↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Medical Vision-Language Pretraining (MedVLP) shows promise in analyzing medical images and reports, but lacks a unified evaluation standard, hindering fair comparisons of different methods. Existing MedVLP methods vary in terms of datasets, preprocessing steps and finetuning protocols making it challenging to evaluate their generalization capabilities.
To address these issues, researchers introduce BenchX, a unified benchmark framework that standardizes data preprocessing, train-test splits, and evaluation protocols for MedVLP methods. They evaluated nine state-of-the-art MedVLP models across nine datasets and four medical tasks, finding that some earlier methods, with proper configurations, outperformed more recent methods. BenchX provides a valuable tool for future research in this field by enabling more robust and reliable comparisons between MedVLP methods. This work promotes standardization, improving reproducibility, and accelerating progress in the field.
Key Takeaways#
Why does it matter?#
This paper is crucial because it addresses the lack of standardized benchmarks in medical vision-language pretraining (MedVLP). Its unified framework, BenchX, enables fair comparison of MedVLP methods, fostering better evaluation and accelerating progress in this rapidly developing field. The findings challenge existing conclusions by showing that seemingly outdated MedVLP methods can still be highly competitive with proper finetuning and configuration.
Visual Insights#

🔼 This figure illustrates how different MedVLP (Medical Vision-Language Pretraining) models are adapted for three downstream medical tasks: classification, segmentation, and report generation. It highlights the unification of adaptation pipelines, showing how heterogeneous MedVLP model architectures (ResNet, ViT, Swin) are integrated with task-specific heads (linear classifier, UperNet, R2Gen) for consistent evaluation. This addresses the challenge of incompatible model architectures in existing MedVLP methods.
read the caption
Figure 1: The illustrative tasks adaptation pipeline.
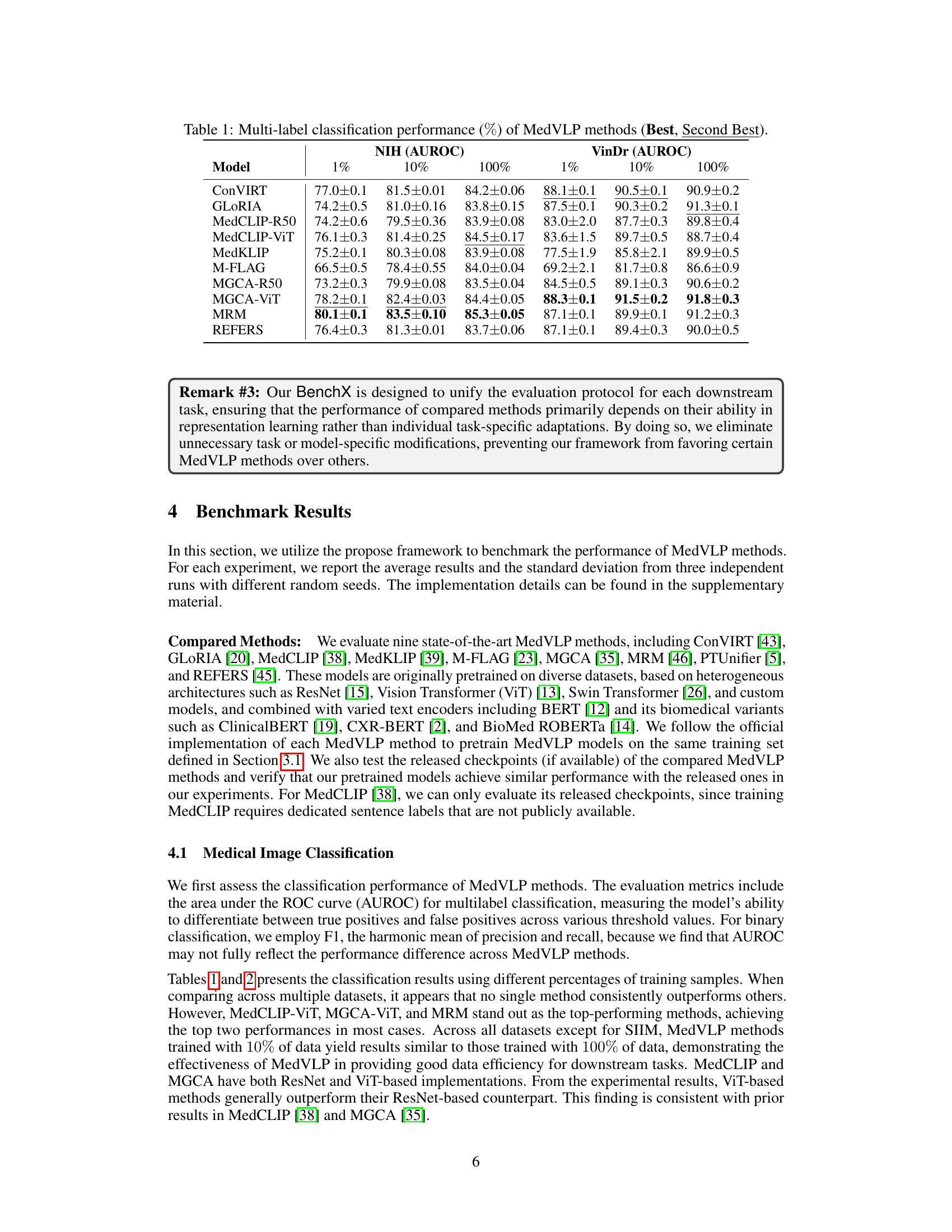
| Model | NIH (AUROC) | VinDr (AUROC) | ||||
|---|---|---|---|---|---|---|
| 1% | 10% | 100% | 1% | 10% | 100% | |
| ConVIRT | 77.0 ± 0.1 | 81.5 ± 0.01 | 84.2 ± 0.06 | 88.1 ± 0.1 | 90.5 ± 0.1 | 90.9 ± 0.2 |
| GLoRIA | 74.2 ± 0.5 | 81.0 ± 0.16 | 83.8 ± 0.15 | 87.5 ± 0.1 | 90.3 ± 0.2 | 91.3 ± 0.1 |
| MedCLIP-R50 | 74.2 ± 0.6 | 79.5 ± 0.36 | 83.9 ± 0.08 | 83.0 ± 2.0 | 87.7 ± 0.3 | 89.8 ± 0.4 |
| MedCLIP-ViT | 76.1 ± 0.3 | 81.4 ± 0.25 | 84.5 ± 0.17 | 83.6 ± 1.5 | 89.7 ± 0.5 | 88.7 ± 0.4 |
| MedKLIP | 75.2 ± 0.1 | 80.3 ± 0.08 | 83.9 ± 0.08 | 77.5 ± 1.9 | 85.8 ± 2.1 | 89.9 ± 0.5 |
| M-FLAG | 66.5 ± 0.5 | 78.4 ± 0.55 | 84.0 ± 0.04 | 69.2 ± 2.1 | 81.7 ± 0.8 | 86.6 ± 0.9 |
| MGCA-R50 | 73.2 ± 0.3 | 79.9 ± 0.08 | 83.5 ± 0.04 | 84.5 ± 0.5 | 89.1 ± 0.3 | 90.6 ± 0.2 |
| MGCA-ViT | 78.2 ± 0.1 | 82.4 ± 0.03 | 84.4 ± 0.05 | 88.3 ± 0.1 | 91.5 ± 0.2 | 91.8 ± 0.3 |
| MRM | 80.1 ± 0.1 | 83.5 ± 0.10 | 85.3 ± 0.05 | 87.1 ± 0.1 | 89.9 ± 0.1 | 91.2 ± 0.3 |
| REFERS | 76.4 ± 0.3 | 81.3 ± 0.01 | 83.7 ± 0.06 | 87.1 ± 0.1 | 89.4 ± 0.3 | 90.0 ± 0.5 |
🔼 This table presents the results of a multi-label image classification task, comparing the performance of various Medical Vision-Language Pretraining (MedVLP) models. The performance is measured using the Area Under the Receiver Operating Characteristic curve (AUROC), a common metric for evaluating the effectiveness of classification models in distinguishing between multiple classes. Results are shown for three different training data sizes (1%, 10%, and 100%), highlighting the impact of data availability on model performance. The table indicates the best and second-best AUROC scores achieved by each MedVLP model on two benchmark datasets, NIH and VinDr.
