↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Radiology report generation (RRG) is crucial but challenging due to the complexity of medical images and reports. Existing AI methods primarily focus on model architecture improvements, neglecting the detailed organ-regional information crucial for accurate diagnoses. This often leads to inaccurate or incomplete reports, increasing radiologists’ workload.
This paper introduces a novel Organ-Regional Information Driven (ORID) framework to address these issues. ORID effectively integrates multi-modal data (radiology images and organ-specific descriptions) using a cross-modal fusion module. It also incorporates an organ importance coefficient analysis module to filter out noise from unrelated organs. Experiments show that ORID significantly outperforms existing methods across various evaluation metrics, proving its effectiveness in generating more accurate and comprehensive radiology reports.
Key Takeaways#
Why does it matter?#
This paper is important because it addresses the limitations of existing radiology report generation methods by incorporating organ-regional information, improving accuracy and efficiency. It introduces a novel framework, offers valuable insights into multimodal learning for medical image analysis, and opens avenues for future research in improving medical report generation.
Visual Insights#

🔼 This figure visualizes how organ-regional information is used in radiology report generation. It shows a chest X-ray image divided into sections representing different organs (lung, pleural, heart, bone, mediastinum). Each organ section is accompanied by a textual description from a diagnostic report. The descriptions highlight key findings relevant to each organ. To illustrate the connection between these pieces of information and the final generated report, colored boxes highlight sections of the image and corresponding text that contribute to specific sentences in a sample radiology report. This demonstrates the system’s ability to integrate multi-modal information from different parts of the image and descriptions.
read the caption
Figure 1: Visualization of organ-regional radiology image and diagnosis descriptions. Relevant segments associated with the target report have been highlighted using distinct colors.
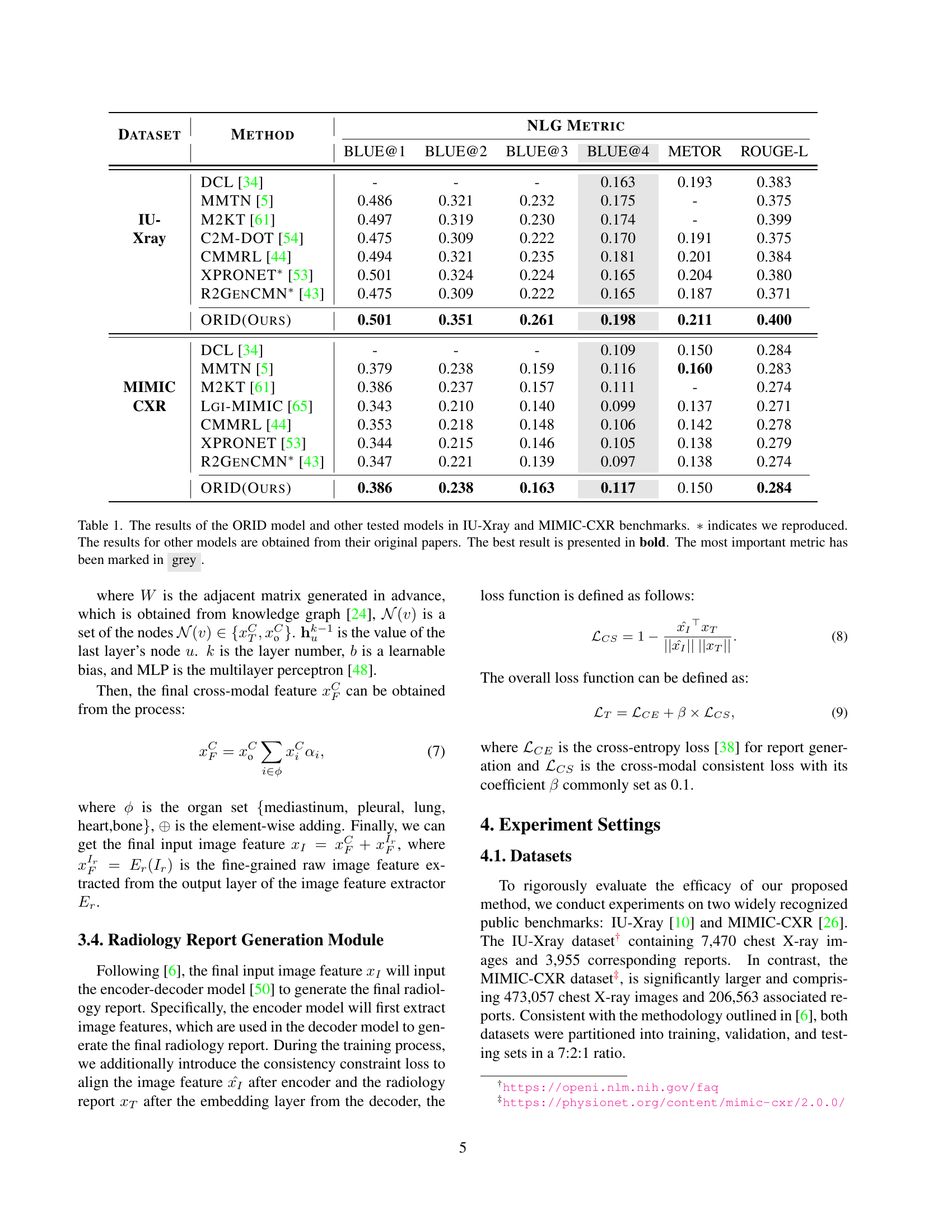
| Dataset | Method | BLUE@1 | BLUE@2 | BLUE@3 | BLUE@4 | METOR | ROUGE-L |
|---|---|---|---|---|---|---|---|
| DCL [34] | - | - | - | 0.163 | 0.193 | 0.383 | |
| MMTN [5] | 0.486 | 0.321 | 0.232 | 0.175 | - | 0.375 | |
| IU- | M2KT [61] | 0.497 | 0.319 | 0.230 | 0.174 | - | 0.399 |
| Xray | C2M-DOT [54] | 0.475 | 0.309 | 0.222 | 0.170 | 0.191 | 0.375 |
| CMMRL [44] | 0.494 | 0.321 | 0.235 | 0.181 | 0.201 | 0.384 | |
| XPRONET* [53] | 0.501 | 0.324 | 0.224 | 0.165 | 0.204 | 0.380 | |
| R2GenCMN* [43] | 0.475 | 0.309 | 0.222 | 0.165 | 0.187 | 0.371 | |
| ORID(Ours) | 0.501 | 0.351 | 0.261 | 0.198 | 0.211 | 0.400 | |
| DCL [34] | - | - | - | 0.109 | 0.150 | 0.284 | |
| MMTN [5] | 0.379 | 0.238 | 0.159 | 0.116 | 0.160 | 0.283 | |
| MIMIC | M2KT [61] | 0.386 | 0.237 | 0.157 | 0.111 | - | 0.274 |
| CXR | Lgi-MIMIC [65] | 0.343 | 0.210 | 0.140 | 0.099 | 0.137 | 0.271 |
| CMMRL [44] | 0.353 | 0.218 | 0.148 | 0.106 | 0.142 | 0.278 | |
| XPRONET [53] | 0.344 | 0.215 | 0.146 | 0.105 | 0.138 | 0.279 | |
| R2GenCMN* [43] | 0.347 | 0.221 | 0.139 | 0.097 | 0.138 | 0.274 | |
| ORID(Ours) | 0.386 | 0.238 | 0.163 | 0.117 | 0.150 | 0.284 |
