 55.0 | 40.3 | 50.2 | |
| InternVL2 | 8B | ✗ | 40.2 | 57.9 | 53.2 | 64.0 | 59.1 | 58.1 | 49.1 | 54.5 | |
| Llama-3.2 | 11B | ✗ | 37.6 | 55.2 | 71.4 | 82.1 | 62.5 | 65.2 | 68.6 | 63.2 | |
| Comp. & Gen. | Show-o | 1.3B | ✗ | 29.0 | 50.4 | 30.9 | 22.0 | 30.8 | 34.2 | 33.8 | 33.0 |
| Unified-IO 2 | 7B | ✗ | 10.8 | 37.7 | 12.3 | 25.3 | 32.6 | 30.9 | 37.7 | 26.8 | |
| Janus | 1.3B | ✗ | 24.9 | 54.8 | 35.9 | 62.7 | 54.2 | 50.7 | 36.8 | 45.7 | |
| HealthGPT-M3 | 3.8B | ✓ | 35.3 | 81.9 | 54.6 | 88.2 | 89.3 | 78.5 | 51.4 | 68.5 | |
| HealthGPT-L14 | 14B | ✓ | 39.0 | 86.6 | 64.1 | 88.6 | 99.7 | 80.9 | 62.2 | 74.4 | |
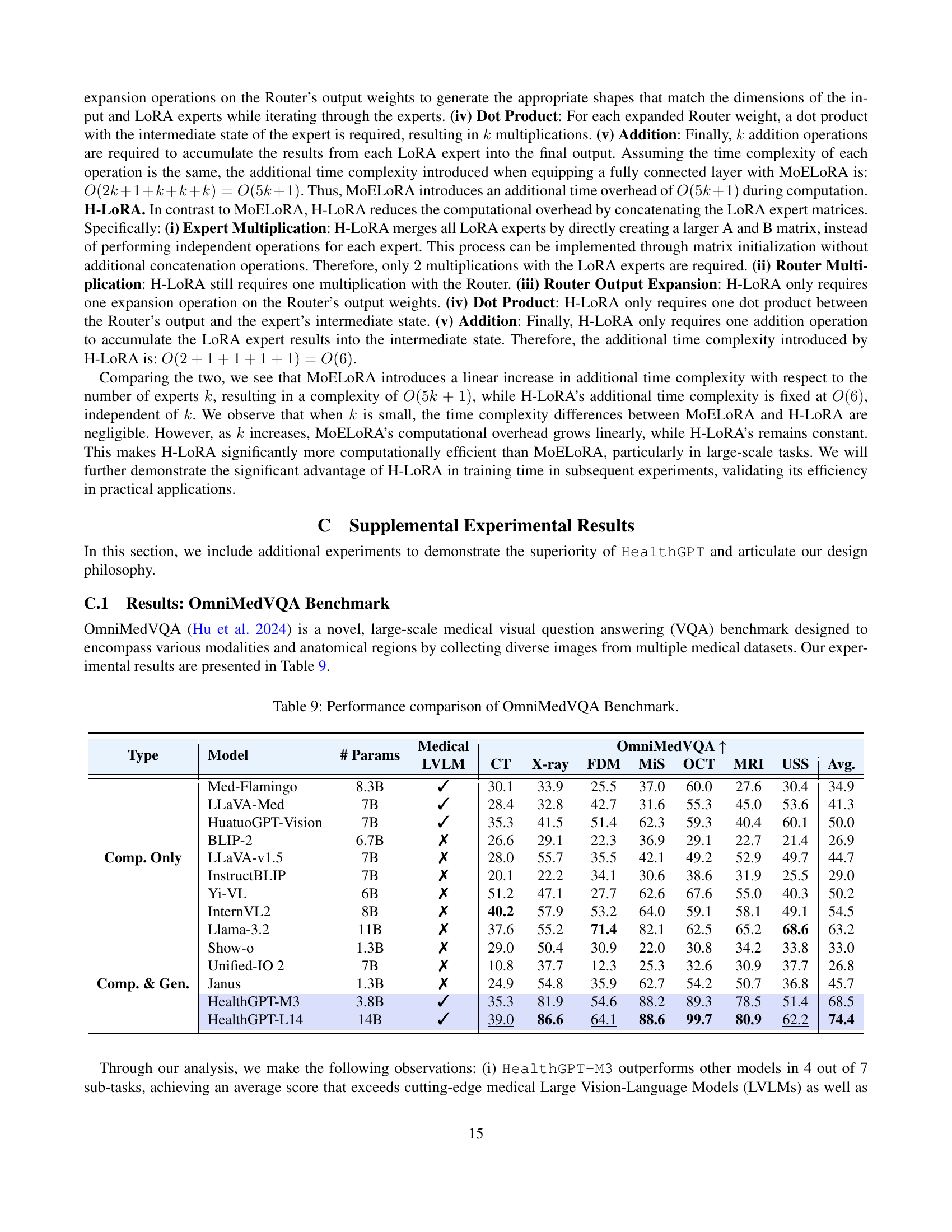
🔼 This table compares the performance of different Large Vision-Language Models (LVLMs) on the OmniMedVQA benchmark. OmniMedVQA is a challenging medical visual question answering benchmark that includes various image modalities and anatomical regions. The table shows the performance of each model across different image modalities (CT, X-ray, FDM, MIS, OCT, MRI, USS), and presents the average performance across all modalities. This allows for a comprehensive comparison of the models’ ability to accurately answer questions about medical images.
read the caption
Table 9: Performance comparison of OmniMedVQA Benchmark.
| n=2 | n=4 | n=8 | n=32 | ||||||||||
| Model | Comp. | Gen. | Time | Comp. | Gen. | Time | Comp. | Gen. | Time | Comp. | Gen. | Time | |
| +MoELoRA | 50.3 | 62.98 | 1.22 | 50.0 | 64.33 | 1.49 | 50.8 | 63.71 | 2.09 | / | / | 5.81 | |
| HealthGPT w/ | +H-LoRA | 51.5 | 63.48 | 0.99 | 52.8 | 64.71 | 1.00 | 53.6 | 64.98 | 0.99 | 53.5 | 64.74 | 1.01 |