read the caption
Table 1: Multi-label classification performance (%percent\%%) of MedVLP methods (Best, Second Best).
In-depth insights#
MedVLP Benchmarking#
The paper introduces BenchX, a novel benchmark framework designed to rigorously evaluate Medical Vision-Language Pretraining (MedVLP) methods. Existing MedVLP evaluations suffer from inconsistencies in datasets, preprocessing, and finetuning, hindering fair comparisons. BenchX addresses these issues by providing a unified framework encompassing diverse, comprehensive datasets, standardized preprocessing and training protocols, and consistent task adaptation pipelines. This allows for head-to-head comparisons of various MedVLP models across different downstream tasks such as classification, segmentation and report generation. By establishing baselines and identifying optimal configurations, BenchX enables a more reliable evaluation of existing and future MedVLP methods, highlighting the importance of standardized methodology for fair comparisons and driving progress in the field. Key findings challenge previous assumptions regarding relative performance and encourage reevaluation of existing conclusions in MedVLP research.
Unified BenchX#
BenchX is a novel unified benchmark framework designed for the head-to-head comparison and systematic analysis of Medical Vision-Language Pretraining (MedVLP) methods on chest X-ray datasets. Its core strength lies in standardizing data preprocessing, training strategies, and finetuning protocols, thus eliminating inconsistencies that hinder fair comparisons among different MedVLP models. This framework employs a comprehensive set of datasets, covering nine datasets and four medical tasks, which helps ensure robust evaluations. BenchX’s standardized evaluation facilitates consistent task adaptation in classification, segmentation, and report generation, allowing for a more accurate assessment of each method’s strengths and weaknesses. By establishing baselines for nine state-of-the-art MedVLP methods, BenchX reveals surprising findings, such as the potential of enhancing early MedVLP models to surpass recent methods, highlighting the need for revisiting conclusions drawn from previous works. The unified nature of BenchX and its publicly available codebase promote reproducibility and contribute to the creation of a more robust and reliable evaluation environment for the advancement of MedVLP research.
MedVLP Baselines#
The paper establishes baselines for nine state-of-the-art MedVLP methods using a unified benchmark framework called BenchX. BenchX ensures fair comparison by standardizing data preprocessing, training, and evaluation protocols across various datasets and tasks. The results reveal inconsistencies in the relative performance of different MedVLP models across tasks, highlighting the importance of robust evaluation methodologies. Surprisingly, older models like ConVIRT demonstrated strong performance when appropriately tuned, surpassing some more recent methods. This underscores the need for comprehensive analysis and careful consideration of hyperparameters when evaluating MedVLP methods. The unified evaluation protocols in BenchX greatly enhance the reliability and reproducibility of MedVLP research.
Task Adaptation#
The research paper section on ‘Task Adaptation’ highlights the challenges in directly applying pre-trained Medical Vision-Language Pretraining (MedVLP) models to downstream tasks due to heterogeneous model architectures and inconsistent finetuning protocols. The authors address these issues by proposing unified task adaptation pipelines for classification, segmentation, and report generation. For classification, a simple linear classifier is added, enabling consistent evaluation across different MedVLP models. Segmentation uses a unified UperNet architecture to accommodate various backbones, avoiding bias from using different segmentation networks. Report generation leverages the adaptable R2Gen framework. Standardized protocols ensure consistent performance evaluation, irrespective of the original MedVLP model architecture, thus enabling fair comparison and analysis among diverse methods. This approach allows for a more robust and reliable evaluation of MedVLP methods by minimizing the influence of task-specific adaptations on the overall performance. The authors emphasize the importance of consistent evaluation methodologies for accurate benchmarking and understanding of the MedVLP advancements.
Future Work#
The provided text does not contain a section explicitly titled “Future Work.” Therefore, I cannot provide a summary of such a section. To generate the requested summary, please provide the relevant text from the research paper’s “Future Work” section.
