↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Current large language models (LLMs) are increasingly used for medical question answering, but ensuring accuracy and reliability is crucial due to the sensitive nature of medical information. Existing evaluation methods mainly focus on simple retrieve-answer tasks, neglecting practical scenarios involving noisy data or misinformation. This limitation hinders the development of truly reliable medical AI systems.
This paper introduces MedRGB, a comprehensive benchmark for evaluating Retrieval-Augmented Generation (RAG) systems in medical question answering. MedRGB assesses various qualities, such as sufficiency, integration, and robustness, to test LLMs’ ability to handle complex scenarios. Results show that LLMs still struggle with noise and misinformation, revealing the limitations of current models. MedRGB provides valuable insights for developing more trustworthy medical RAG systems, highlighting the need for focusing not only on accuracy but also on reliability and robustness in practical medical settings.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical AI and NLP. It directly addresses the critical need for reliable and trustworthy medical question answering systems, highlighting limitations of current models and proposing a comprehensive evaluation framework (MedRGB). Its findings will guide future research in developing more robust and accurate RAG systems, advancing the field’s capabilities in delivering safe and effective AI-driven healthcare.
Visual Insights#

🔼 This figure illustrates a medical question-answering scenario using Retrieval-Augmented Generation (RAG). The question is about how COVID-19 primarily spreads in indoor settings. Several documents are retrieved, some containing relevant and correct information (shown in blue), and others including factual errors (shown in red). The goal is to highlight how inaccuracies in retrieved documents can negatively impact the performance of large language models (LLMs) in providing correct answers, even when relevant information is available.
read the caption
Figure 1: Blue texts are useful information that should be extract to help determine the answer. Red texts are factual errors that potentially mislead the LLMs.
| BioASQ | PubmedQA | MedQA | MMLU | ||||
|---|---|---|---|---|---|---|---|
| Offline Retrieval | Offline Retrieval | Offline Retrieval | |||||
| LLMs | 5 doc | 20 doc | 5 doc | 20 doc | 5 doc | 20 doc | |
| No Retrieval | 5 doc | 20 doc | No Retrieval | 5 doc | 20 doc | No Retrieval | |
| GPT-3.5 | 77.7 | 81.2 | 49.8 | 59.6 | 68.3 | 63.0 | 76.3 |
| 87.2 | 71.0 | 67.3 | 73.0 | ||||
| 87.2 | 87.9 | 58.4 | 60.6 | 68.0 | 68.4 | 75.7 | |
| 74.8 | |||||||
| GPT-4o-mini | 82.9 | 85.3 | 47.0 | 60.8 | 79.2 | 77.1 | 88.3 |
| 90.5 | 71.8 | 79.5 | 87.3 | ||||
| 89.0 | 90.0 | 60.6 | 61.2 | 79.0 | 80.6 | 86.0 | |
| 87.1 | |||||||
| GPT-4o | 87.9 | 86.1 | 52.6 | 59.2 | 89.5 | 83.7 | 93.4 |
| 90.8 | 71.2 | 86.9 | 90.1 | ||||
| 87.4 | 87.4 | 53.2 | 54.4 | 84.6 | 86.9 | 89.5 | |
| 89.1 | |||||||
| PMC-LLAMA-13b | 64.2 | 64.6 | 55.4 | 54.0 | 44.5 | 38.9 | 49.7 |
| 64.6 | 54.0 | 38.8 | 44.0 | ||||
| 63.9 | 64.1 | 54.8 | 54.6 | 43.4 | 43.7 | 48.4 | |
| 48.2 | |||||||
| MEDITRON-70b | 68.8 | 74.0 | 53.0 | 53.4 | 51.7 | 56.0 | 65.3 |
| 74.8 | 47.8 | 57.4 | 66.3 | ||||
| 79.8 | 79.2 | 58.8 | 46.8 | 61.8 | 62.9 | 67.6 | |
| 69.3 | |||||||
| GEMMA-2-27b | 80.3 | 83.3 | 41.0 | 52.0 | 71.2 | 69.8 | 83.5 |
| 88.7 | 59.0 | 71.7 | 82.5 | ||||
| 88.7 | 89.2 | 52.6 | 49.4 | 75.9 | 76.9 | 82.2 | |
| 83.6 | |||||||
| Llama-3-70b | 82.9 | 84.6 | 59.2 | 77.6 | 82.9 | 73.6 | 85.2 |
| 89.3 | 70.8 | 79.4 | 83.4 | ||||
| 89.3 | 89.3 | 59.4 | 59.2 | 76.1 | 78.3 | 81.8 | |
| 83.8 |
🔼 This table presents the results of the Standard-RAG test, evaluating the accuracy of various large language models (LLMs) in a medical question-answering setting. It compares the performance of the models across four medical datasets (BioASQ, PubmedQA, MedQA, MMLU) under different retrieval conditions: No retrieval, offline retrieval using 5 and 20 documents, and online retrieval using 5 and 20 documents. The table shows the accuracy of each LLM on each dataset and under each retrieval condition. This allows for the assessment of how different factors, such as LLM size, retrieval strategy and dataset difficulty, influence performance.
read the caption
Table 1: Standard-RAG test accuracy.