🔼 This table presents a comparison of the performance of the proposed ORID model against several state-of-the-art models on two benchmark datasets: IU-Xray and MIMIC-CXR. The evaluation metrics used are BLEU (at various n-gram levels), METEOR, and ROUGE-L, which are standard metrics for evaluating natural language generation. The results for the ORID model are directly from the authors’ experiments. Results for other models were taken from their respective papers. The best score for each metric is highlighted in bold, and the most important metric (ROUGE-L) is shown in gray.
read the caption
Table 1: The results of the ORID model and other tested models in IU-Xray and MIMIC-CXR benchmarks. ∗*∗ indicates we reproduced. The results for other models are obtained from their original papers. The best result is presented in bold. The most important metric has been marked in grey.
In-depth insights#
ORID Framework#
The ORID (Organ-Regional Information Driven) framework presents a novel approach to radiology report generation. It cleverly integrates multi-modal information from radiological images and organ-specific diagnostic descriptions. A key strength lies in its ability to reduce noise from irrelevant organs, improving the accuracy and relevance of the generated report. This is achieved through a sophisticated architecture incorporating an organ-based cross-modal fusion module and an organ importance coefficient analysis module which uses Graph Neural Networks (GNNs) to analyze organ interconnections and assign importance weights. The framework’s foundation involves instruction-tuning of LLaVA-Med to create LLaVA-Med-RRG, enhancing organ-regional diagnostic capabilities. Overall, ORID demonstrates a significant advancement over existing methods by leveraging the detailed organ-regional information inherent in radiology, resulting in more accurate and comprehensive reports. The results show promising performance improvements across various evaluation metrics, highlighting the method’s potential to improve both the efficiency and reliability of radiology report generation.
LLaVA-Med Enhancement#
The LLaVA-Med Enhancement section would detail how the authors adapted the LLaVA-Med model, a large language and vision assistant, for radiology report generation. This likely involved fine-tuning LLaVA-Med on a new dataset of radiology images and their corresponding reports, specifically focusing on organ-regional information. This dataset would probably be curated to improve the model’s ability to identify and describe findings within specific organs, reducing noise from irrelevant regions. The enhancement might also focus on the model architecture, possibly by incorporating modules for multi-modal fusion of image and textual data, or by integrating techniques to weigh the importance of different organ regions within a report, thereby improving the overall accuracy and coherence of generated reports. Ultimately, the success of this enhancement would be judged by its ability to surpass the performance of existing radiology report generation models on established benchmark datasets, demonstrated by improvements in metrics such as BLEU, ROUGE-L, and METEOR, in addition to clinical evaluation metrics that assess the accuracy and relevance of the reports from a medical perspective.
Cross-Modal Fusion#