More visual insights#
More on tables
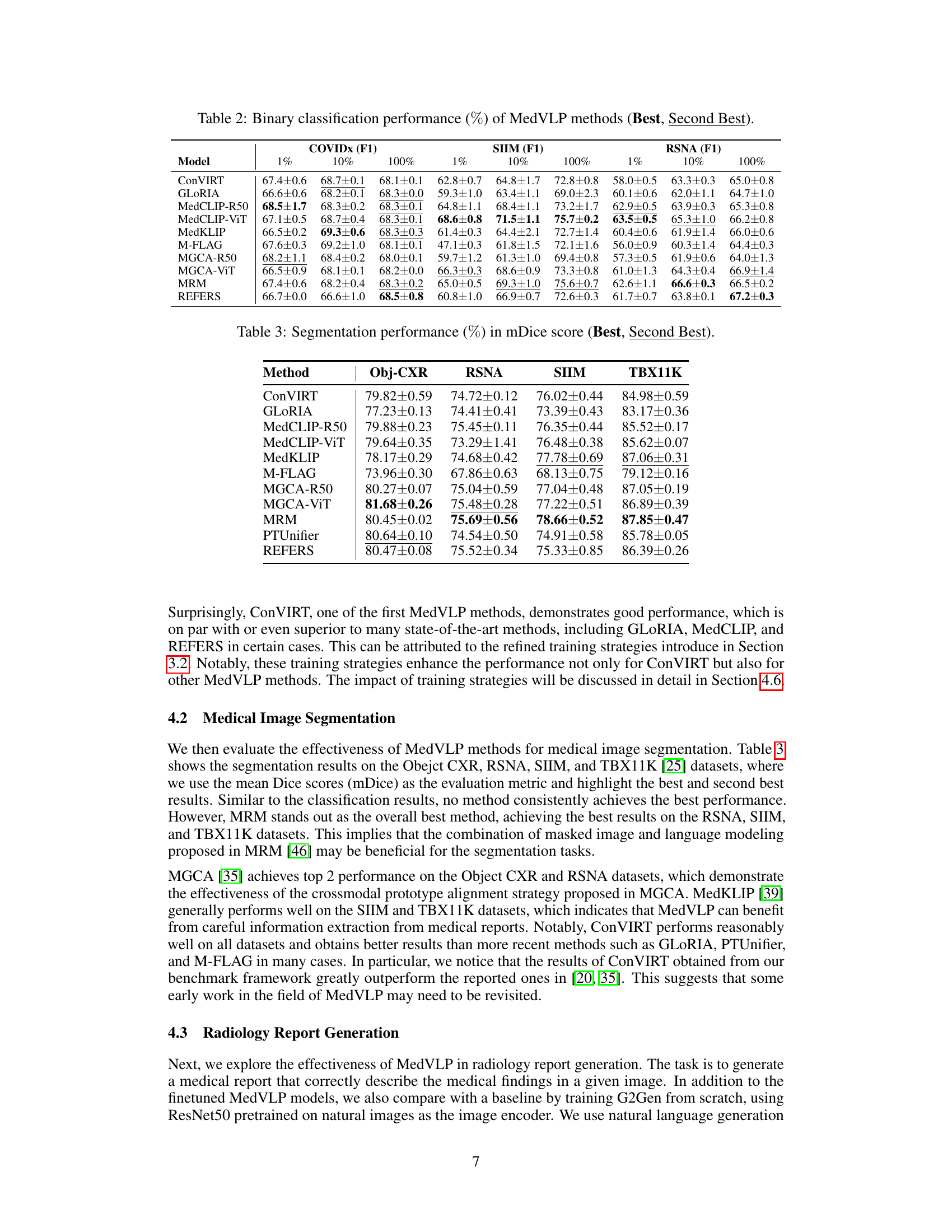
| Model | 1% | 10% | 100% | SIIM (F1) | 1% | 10% | 100% | RSNA (F1) | 1% | 10% | 100% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ConVIRT | 67.4±0.6 | 68.7±0.1 | 68.1±0.1 | 62.8±0.7 | 64.8±1.7 | 72.8±0.8 | 58.0±0.5 | 63.3±0.3 | 65.0±0.8 | ||
| GLoRIA | 66.6±0.6 | 68.2±0.1 | 68.3±0.0 | 59.3±1.0 | 63.4±1.1 | 69.0±2.3 | 60.1±0.6 | 62.0±1.1 | 64.7±1.0 | ||
| MedCLIP-R50 | 68.5±1.7 | 68.3±0.2 | 68.3±0.1 | 64.8±1.1 | 68.4±1.1 | 73.2±1.7 | 62.9±0.5 | 63.9±0.3 | 65.3±0.8 | ||
| MedCLIP-ViT | 67.1±0.5 | 68.7±0.4 | 68.3±0.1 | 68.6±0.8 | 71.5±1.1 | 75.7±0.2 | 63.5±0.5 | 65.3±1.0 | 66.2±0.8 | ||
| MedKLIP | 66.5±0.2 | 69.3±0.6 | 68.3±0.3 | 61.4±0.3 | 64.4±2.1 | 72.7±1.4 | 60.4±0.6 | 61.9±1.4 | 66.0±0.6 | ||
| M-FLAG | 67.6±0.3 | 69.2±1.0 | 68.1±0.1 | 47.1±0.3 | 61.8±1.5 | 72.1±1.6 | 56.0±0.9 | 60.3±1.4 | 64.4±0.3 | ||
| MGCA-R50 | 68.2±1.1 | 68.4±0.2 | 68.0±0.1 | 59.7±1.2 | 61.3±1.0 | 69.4±0.8 | 57.3±0.5 | 61.9±0.6 | 64.0±1.3 | ||
| MGCA-ViT | 66.5±0.9 | 68.1±0.1 | 68.2±0.0 | 66.3±0.3 | 68.6±0.9 | 73.3±0.8 | 61.0±1.3 | 64.3±0.4 | 66.9±1.4 | ||
| MRM | 67.4±0.6 | 68.2±0.4 | 68.3±0.2 | 65.0±0.5 | 69.3±1.0 | 75.6±0.7 | 62.6±1.1 | 66.6±0.3 | 66.5±0.2 | ||
| REFERS | 66.7±0.0 | 66.6±1.0 | 68.5±0.8 | 60.8±1.0 | 66.9±0.7 | 72.6±0.3 | 61.7±0.7 | 63.8±0.1 | 67.2±0.3 |
🔼 This table presents the results of binary classification experiments using various Medical Vision-Language Pretraining (MedVLP) methods. It shows the performance, measured as the F1 score (%), across three different datasets: COVIDx, RSNA, and SIIM. Results are presented for three training set sizes (1%, 10%, and 100%) to illustrate the effect of data availability. The best and second-best performing models are highlighted for each dataset and training set size.
read the caption
Table 2: Binary classification performance (%percent\%%) of MedVLP methods (Best, Second Best).