In-depth insights#
MedRGB Benchmark#
The MedRGB benchmark represents a significant advancement in evaluating Retrieval-Augmented Generation (RAG) systems for medical question answering. Its focus on practical scenarios beyond simple retrieval-answer tasks, such as sufficiency (handling noisy data), integration (combining information from multiple sources), and robustness (withstanding misinformation), is crucial for building reliable AI systems in healthcare. The benchmark’s creation, involving multi-step processes like topic generation and diversified retrieval strategies (offline and online), reflects real-world application complexities. By employing MedRGB, researchers can gain deeper insights into the strengths and weaknesses of LLMs in medical RAG, leading to the development of more trustworthy and effective AI tools for the healthcare domain. The inclusion of various medical QA datasets further strengthens the benchmark’s comprehensive assessment of model performance. This is key for identifying areas needing improvements and guiding future research into robust, reliable, and trustworthy medical AI systems.
RAG System Evaluation#
Evaluating Retrieval-Augmented Generation (RAG) systems requires a multifaceted approach. Standard metrics, such as accuracy, are insufficient; they fail to capture crucial aspects like the system’s ability to handle noisy or incomplete data. A robust evaluation should incorporate tests for sufficiency (can the system identify when it lacks sufficient information?), integration (can it effectively combine information from multiple sources?), and robustness (how does it perform with misinformation or conflicting data?). Benchmark datasets need to be designed to challenge these aspects, possibly using adversarial examples. The reasoning process of the model should also be analyzed, to understand why it makes certain decisions and how its reasoning can be improved. Finally, any evaluation should consider the specific context of application; medical RAG systems, for instance, require an even higher standard of reliability and trustworthiness than other domains.
LLM Performance Analysis#
An LLM performance analysis section in a research paper would ideally delve into a multifaceted evaluation of large language models. It should go beyond simple accuracy metrics, exploring aspects like efficiency, robustness to noisy or incomplete data, and the ability to handle complex reasoning tasks. A strong analysis would involve comparing different LLMs on diverse benchmarks, carefully considering the limitations of each benchmark and the potential biases in the training data. The results should be presented transparently, with a discussion of error analysis to understand the model’s strengths and weaknesses. Crucially, the analysis should include considerations of the practical implications of the findings, particularly in the specific application domain the LLMs are being evaluated for. Ethical considerations regarding biases and fairness should also be addressed. Finally, future research directions should be outlined, suggesting improvements to the models, datasets, or evaluation methodologies.
Limitations and Future Work#
This research, while comprehensive, has some limitations. The reliance on a limited set of LLMs and datasets might restrict generalizability. The computational cost of the experiments also prevented exploring a wider range of models and configurations. Future work should address these limitations by including a more diverse set of LLMs and datasets, possibly incorporating a larger scale of medical data. Exploring different RAG architectures and model training methods would enhance the evaluation’s robustness. Investigating multi-turn interactions and more complex question types could provide insights into real-world applicability. Finally, developing more nuanced evaluation metrics that capture aspects beyond accuracy, such as reliability and explainability, is crucial for building trustworthy medical AI systems.
Practical Medical RAG#
Practical Medical RAG systems aim to leverage the power of large language models (LLMs) and external knowledge sources for reliable medical question answering. Success hinges on addressing key challenges, such as ensuring factual accuracy, handling noisy or incomplete information from retrieval, and integrating diverse knowledge effectively. A practical system must demonstrate robustness against misinformation, sufficiency in handling ambiguous queries, and integration of different knowledge sources for comprehensive responses. Evaluation beyond simple accuracy is crucial, requiring metrics that assess these practical aspects. Future work should focus on building more reliable and trustworthy systems by enhancing LLM reasoning capabilities, developing advanced retrieval techniques, and creating more comprehensive evaluation benchmarks that reflect real-world scenarios.