The effectiveness of radiology report generation hinges on effectively integrating information from multiple modalities, such as images and textual descriptions. Cross-modal fusion is the crucial step in achieving this integration. The paper explores organ-based cross-modal fusion, a method that processes image and text features from individual organs separately. This strategy is particularly advantageous as it reduces the influence of noise from unrelated organs, a significant challenge in handling complex medical images. By focusing on specific organ regions, the fusion process can better isolate relevant image features pertinent to disease characteristics within each organ. This approach likely improves the precision and accuracy of the generated radiology report, potentially leading to better clinical decision making. The method also incorporates a coarse-grained fusion which adds all organ-level features together to account for diseases that affect multiple organs, which standard methods might not fully capture. This multi-level approach is a key strength, striking a balance between organ-specific detail and holistic analysis of disease patterns across the whole image.
Organ Importance#
The concept of ‘Organ Importance’ in radiology report generation is crucial for improving the accuracy and efficiency of automated systems. The research highlights how some organs are more critical to a diagnosis than others and proposes a method to quantify this importance. This is achieved by using a Graph Neural Network (GNN) to analyze the interconnections of multi-modal information (image and text) for each organ. This innovative approach effectively filters out noise from less relevant organs, leading to more focused and precise reports. The GNN’s ability to model complex relationships between different organ regions allows the system to prioritize information relevant to a diagnosis. By weighting the contribution of each organ based on its importance, the system can reduce the influence of irrelevant details and focus on the most critical aspects for a comprehensive report. This method improves the accuracy of disease detection and the relevance of the generated text, improving the quality of automatic radiology report generation and significantly enhancing radiologist workflows.
Ablation Study#
The ablation study systematically evaluates the contribution of each module within the proposed ORID framework. By removing components one at a time (e.g., the Organ-based Cross-modal Fusion module, the Organ Importance Coefficient Analysis module), the researchers assessed the impact on performance. The results reveal a significant performance boost with the addition of the cross-modal fusion module, indicating its importance in integrating image and textual information for accurate report generation. Furthermore, including both fine-grained and coarse-grained analysis enhances the model’s ability to capture nuanced organ-level details. The ablation study’s findings strongly support the design choices within ORID, highlighting the synergistic effect of these modules in achieving superior results compared to simpler baseline models. The methodical approach of the ablation study strengthens the overall validity and trustworthiness of the proposed framework. The study also suggests a balance between the inclusion of relevant detail and the filtering out of noise from less important areas. Finally, this process provides valuable insights into the individual contributions of each component and confirms the overall effectiveness of the ORID architecture.
More visual insights#
More on figures