| Method | Obj-CXR | RSNA | SIIM | TBX11K |
|---|---|---|---|---|
| ConVIRT | 79.82 ± 0.59 | 74.72 ± 0.12 | 76.02 ± 0.44 | 84.98 ± 0.59 |
| GLoRIA | 77.23 ± 0.13 | 74.41 ± 0.41 | 73.39 ± 0.43 | 83.17 ± 0.36 |
| MedCLIP-R50 | 79.88 ± 0.23 | 75.45 ± 0.11 | 76.35 ± 0.44 | 85.52 ± 0.17 |
| MedCLIP-ViT | 79.64 ± 0.35 | 73.29 ± 1.41 | 76.48 ± 0.38 | 85.62 ± 0.07 |
| MedKLIP | 78.17 ± 0.29 | 74.68 ± 0.42 | 77.78 ± 0.69 | 87.06 ± 0.31 |
| M-FLAG | 73.96 ± 0.30 | 67.86 ± 0.63 | 68.13 ± 0.75 | 79.12 ± 0.16 |
| MGCA-R50 | 80.27 ± 0.07 | 75.04 ± 0.59 | 77.04 ± 0.48 | 87.05 ± 0.19 |
| MGCA-ViT | 81.68 ± 0.26 | 75.48 ± 0.28 | 77.22 ± 0.51 | 86.89 ± 0.39 |
| MRM | 80.45 ± 0.02 | 75.69 ± 0.56 | 78.66 ± 0.52 | 87.85 ± 0.47 |
| PTUnifier | 80.64 ± 0.10 | 74.54 ± 0.50 | 74.91 ± 0.58 | 85.78 ± 0.05 |
| REFERS | 80.47 ± 0.08 | 75.52 ± 0.34 | 75.33 ± 0.85 | 86.39 ± 0.26 |
🔼 This table presents the performance of various Medical Vision-Language Pretraining (MedVLP) models on medical image segmentation tasks. The mDice score, a common metric for evaluating segmentation accuracy, is reported for each model on four different chest X-ray datasets (Obj-CXR, RSNA, SIIM, and TBX11K). The table shows the best and second-best performing models for each dataset, providing a detailed comparison of the MedVLP methods’ ability to perform accurate medical image segmentation.
read the caption
Table 3: Segmentation performance (%percent\%%) in mDice score (Best, Second Best).
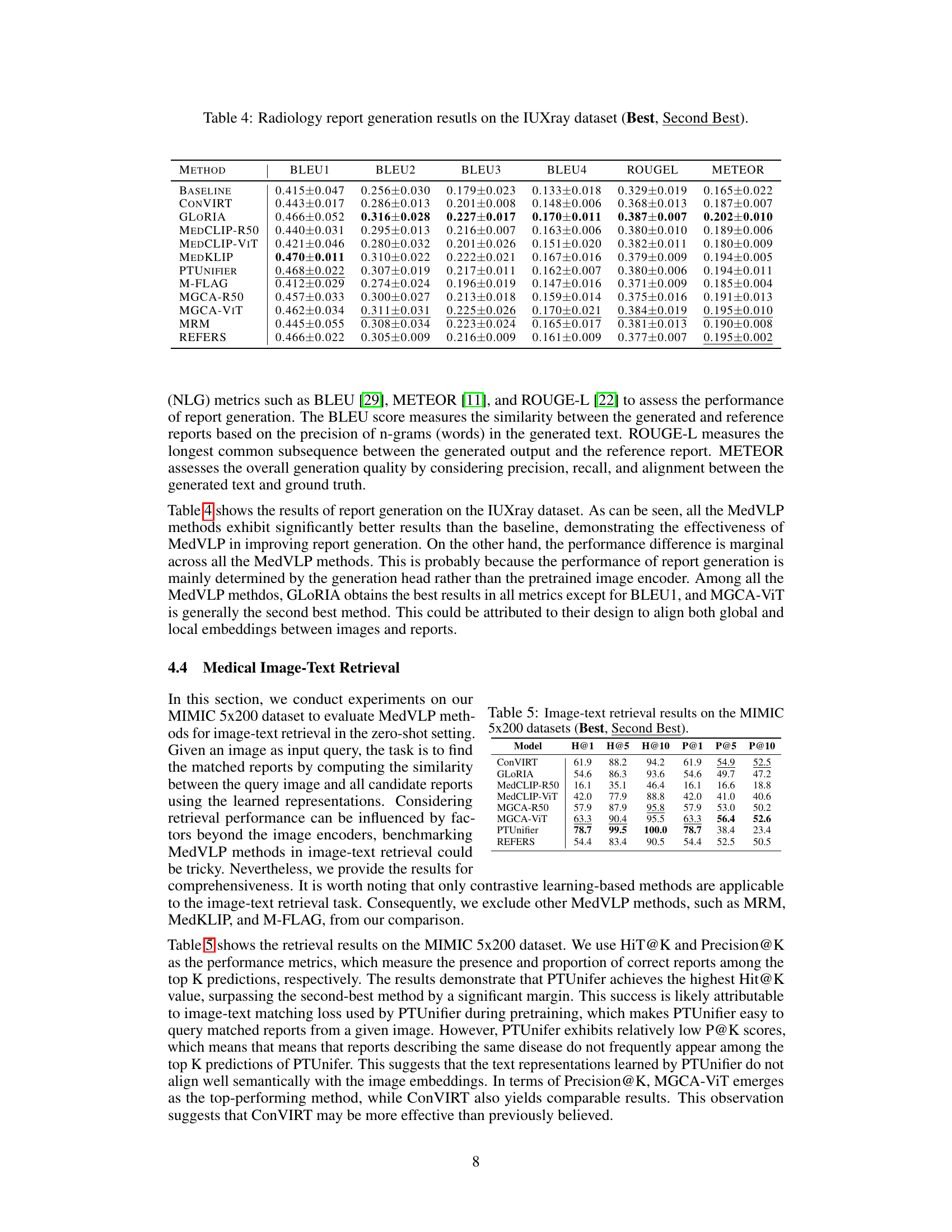
| Method | BLEU1 | BLEU2 | BLEU3 | BLEU4 | ROUGEL | METEOR |
|---|---|---|---|---|---|---|
| Baseline | 0.415 ± 0.047 | 0.256 ± 0.030 | 0.179 ± 0.023 | 0.133 ± 0.018 | 0.329 ± 0.019 | 0.165 ± 0.022 |
| ConVIRT | 0.443 ± 0.017 | 0.286 ± 0.013 | 0.201 ± 0.008 | 0.148 ± 0.006 | 0.368 ± 0.013 | 0.187 ± 0.007 |
| GLoRIA | 0.466 ± 0.052 | 0.316 ± 0.028 | 0.227 ± 0.017 | 0.170 ± 0.011 | 0.387 ± 0.007 | 0.202 ± 0.010 |
| MedCLIP-R50 | 0.440 ± 0.031 | 0.295 ± 0.013 | 0.216 ± 0.007 | 0.163 ± 0.006 | 0.380 ± 0.010 | 0.189 ± 0.006 |
| MedCLIP-ViT | 0.421 ± 0.046 | 0.280 ± 0.032 | 0.201 ± 0.026 | 0.151 ± 0.020 | 0.382 ± 0.011 | 0.180 ± 0.009 |
| MedKLIP | 0.470 ± 0.011 | 0.310 ± 0.022 | 0.222 ± 0.021 | 0.167 ± 0.016 | 0.379 ± 0.009 | 0.194 ± 0.005 |
| PTUnifier | 0.468 ± 0.022 | 0.307 ± 0.019 | 0.217 ± 0.011 | 0.162 ± 0.007 | 0.380 ± 0.006 | 0.194 ± 0.011 |
| M-FLAG | 0.412 ± 0.029 | 0.274 ± 0.024 | 0.196 ± 0.019 | 0.147 ± 0.016 | 0.371 ± 0.009 | 0.185 ± 0.004 |
| MGCA-R50 | 0.457 ± 0.033 | 0.300 ± 0.027 | 0.213 ± 0.018 | 0.159 ± 0.014 | 0.375 ± 0.016 | 0.191 ± 0.013 |
| MGCA-ViT | 0.462 ± 0.034 | 0.311 ± 0.031 | 0.225 ± 0.026 | 0.170 ± 0.021 | 0.384 ± 0.019 | 0.195 ± 0.010 |
| REFERS | 0.466 ± 0.022 | 0.305 ± 0.009 | 0.216 ± 0.009 | 0.161 ± 0.009 | 0.377 ± 0.007 | 0.195 ± 0.002 |
🔼 This table presents the quantitative results of radiology report generation on the IUXray dataset. It compares the performance of various Medical Vision-Language Pretraining (MedVLP) models against a baseline method. The evaluation metrics used are BLEU (1-4), ROUGE-L, and METEOR, all commonly used in Natural Language Generation (NLG) to assess the quality and similarity of generated text to reference text. The ‘Best’ and ‘Second Best’ columns indicate the top-performing MedVLP models for each metric.