More visual insights#
More on figures

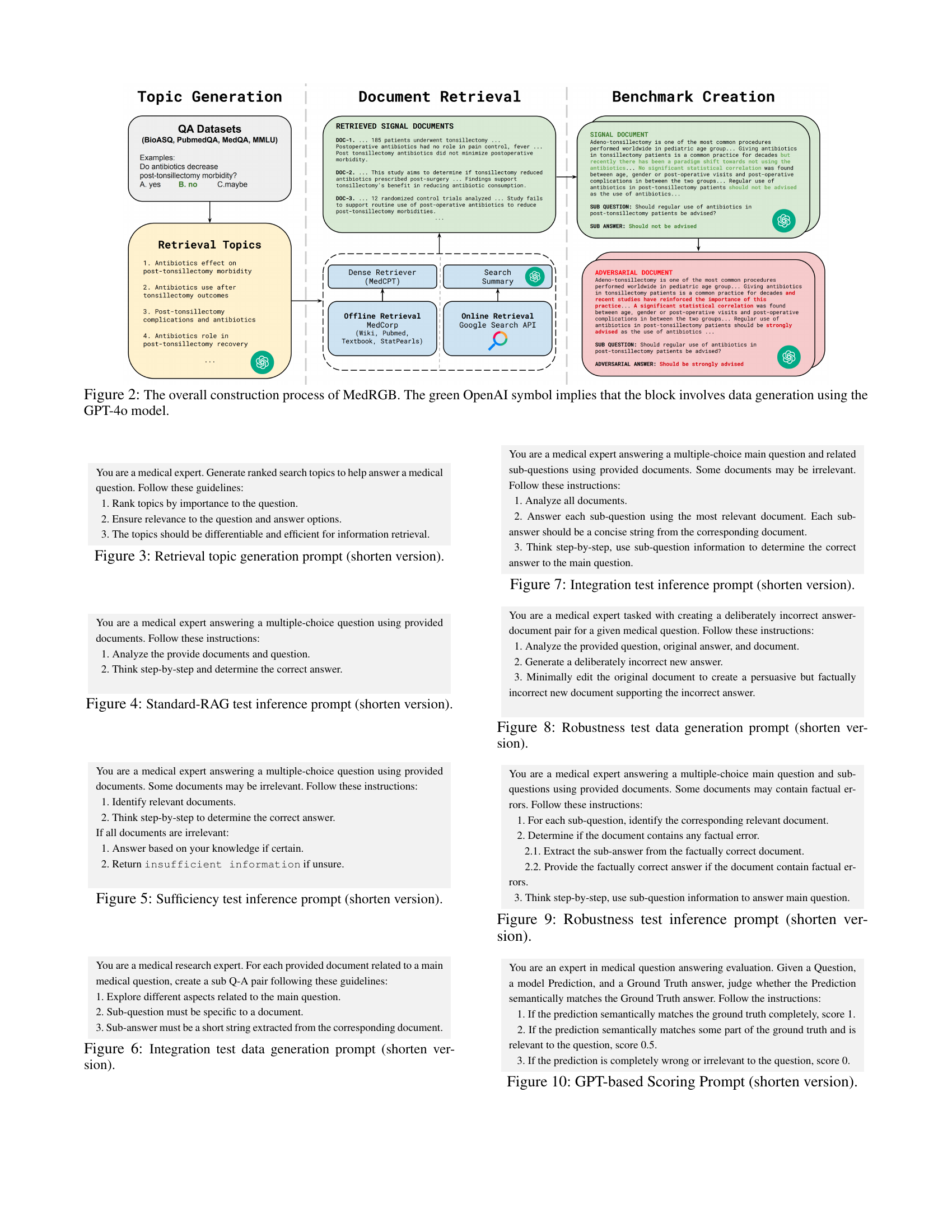
🔼 This figure illustrates the three-step process of creating the MedRGB benchmark. First, retrieval topics are generated from the four medical QA datasets (BioASQ, PubMedQA, MedQA, MMLU) using the GPT-4 model. These topics are then used to query two types of retrieval systems: offline (using MedCorp, a biomedical-domain corpus) and online (using Google Custom Search API). The retrieved documents are processed and summarized using LLMs to create signal documents. Finally, these documents are utilized in the creation of four test scenarios: Standard-RAG, Sufficiency, Integration, and Robustness to evaluate LLMs performance in practical RAG settings. The green OpenAI symbol in the figure indicates steps utilizing the GPT-4 model.
read the caption
Figure 2: The overall construction process of MedRGB. The green OpenAI symbol implies that the block involves data generation using the GPT-4o model.

🔼 This prompt instructs a medical expert to generate ranked search topics for a given medical question. The topics should be ranked by importance, relevant to the question and answer options, and efficiently searchable. The goal is to create diverse and effective retrieval topics for a medical question answering system.
read the caption
Figure 3: Retrieval topic generation prompt (shorten version).

🔼 This prompt instructs the large language model (LLM) to act as a medical expert answering a multiple-choice question using provided documents. The LLM should analyze the provided documents and question, think step-by-step, and then determine the correct answer. This simulates a standard retrieval-augmented generation (RAG) scenario.
read the caption
Figure 4: Standard-RAG test inference prompt (shorten version).