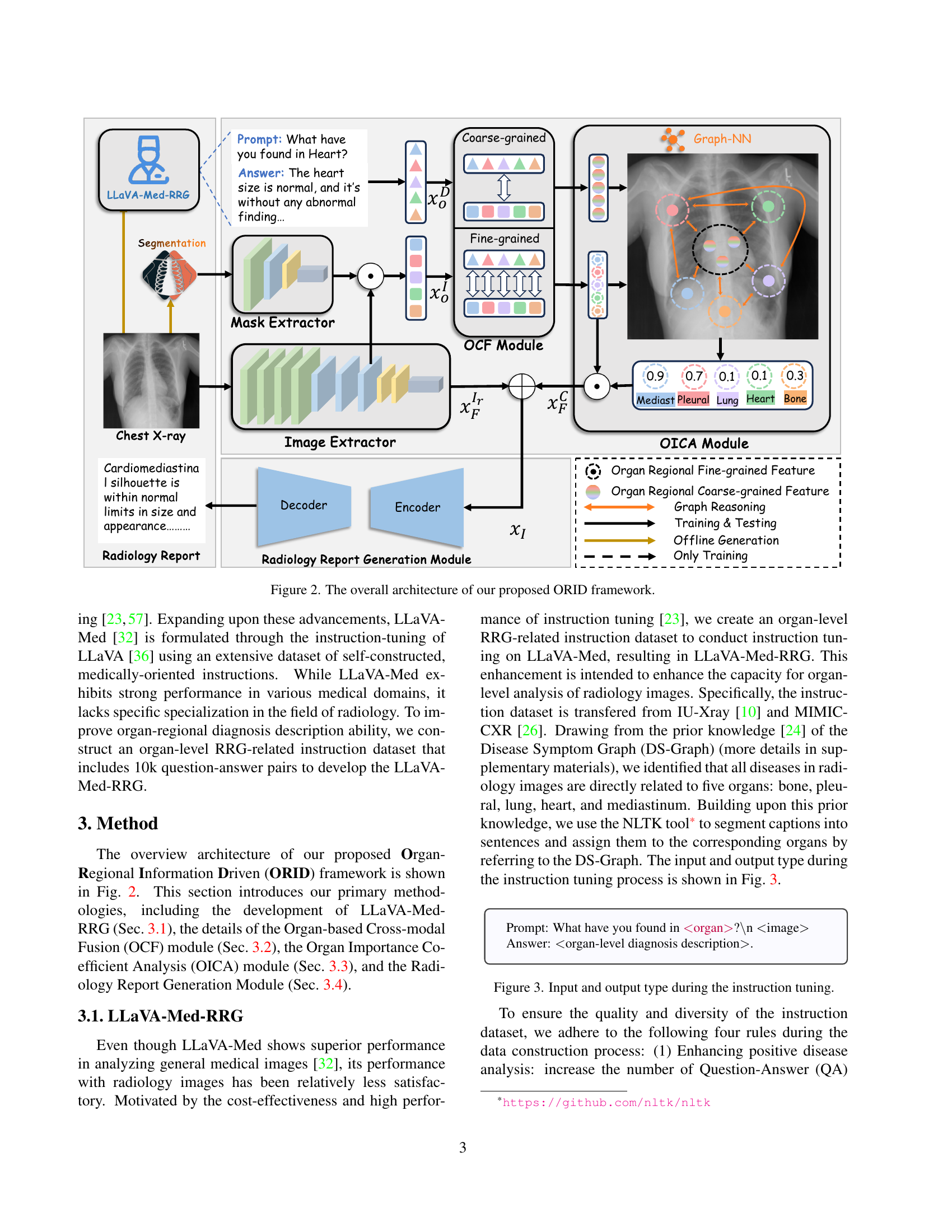
🔼 The figure illustrates the architecture of the Organ-Regional Information Driven (ORID) framework for radiology report generation. The framework consists of four key modules: 1) LLaVA-Med-RRG, which generates organ-regional descriptions from radiology images; 2) an Organ-based Cross-modal Fusion (OCF) module that combines the organ-regional descriptions with image features; 3) an Organ Importance Coefficient Analysis (OICA) module which uses graph neural networks to determine the importance of different organ regions; and 4) a Radiology Report Generation Module which produces the final report. The figure shows the data flow between these modules and highlights the integration of multi-modal information for improved report accuracy.
read the caption
Figure 2: The overall architecture of our proposed ORID framework.