🔼 This table presents a comparison of the performance and training time of MoELoRA and H-LoRA models with varying numbers of LoRA experts (n). It demonstrates that as the number of experts increases, MoELoRA’s training time grows significantly, while H-LoRA’s training time remains relatively stable. The experiment highlights the computational efficiency of H-LoRA compared to MoELoRA, especially when a large number of experts are used. At n=32, MoELoRA failed to complete training, further illustrating the scalability advantage of H-LoRA.
read the caption
Table 10: We explored the performance of MoELoRA and H-LoRA with different numbers of LoRA experts. At n=32𝑛32n=32italic_n = 32, MoELoRA was unable to complete training.
| CT(Brain) | CT(Pelvis) | MRI (Brain) | MRI(Pelvis) | |||||||||
| Model | SSIM | PSNR | MSE | SSIM | PSNR | MSE | SSIM | PSNR | MSE | SSIM | PSNR | MSE |
| SEED-X | 20.18 | 27.66 | 112.11 | 21.53 | 28.02 | 102.87 | 4.90 | 27.62 | 112.86 | 6.31 | 27.89 | 106.21 |
| Unified-IO 2 | 83.93 | 36.09 | 17.95 | 85.36 | 35.10 | 25.46 | 87.50 | 34.25 | 25.47 | 86.31 | 33.53 | 29.80 |
| HealthGPT-M3 | 91.73 | 36.42 | 15.46 | 94.26 | 37.30 | 12.53 | 88.76 | 33.97 | 27.05 | 84.40 | 33.11 | 32.62 |
🔼 This table presents a quantitative comparison of HealthGPT’s performance against other state-of-the-art models on four medical image reconstruction tasks. These tasks involve converting between different imaging modalities (CT to MRI and MRI to CT) and for brain and pelvic regions. The metrics used to evaluate performance are Structural Similarity Index (SSIM), Peak Signal-to-Noise Ratio (PSNR), and Mean Squared Error (MSE). Lower MSE values indicate better performance. The table allows readers to directly compare the quantitative results of different models on each specific task, highlighting HealthGPT’s capabilities in handling these complex medical image transformations.
read the caption
Table 11: The experimental results for the four reconstruction tasks.
Full paper#