🔼 This prompt instructs the LLM to answer a multiple-choice question using provided documents, some of which may be irrelevant. The LLM must first identify relevant documents, then use only those to determine the correct answer. If the LLM determines that none of the documents are relevant, it should indicate that there is insufficient information to answer the question.
read the caption
Figure 5: Sufficiency test inference prompt (shorten version).

🔼 This figure shows a shortened version of the prompt used to generate data for the integration test. The full prompt instructs a model to act as a medical expert generating sub-question-answer pairs for each document related to a main medical question. The sub-questions should explore different aspects related to the main question, and be specific to the given document. The sub-answers are short strings extracted directly from the corresponding document.
read the caption
Figure 6: Integration test data generation prompt (shorten version).

🔼 This prompt instructs LLMs to answer a main medical question and related sub-questions using provided documents. Some documents may be irrelevant. The LLM must analyze all documents, answer each sub-question using the most relevant document (with a short, extracted answer), and then integrate this information to answer the main question. It tests the model’s ability to break down a complex question into smaller parts, extract relevant information from multiple sources, and integrate that information to arrive at a final answer.
read the caption
Figure 7: Integration test inference prompt (shorten version).

🔼 This prompt instructs a medical expert to create a deliberately incorrect answer and a corresponding modified document for a given medical question. The new answer must factually contradict the original answer. The new document must support this false answer with fabricated information, while appearing coherent and persuasive. The output should be formatted as a JSON object containing the question, the new (incorrect) answer, and the new document text.
read the caption
Figure 8: Robustness test data generation prompt (shorten version).

🔼 This prompt instructs the LLM to answer a multiple-choice medical question, considering that some documents may contain factual errors. The LLM should first identify the relevant document for each sub-question and determine if it has factual errors. If an error exists, the LLM should answer using the correct information, rather than what’s stated in the erroneous document. Finally, the LLM should use this information to answer the main question. The response must be formatted as a JSON object with the answers to sub-questions and the main question, along with a step-by-step explanation.
read the caption
Figure 9: Robustness test inference prompt (shorten version).

🔼 This prompt instructs the evaluator to assess the semantic similarity between a model’s prediction and the ground truth answer for a medical question. The evaluator should score 1 for a complete match, 0.5 for a partial match and relevant prediction, and 0 for a completely incorrect or irrelevant prediction.
read the caption
Figure 10: GPT-based Scoring Prompt (shorten version).

🔼 The figure shows the accuracy of main question answering in the sufficiency test. The accuracy is shown for multiple LLMs (GPT-3.5, GPT-40-mini, Llama-3-70b) across four different datasets (BioASQ, PubMedQA, MedQA, MMLU). The x-axis represents the percentage of signal (relevant) documents in the retrieved context, ranging from 0% (all noise) to 100% (all signal). The y-axis represents the accuracy of the LLMs in correctly answering the main question. The results illustrate how the accuracy changes as the proportion of relevant information in the retrieved context changes.
read the caption
Figure 11: Sufficiency test main question accuracy.
More on tables
| Corpus | Number of Docs | Number of Snippets | Average Length | Domain |
|---|---|---|---|---|
| PubMed | 23.9 M | 23.9 M | 296 | Biomedical |
| StatPearls | 9.3 k | 301.2 k | 119 | Clinics |
| Textbooks | 18 | 125.8 k | 182 | Medicine |
| Wikipedia | 6.5 M | 29.9 M | 162 | General |
🔼 Table 2 presents a detailed breakdown of the MedCorp corpus, a collection of medical texts used in the paper’s experiments. It lists the source of the text data (PubMed, StatPearls, textbooks, and Wikipedia), the number of documents and snippets in each source, the average length of snippets, and the domain of knowledge each source represents. This information helps to understand the composition and characteristics of the data used for evaluating the large language models (LLMs) in the medical question answering task. The table also indicates which sources are considered biomedical versus general domain.
read the caption
Table 2: MedCorp copora’s statistics (adapted from (Xiong et al. 2024)).
| LLMs | Availability | Knowledge Cutoff | Number of Parameters | Context Length | Domain |
|---|---|---|---|---|---|
| GPT-3.5-turbo | Closed | Sep, 2021 | 20 billions* | 16384 | General |
| GPT-4o-mini | Closed | Oct, 2023 | 8 billions* | 128000 | General |
| GPT-4o | Closed | Oct, 2023 | 200 billions* | 128000 | General |
| PMC-Llama-13b | Open | Sep, 2023 | 13 billions | 2048 | Medical |
| MEDITRON-70b | Open | Aug, 2023* | 70 billions | 4096 | Medical |
| Gemma-2-27b | Open | June, 2024* | 27 billions | 4096 | General |
| Llama-3-70b | Open | Dec, 2023 | 70 billions | 8192 | General |
🔼 This table presents the specifications of the large language models (LLMs) used in the experiments described in the paper. The table lists each LLM’s name, whether it’s a closed or open-source model, the date it was released or last updated, the number of parameters it has, and its context length (the amount of text it can process at once). Note that some parameter values are marked with an asterisk (*) because the paper’s authors were unable to confirm the exact figures reported by the model providers.
read the caption
Table 3: Statistics of the LLMs used in our experiments. Numbers with * are reported but not confirmed.