🔼 This figure illustrates the input and output format used during the instruction tuning phase of the LLaVA-Med-RRG model. The input consists of a prompt in the form of a question about a specific organ (‘What have you found in
?’) followed by the corresponding radiology image. The output is an organ-level diagnosis description that answers the prompt based on the input image. read the caption
Figure 3: Input and output type during the instruction tuning.

🔼 This figure compares the organ-regional diagnostic descriptions generated by LLaVA-Med and LLaVA-Med-RRG models. LLaVA-Med-RRG is a modified version of LLaVA-Med specifically trained for radiology report generation. The figure shows an example of a chest X-ray image and the respective descriptions. Sentences in the generated reports that match or are closely related to the ground truth (target report) are highlighted in green, while those that do not match are marked in red. This visualization highlights the improvement in accuracy and relevance of organ-level diagnostic descriptions achieved by the LLaVA-Med-RRG model compared to the original LLaVA-Med model.
read the caption
Figure 4: An example of LLaVA-Med’s organ-reional diagnosis description compare with that of LLaVA-Med-RRG. The sentences that are correct or highly-related with target reports have been marked in green, otherwise have been marked in red.

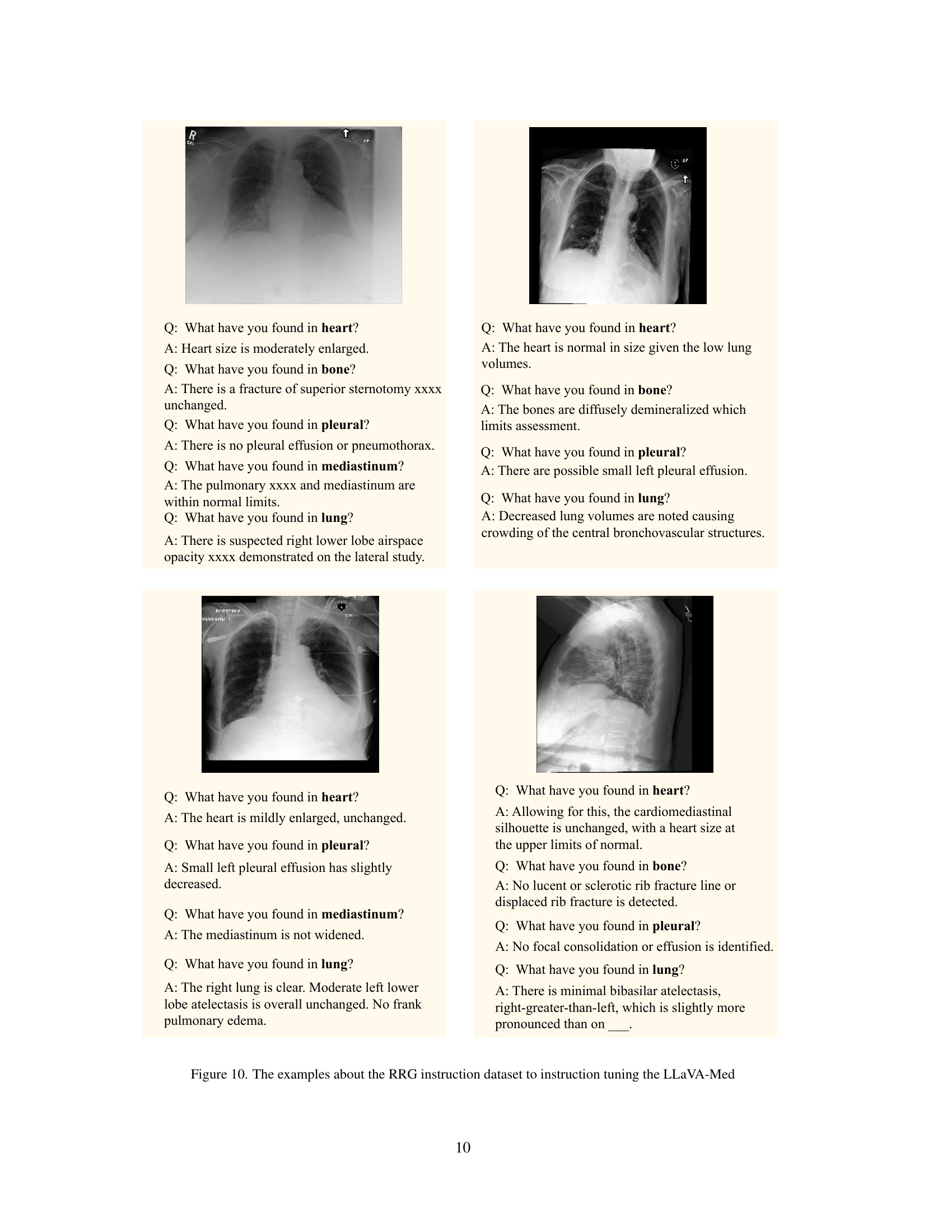
🔼 This figure presents a statistical analysis of the dataset used for instruction tuning of the LLaVA-Med model for radiology report generation. It shows the number of question-answer pairs and the average token length for each of the five organs considered: lung, pleural, heart, bone, and mediastinum. This visualization helps understand the distribution of data across different organs and the complexity of the language descriptions associated with them.
read the caption
Figure 5: Statistical analysis of question-answer pairs and average token length for each organ.