read the caption
Table 4: Radiology report generation resutls on the IUXray dataset (Best, Second Best).
| Model | H@1 | H@5 | H@10 | P@1 | P@5 | P@10 |
|---|---|---|---|---|---|---|
| ConVIRT | 61.9 | 88.2 | 94.2 | 61.9 | 54.9 | 52.5 |
| GLoRIA | 54.6 | 86.3 | 93.6 | 54.6 | 49.7 | 47.2 |
| MedCLIP-R50 | 16.1 | 35.1 | 46.4 | 16.1 | 16.6 | 18.8 |
| MedCLIP-ViT | 42.0 | 77.9 | 88.8 | 42.0 | 41.0 | 40.6 |
| MGCA-R50 | 57.9 | 87.9 | 95.8 | 57.9 | 53.0 | 50.2 |
| MGCA-ViT | 63.3 | 90.4 | 95.5 | 63.3 | 56.4 | 52.6 |
| PTUnifier | 78.7 | 99.5 | 100.0 | 78.7 | 38.4 | 23.4 |
| REFERS | 54.4 | 83.4 | 90.5 | 54.4 | 52.5 | 50.5 |
🔼 This table presents the results of image-text retrieval experiments conducted on the MIMIC 5x200 dataset. The MIMIC 5x200 dataset is a subset of the larger MIMIC-CXR dataset, specifically focusing on 5 different medical findings (Atelectasis, Cardiomegaly, Edema, Pleural Effusion, and Consolidation). The task involves using an image as a query and retrieving the most relevant text reports describing that image. The table shows the performance of various MedVLP (Medical Vision-Language Pretraining) models, measured using two metrics: Hit@K (the percentage of correctly retrieved reports within the top K results) and Precision@K (the proportion of correctly retrieved reports among the top K results). The results are presented for K=1, 5, and 10. The table highlights the best and second-best performing models for each metric.
read the caption
Table 5: Image-text retrieval results on the MIMIC 5x200 datasets (Best, Second Best).
| Method | None | +DLR | +DLR+LN | All |
|---|---|---|---|---|
| ConVIRT | 71.7 | 76.9 ↑ | 74.5 ↓ | 77.0 ↑ |
| GLoRIA | 72.8 | 74.2 ↑ | 70.6 ↓ | 74.9 ↑ |
| MedCLIP-R50 | 74.1 | 73.7 ↓ | 74.2 ↑ | 73.8 ↓ |
| MedCLIP-ViT | 75.5 | 75.7 ↑ | 75.9 ↑ | 70.7 ↓ |
| MedKLIP | 74.4 | 71.9 ↓ | 75.2 ↑ | 73.7 ↓ |
| MGCA-R50 | 72.8 | 73.0 ↑ | 69.6 ↓ | 73.8 ↑ |
| MGCA-ViT | 77.7 | 78.1 ↑ | 78.2 ↑ | 78.2 = |
| MRM | 77.9 | 80.0 ↑ | 79.5 ↓ | 80.1 ↑ |
| REFERS | 76.8 | 75.9 ↓ | 76.2 ↓ | 75.6 ↓ |
🔼 This table presents the Area Under the Receiver Operating Characteristic Curve (AUROC) scores for different medical vision-language pretraining (MedVLP) models on the NIH Chest X-ray dataset. The models are evaluated using only 1% of the training data. Crucially, it showcases the impact of three different training strategies: Layer Normalization (LN), Truncated Normal Initialization (TNI), and Discriminative Learning Rates (DLR). By comparing AUROC scores across various combinations of these strategies, the table quantifies the impact of training choices on MedVLP model performance.
read the caption
Table 6: Classification results (AUROC) with different training strategies on the NIH dataset with 1%percent11\%1 % training data.