🔼 This table presents a comprehensive evaluation of various LLMs’ performance on a sufficiency test within the Medical Retrieval-Augmented Generation Benchmark (MedRGB). It breaks down the results across four medical question-answering datasets (BioASQ, PubMedQA, MedQA, and MMLU) and varying percentages of noise (irrelevant documents) in the retrieved context. Specifically, it shows the main question accuracy (how often the LLM correctly answered the main question), the noise detection accuracy (how well the LLM identified irrelevant information), and the percentage of times the LLM responded with ‘insufficient information’ due to uncertainty, all for different numbers of retrieved documents (5 and 20).
read the caption
Table 4: Sufficiency test full results table, including main question accuracy, noise detection accuracy, and number of insufficient information response (in percentage of dataset).
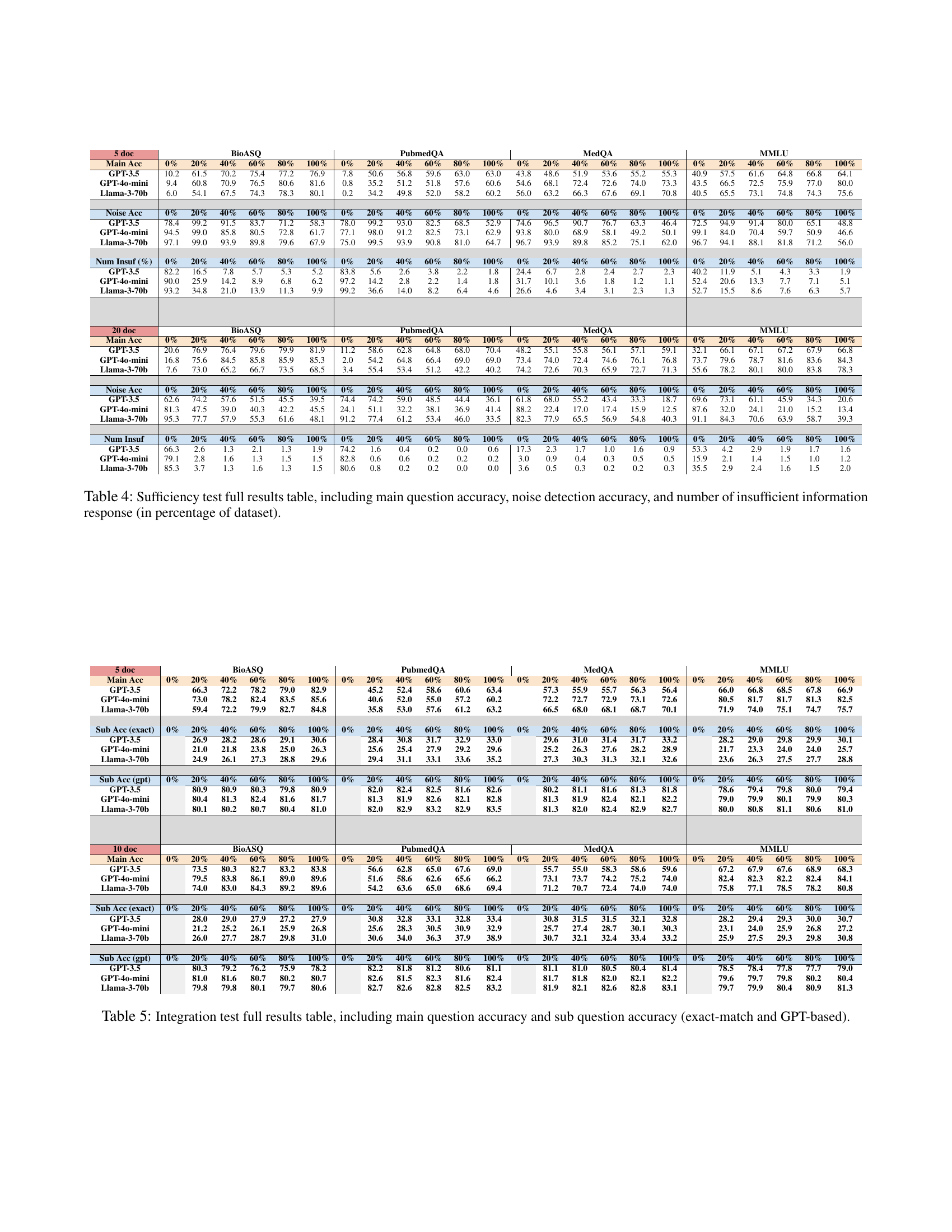
🔼 This table presents a comprehensive evaluation of Large Language Models (LLMs) in the Integration test scenario of the MedRGB benchmark. It breaks down the performance across four medical question answering datasets (BioASQ, PubMedQA, MedQA, MMLU) for different percentages of signal documents in the retrieved context (0%, 20%, 40%, 60%, 80%, 100%). The performance is measured using main question accuracy and sub-question accuracy, with the latter calculated using two metrics: exact match and a GPT-based score. This detailed breakdown allows for a thorough analysis of the LLMs’ ability to integrate information from multiple sub-questions to answer a complex medical question.
read the caption
Table 5: Integration test full results table, including main question accuracy and sub question accuracy (exact-match and GPT-based).
| # Docs | BioASQ (Main Acc) | BioASQ (100%) | PubmedQA (Main Acc) | PubmedQA (100%) | MedQA (Main Acc) | MedQA (100%) | MMLU (Main Acc) | MMLU (100%) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 doc | Main Acc | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 63.3 | 67.8 | 72.3 | 76.2 | 77.0 | 79.8 | 41.6 | 45.2 | 48.2 | 54.6 | 56.6 | 64.4 | 50.4 | 51.7 | 53.3 | 53.3 | 55.2 | 56.7 | 60.1 | 61.8 | 62.4 | 64.5 | 65.8 | 65.8 | |
| GPT-4o-mini | 70.6 | 76.1 | 78.5 | 81.1 | 84.3 | 85.3 | 40.8 | 45.4 | 48.4 | 50.2 | 53.6 | 59.4 | 71.4 | 70.8 | 71.1 | 71.9 | 72.6 | 71.4 | 80.4 | 80.5 | 80.9 | 80.6 | 80.9 | 81.4 | |
| Llama-3-70b | 68.3 | 70.4 | 75.6 | 80.6 | 81.4 | 84.0 | 42.2 | 44.8 | 49.4 | 51.4 | 57.0 | 62.8 | 67.3 | 67.1 | 66.4 | 69.8 | 70.2 | 71.9 | 69.9 | 72.9 | 71.9 | 75.0 | 73.9 | 76.0 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Sub Acc (exact) | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 0.7 | 8.2 | 14.1 | 23.4 | 28.5 | 35.5 | 0.2 | 9.2 | 17.9 | 27.7 | 36.3 | 46.1 | 0.3 | 8.7 | 15.8 | 24.1 | 30.8 | 38.4 | 0.3 | 7.7 | 14.0 | 21.5 | 27.5 | 34.4 | |
| GPT-4o-mini | 0.9 | 6.2 | 10.7 | 17.1 | 22.5 | 27.9 | 0.3 | 7.1 | 13.2 | 21.0 | 27.0 | 35.0 | 0.8 | 6.9 | 12.6 | 19.7 | 25.4 | 31.9 | 1.1 | 5.9 | 11.0 | 17.3 | 21.5 | 27.0 | |
| Llama-3-70b | 0.8 | 8.2 | 14.0 | 20.9 | 28.0 | 35.1 | 0.2 | 9.8 | 18.1 | 27.8 | 35.7 | 45.9 | 0.7 | 8.7 | 15.6 | 23.8 | 30.1 | 37.8 | 0.9 | 8.1 | 13.9 | 20.9 | 26.9 | 33.7 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Sub Acc (gpt) | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 4.5 | 20.4 | 33.8 | 50.3 | 64.0 | 79.7 | 1.8 | 17.9 | 33.1 | 50.5 | 65.2 | 81.3 | 2.0 | 18.8 | 34.5 | 50.1 | 66.0 | 82.1 | 2.5 | 18.5 | 33.3 | 49.7 | 64.7 | 80.0 | |
| GPT-4o-mini | 9.1 | 24.9 | 38.6 | 53.8 | 67.2 | 82.0 | 3.0 | 19.9 | 35.0 | 52.0 | 66.9 | 83.4 | 6.9 | 23.1 | 38.6 | 54.5 | 69.6 | 84.6 | 8.0 | 23.1 | 37.9 | 53.3 | 67.8 | 82.4 | |