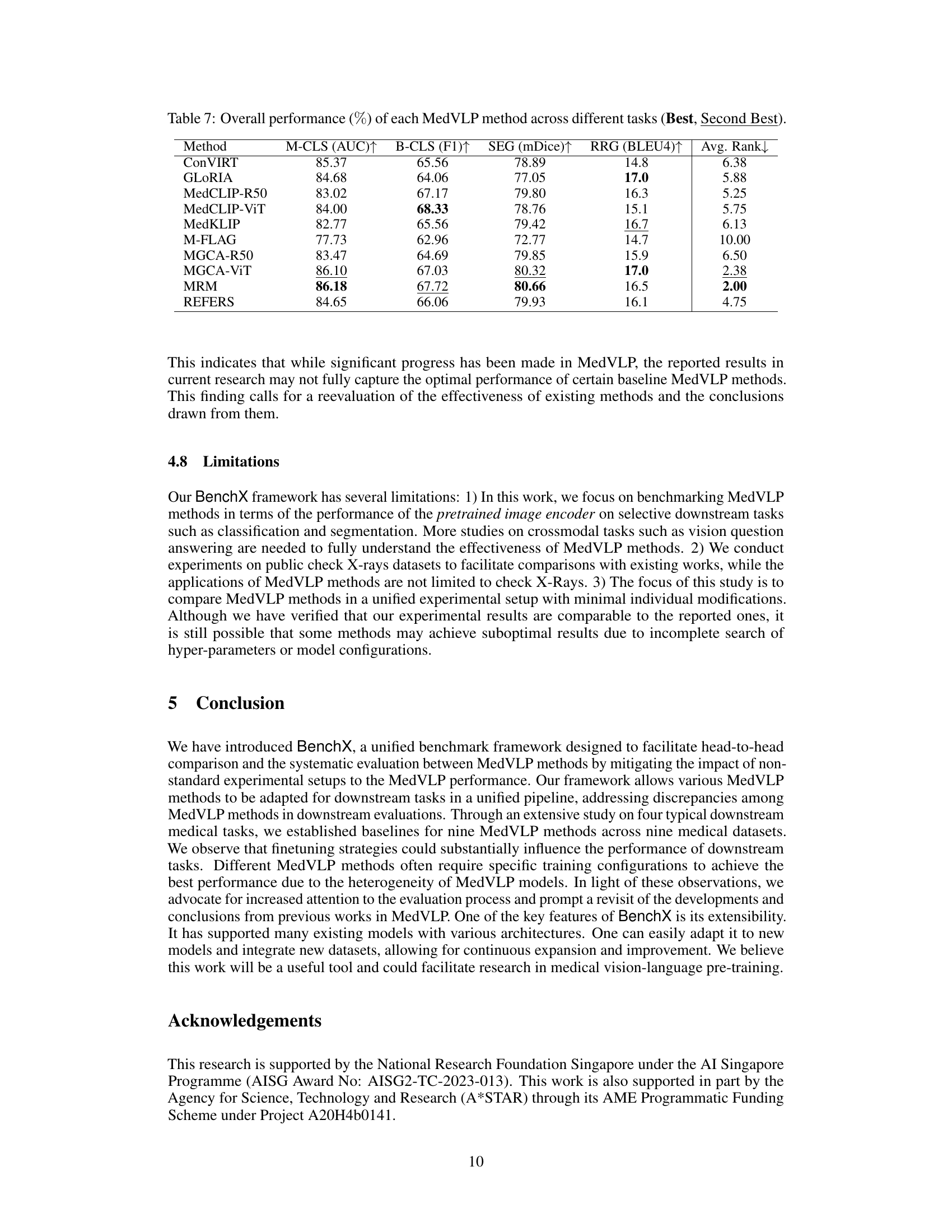
| Method | M-CLS (AUC) ↑ | B-CLS (F1) ↑ | SEG (mDice) ↑ | RRG (BLEU4) ↑ | Avg. Rank ↓ |
|---|---|---|---|---|---|
| ConVIRT | 85.37 | 65.56 | 78.89 | 14.8 | 6.38 |
| GLoRIA | 84.68 | 64.06 | 77.05 | 17.0 | 5.88 |
| MedCLIP-R50 | 83.02 | 67.17 | 79.80 | 16.3 | 5.25 |
| MedCLIP-ViT | 84.00 | 68.33 | 78.76 | 15.1 | 5.75 |
| MedKLIP | 82.77 | 65.56 | 79.42 | 16.7 | 6.13 |
| M-FLAG | 77.73 | 62.96 | 72.77 | 14.7 | 10.00 |
| MGCA-R50 | 83.47 | 64.69 | 79.85 | 15.9 | 6.50 |
| MGCA-ViT | 86.10 | 67.03 | 80.32 | 17.0 | 2.38 |
| MRM | 86.18 | 67.72 | 80.66 | 16.5 | 2.00 |
| REFERS | 84.65 | 66.06 | 79.93 | 16.1 | 4.75 |
🔼 This table presents a comprehensive comparison of nine Medical Vision-Language Pretraining (MedVLP) models across four distinct downstream medical tasks: multi-label classification, binary classification, segmentation, and radiology report generation. For each task, the table shows the average performance of each MedVLP model, expressed as a percentage, based on the best and second-best results achieved. The models are ranked based on their overall performance across all four tasks, offering insights into their relative strengths and weaknesses in handling different types of medical image analysis.
read the caption
Table 7: Overall performance (%percent\%%) of each MedVLP method across different tasks (Best, Second Best).
| Dataset | Image Size | Dataset Size | Task | Annotation |
|---|---|---|---|---|
| NIH ChestX-ray 14 | 224x224 | 112,120 | CLS | 14 Classes |
| VinDr-CXR | 512x640 | 18,000 | CLS | 28 classes, BBoxes |
| COVIDx CXR-4 | 1024x1024 | 84,818 | CLS | 2 Classes |
| SIIM-ACR PTX | 512x512 | 12,047 | CLS, SEG | 2 Classes, Masks |
| RSNA Pneumonia | 1024x1024 | 26,684 | CLS, SEG | BBoxes |
| IU-Xray | 512x640 | 3,955 | RRG | Image-Report Pairs |
| Object CXR | 2048x2624 | 10,000 | DET | BBoxes, Ellipse, Polygons |
| TBX11K | 512x512 | 11,200 | CLS, SEG | 3 classes, BBoxes |
| MIMIC 5x200 | 512x512 | 1,000 | RET | Image-Report Pairs |
🔼 This table presents a summary of the nine chest X-ray datasets used for evaluating the performance of various Medical Vision-Language Pretraining (MedVLP) methods. For each dataset, it lists the image size, the number of images, the type of task(s) it is used for (classification, segmentation, report generation, or image-text retrieval), and the type of annotations available (e.g., class labels, bounding boxes, masks, or image-report pairs).
read the caption
Table 8: Statistics of the test datasets.