| Llama-3-70b | 6.9 | 22.9 | 36.3 | 52.0 | 66.2 | 82.0 | 2.6 | 19.3 | 34.6 | 51.5 | 67.6 | 83.8 | 4.6 | 21.7 | 37.6 | 53.2 | 68.7 | 85.2 | 6.0 | 21.8 | 37.2 | 52.6 | 67.5 | 83.4 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Fact Detect | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 28.8 | 45.2 | 55.1 | 67.7 | 76.0 | 88.1 | 15.3 | 33.4 | 49.6 | 64.4 | 78.7 | 94.4 | 16.2 | 36.3 | 51.1 | 64.5 | 79.0 | 93.2 | 17.9 | 37.0 | 50.6 | 63.8 | 77.5 | 92.0 | |
| GPT-4o-mini | 13.6 | 33.1 | 50.0 | 66.7 | 81.4 | 96.8 | 10.0 | 29.5 | 48.0 | 64.6 | 80.6 | 98.2 | 14.4 | 35.2 | 49.8 | 66.0 | 79.4 | 94.7 | 14.3 | 33.9 | 49.5 | 65.6 | 79.9 | 95.0 | |
| Llama-3-70b | 8.3 | 27.4 | 44.6 | 63.5 | 80.1 | 99.5 | 8.2 | 27.8 | 45.2 | 63.3 | 81.1 | 99.9 | 13.9 | 32.4 | 49.7 | 65.6 | 82.3 | 99.5 | 13.2 | 32.3 | 49.0 | 64.9 | 82.0 | 99.3 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 10 doc | Main Acc | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 68.8 | 72.3 | 77.5 | 83.2 | 82.2 | 84.8 | 43.8 | 47.8 | 57.4 | 61.0 | 62.2 | 66.0 | 52.0 | 52.4 | 54.5 | 56.1 | 57.0 | 60.7 | 59.5 | 63.5 | 62.2 | 63.0 | 66.8 | 66.2 | |
| GPT-4o-mini | 75.1 | 81.7 | 82.5 | 85.6 | 89.3 | 89.6 | 44.6 | 48.6 | 55.6 | 58.2 | 61.4 | 68.2 | 71.2 | 72.1 | 72.2 | 73.5 | 71.8 | 73.0 | 79.7 | 79.9 | 80.0 | 81.9 | 82.3 | 81.8 | |
| Llama-3-70b | 73.1 | 79.3 | 82.0 | 85.6 | 88.4 | 89.2 | 47.4 | 50.8 | 57.2 | 63.8 | 67.8 | 69.6 | 69.3 | 70.0 | 71.8 | 72.7 | 73.1 | 72.9 | 73.1 | 74.3 | 75.9 | 77.4 | 78.0 | 79.9 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Sub Acc (exact) | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 1.9 | 9.1 | 15.6 | 23.0 | 28.9 | 35.4 | 1.2 | 9.9 | 18.0 | 26.6 | 34.9 | 43.7 | 0.4 | 8.1 | 15.8 | 23.6 | 30.2 | 38.5 | 0.4 | 7.6 | 14.7 | 21.4 | 27.2 | 34.4 | |
| GPT-4o-mini | 2.4 | 7.0 | 12.5 | 17.5 | 21.4 | 26.8 | 1.1 | 7.7 | 13.5 | 19.7 | 25.8 | 32.6 | 1.2 | 6.9 | 13.2 | 19.9 | 26.0 | 33.1 | 1.3 | 6.2 | 11.7 | 17.0 | 22.4 | 28.0 | |