| Method | Learning Rate | Batch Size | Optimizer | LN | DLR |
|---|---|---|---|---|---|
| ConVIRT | 1e-4 | 64 | Adam | Yes | Yes |
| GLoRIA | 1e-4 | 64 | Adam | Yes | Yes |
| MedCLIP-R50 | 1e-5 | 64 | Adam | No | No |
| MedCLIP-ViT | 1e-5 | 32 | Adam | No | No |
| MedKLIP | 1e-4 | 128 | Adam | No | Yes |
| M-FLAG | 1e-4 | 32 | Adam | Yes | No |
| MGCA-R50 | 1e-5 | 32 | Adam | Yes | No |
| MGCA-ViT | 1e-2 | 64 | SGD | Yes | Yes |
| MRM | 3e-2 | 64 | SGD | Yes | Yes |
| REFERS | 3e-2 | 32 | SGD | Yes | No |
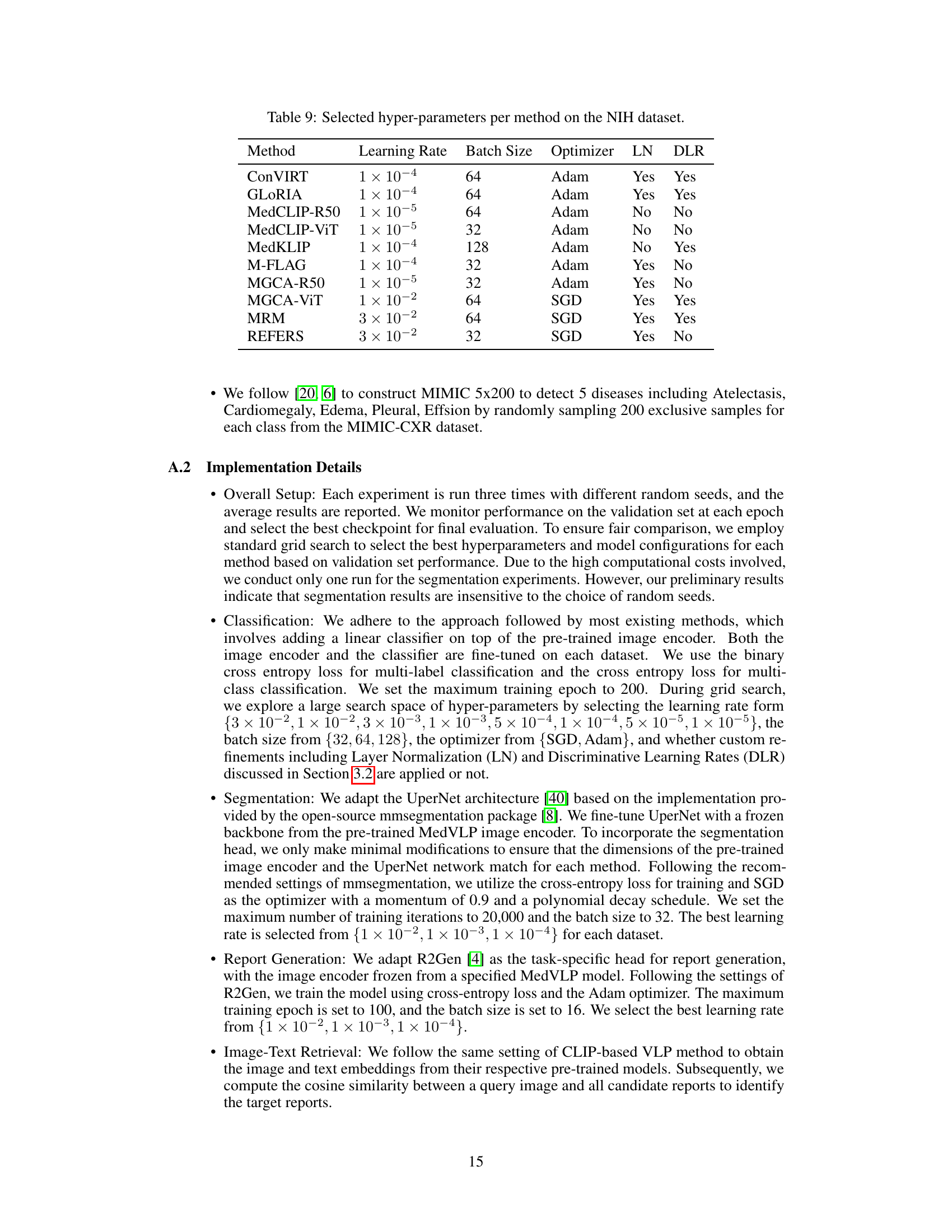
🔼 This table lists the hyperparameters used for each of the nine MedVLP methods evaluated on the NIH ChestX-Ray dataset. For each method, it shows the learning rate, batch size, optimizer used (Adam or SGD), whether layer normalization (LN) was applied, and whether discriminative learning rates (DLR) were used. These hyperparameters were chosen to optimize performance on the NIH dataset during the experiments.
read the caption
Table 9: Selected hyper-parameters per method on the NIH dataset.
| Method | Learning Rate | Batch Size | Optimizer | LN | DLR |
|---|---|---|---|---|---|
| ConVIRT | 5e-05 | 32 | Adam | Yes | Yes |
| GLoRIA | 1e-04 | 64 | Adam | Yes | Yes |
| MedCLIP-R50 | 1e-04 | 128 | Adam | No | No |
| MedCLIP-ViT | 1e-04 | 128 | Adam | No | No |
| MedKLIP | 1e-04 | 64 | Adam | No | Yes |
| M-FLAG | 1e-04 | 64 | Adam | Yes | No |
| MGCA-R50 | 5e-05 | 64 | Adam | Yes | No |
| MGCA-ViT | 0.03 | 64 | SGD | Yes | Yes |
| MRM | 0.01 | 64 | SGD | Yes | Yes |
| REFERS | 0.03 | 128 | SGD | Yes | No |
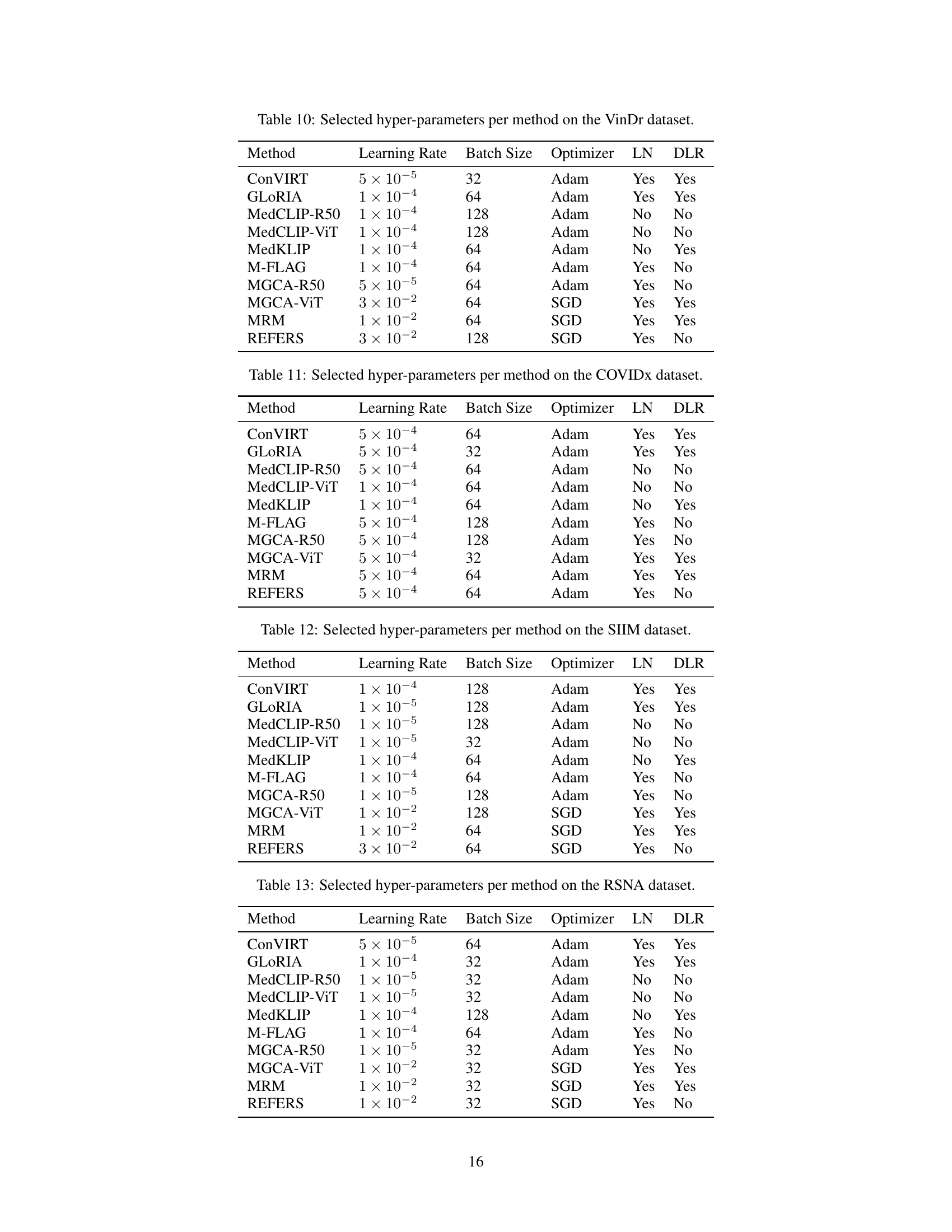
🔼 This table details the hyperparameters used for each of the nine MedVLP methods evaluated on the VinDr dataset. For each method, it lists the learning rate, batch size, optimizer used (Adam or SGD), whether layer normalization (LN) was applied, and whether discriminative learning rates (DLR) were used. This information is crucial for understanding and reproducing the experimental results, showcasing the fine-tuning choices made to optimize each method’s performance on this specific dataset.
read the caption
Table 10: Selected hyper-parameters per method on the VinDr dataset.
| Method | Learning Rate | Batch Size | Optimizer | LN | DLR |
|---|---|---|---|---|---|
| ConVIRT | 5e-04 | 64 | Adam | Yes | Yes |
| GLoRIA | 5e-04 | 32 | Adam | Yes | Yes |
| MedCLIP-R50 | 5e-04 | 64 | Adam | No | No |
| MedCLIP-ViT | 1e-04 | 64 | Adam | No | No |
| MedKLIP | 1e-04 | 64 | Adam | No | Yes |
| M-FLAG | 5e-04 | 128 | Adam | Yes | No |
| MGCA-R50 | 5e-04 | 128 | Adam | Yes | No |
| MGCA-ViT | 5e-04 | 32 | Adam | Yes | Yes |
| MRM | 5e-04 | 64 | Adam | Yes | Yes |
| REFERS | 5e-04 | 64 | Adam | Yes | No |
🔼 This table details the optimal hyperparameters used for each of the nine MedVLP (Medical Vision-Language Pretraining) models evaluated on the COVIDx dataset. The hyperparameters include the learning rate, batch size, optimizer used (Adam or SGD), and whether layer normalization (LN) and discriminative learning rates (DLR) were applied during training. This information is crucial for understanding the experimental setup and reproducibility of the results reported for each MedVLP model on this specific dataset.
read the caption
Table 11: Selected hyper-parameters per method on the COVIDx dataset.
| Method | Learning Rate | Batch Size | Optimizer | LN | DLR |
|---|---|---|---|---|---|
| ConVIRT | 1e-4 | 128 | Adam | Yes | Yes |
| GLoRIA | 1e-5 | 128 | Adam | Yes | Yes |
| MedCLIP-R50 | 1e-5 | 128 | Adam | No | No |
| MedCLIP-ViT | 1e-5 | 32 | Adam | No | No |
| MedKLIP | 1e-4 | 64 | Adam | No | Yes |
| M-FLAG | 1e-4 | 64 | Adam | Yes | No |
| MGCA-R50 | 1e-5 | 128 | Adam | Yes | No |
| MGCA-ViT | 1e-2 | 128 | SGD | Yes | Yes |
| MRM | 1e-2 | 64 | SGD | Yes | Yes |
| REFERS | 3e-2 | 64 | SGD | Yes | No |
🔼 This table details the hyperparameters used for each of the nine MedVLP (Medical Vision-Language Pretraining) methods tested on the SIIM (Society for Imaging Informatics in Medicine) dataset. It lists the learning rate, batch size, optimizer used, and whether layer normalization (LN) and discriminative learning rates (DLR) were applied during training. These settings are crucial for ensuring fair comparison between different MedVLP models on the SIIM dataset’s image segmentation task.
read the caption
Table 12: Selected hyper-parameters per method on the SIIM dataset.
| Method | Learning Rate | Batch Size | Optimizer | LN | DLR |
|---|---|---|---|---|---|
| ConVIRT | 5e-05 | 64 | Adam | Yes | Yes |
| GLoRIA | 1e-04 | 32 | Adam | Yes | Yes |
| MedCLIP-R50 | 1e-05 | 32 | Adam | No | No |
| MedCLIP-ViT | 1e-05 | 32 | Adam | No | No |
| MedKLIP | 1e-04 | 128 | Adam | No | Yes |
| M-FLAG | 1e-04 | 64 | Adam | Yes | No |
| MGCA-R50 | 1e-05 | 32 | Adam | Yes | No |
| MGCA-ViT | 0.01 | 32 | SGD | Yes | Yes |
| MRM | 0.01 | 32 | SGD | Yes | Yes |
| REFERS | 0.01 | 32 | SGD | Yes | No |
🔼 This table details the hyperparameters used for each of the nine MedVLP methods evaluated on the RSNA dataset. It lists the learning rate, batch size, optimizer used (Adam or SGD), and whether layer normalization (LN) and discriminative learning rates (DLR) were employed. This information is crucial for reproducibility and understanding the experimental setup of the study.
read the caption
Table 13: Selected hyper-parameters per method on the RSNA dataset.
Full paper#