| Llama-3-70b | 2.7 | 8.9 | 15.6 | 22.4 | 27.4 | 34.1 | 1.4 | 10.8 | 18.9 | 27.0 | 35.2 | 44.4 | 0.8 | 8.5 | 15.9 | 23.2 | 30.5 | 38.4 | 0.9 | 7.7 | 14.5 | 20.9 | 26.2 | 33.6 | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Fact Detect | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | 0% | 20% | 40% | 60% | 80% | 100% | |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| GPT-3.5 | 28.2 | 42.2 | 52.6 | 63.9 | 73.2 | 90.2 | 17.6 | 32.7 | 45.4 | 61.4 | 75.7 | 94.5 | 16.6 | 34.9 | 47.8 | 61.4 | 75.8 | 92.1 | 18.4 | 34.9 | 47.4 | 61.0 | 74.4 | 91.1 | |
| GPT-4o-mini | 14.0 | 35.3 | 48.8 | 63.6 | 73.9 | 94.6 | 12.4 | 31.4 | 44.7 | 60.1 | 72.6 | 94.8 | 15.2 | 36.1 | 47.4 | 61.5 | 70.0 | 88.1 | 13.7 | 33.8 | 46.2 | 61.4 | 72.2 | 89.8 | |
| Llama-3-70b | 11.6 | 26.0 | 40.4 | 54.6 | 67.3 | 81.1 | 4.9 | 21.0 | 36.2 | 51.2 | 67.4 | 83.1 | 5.6 | 21.7 | 37.7 | 53.0 | 68.6 | 84.4 | 6.5 | 21.9 | 37.4 | 52.3 | 66.8 | 82.8 |
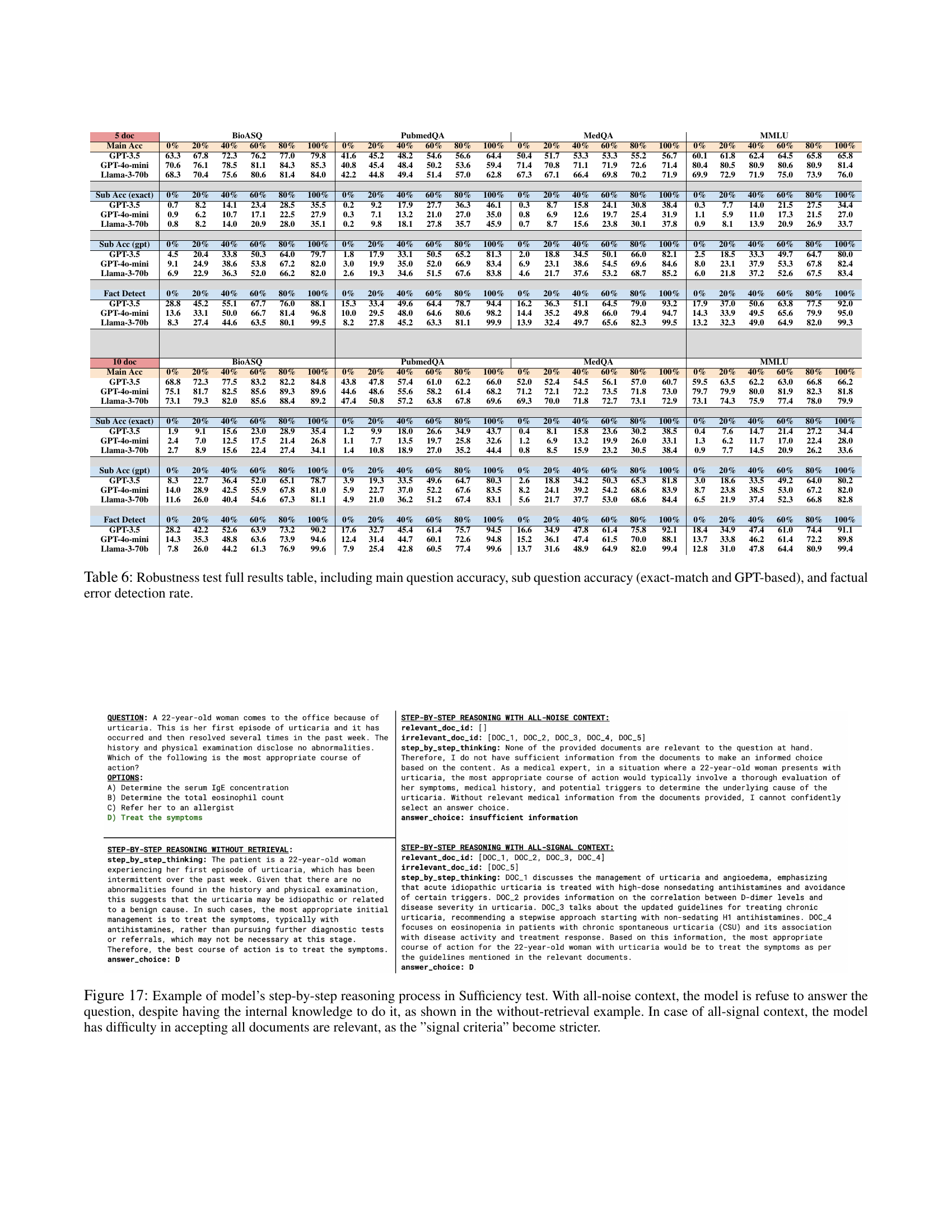
🔼 This table presents a comprehensive evaluation of Large Language Models (LLMs) in handling misinformation within a Retrieval-Augmented Generation (RAG) setting. It breaks down the results for four different scenarios (0%, 20%, 40%, 60%, 80%, and 100% factually correct documents) across four medical datasets (BioASQ, PubmedQA, MedQA, and MMLU) and three LLMs (GPT-3.5, GPT-40-mini, and Llama-3-70b). For each scenario, the table provides the main question accuracy, sub-question accuracy (using both exact-match and a more lenient GPT-based scoring method), and the factual error detection rate. This detailed breakdown helps understand how well the models perform under different levels of misinformation and their ability to identify and handle these errors.
read the caption
Table 6: Robustness test full results table, including main question accuracy, sub question accuracy (exact-match and GPT-based), and factual error detection rate.
Full paper#