🔼 This figure shows a word cloud visualization summarizing the terms frequently used in the lung section of radiology reports. The size of each word reflects its frequency, providing a quick overview of the most common findings and descriptors associated with the lungs in the dataset used for training the radiology report generation model. It helps to understand the model’s focus on certain aspects of lung-related analysis.
read the caption
(a) Lung

🔼 This subfigure shows an example of segmented organ regions from a chest X-ray image. Specifically, it highlights the regions related to the pleural area, which is the thin membrane that surrounds the lungs. Different colors likely represent different sub-regions within the pleural cavity such as different parts of the pleura (visceral and parietal) or areas potentially showing different findings, like pleural thickening or effusion. The image demonstrates the precise segmentation ability crucial for the model’s organ-regional analysis.
read the caption
(b) Pleural

🔼 This image shows a visualization of the mediastinum region from a chest X-ray. The mediastinum is the central compartment of the thorax, containing the heart, great vessels, trachea, esophagus, and other structures. Different image segmentation masks are overlaid to highlight the specific areas of each organ within the mediastinum, helping to illustrate organ-regional information.
read the caption
(c) Mediastinum

🔼 The figure shows a visual representation of heart-related findings from the radiology report generation model’s output. It displays various descriptions from different models highlighting features like ‘mild cardiomegaly,’ ’normal heart size,’ and ’likely normal moderately_enlarged.’ These descriptions represent different levels of precision and accuracy in detecting and characterizing cardiac abnormalities, which demonstrates the impact of different models on radiology report generation. This variability underscores the challenges inherent in automatically generating accurate and detailed radiology reports.
read the caption
(d) Heart

🔼 This subfigure shows several examples of bone-related findings in chest X-ray images. The findings illustrate various conditions that may be detected in bone, such as fractures (acute or chronic), displaced ribs, and general bone abnormalities. These diverse examples highlight the range of bone-related issues that radiologists may encounter when analyzing chest X-rays.
read the caption
(e) Bone

🔼 This figure shows a word cloud visualization summarizing the most frequent terms used in the radiology reports for each organ (lung, pleural, heart, bone, mediastinum) and the overall report. It provides a visual representation of the key terminology associated with different organ systems, highlighting common themes and diagnostic terms present in the dataset.
read the caption
(f) Total

🔼 This figure visualizes the frequency of words related to each organ (lung, pleural, heart, bone, mediastinum) and the overall dataset used for instruction tuning. Word size corresponds to frequency; larger words appeared more often in the dataset. This provides insight into the types of descriptions present in the training data for each organ.
read the caption
Figure 6: The word cloud analysis about each organ and total in instruction-tuning dataset.

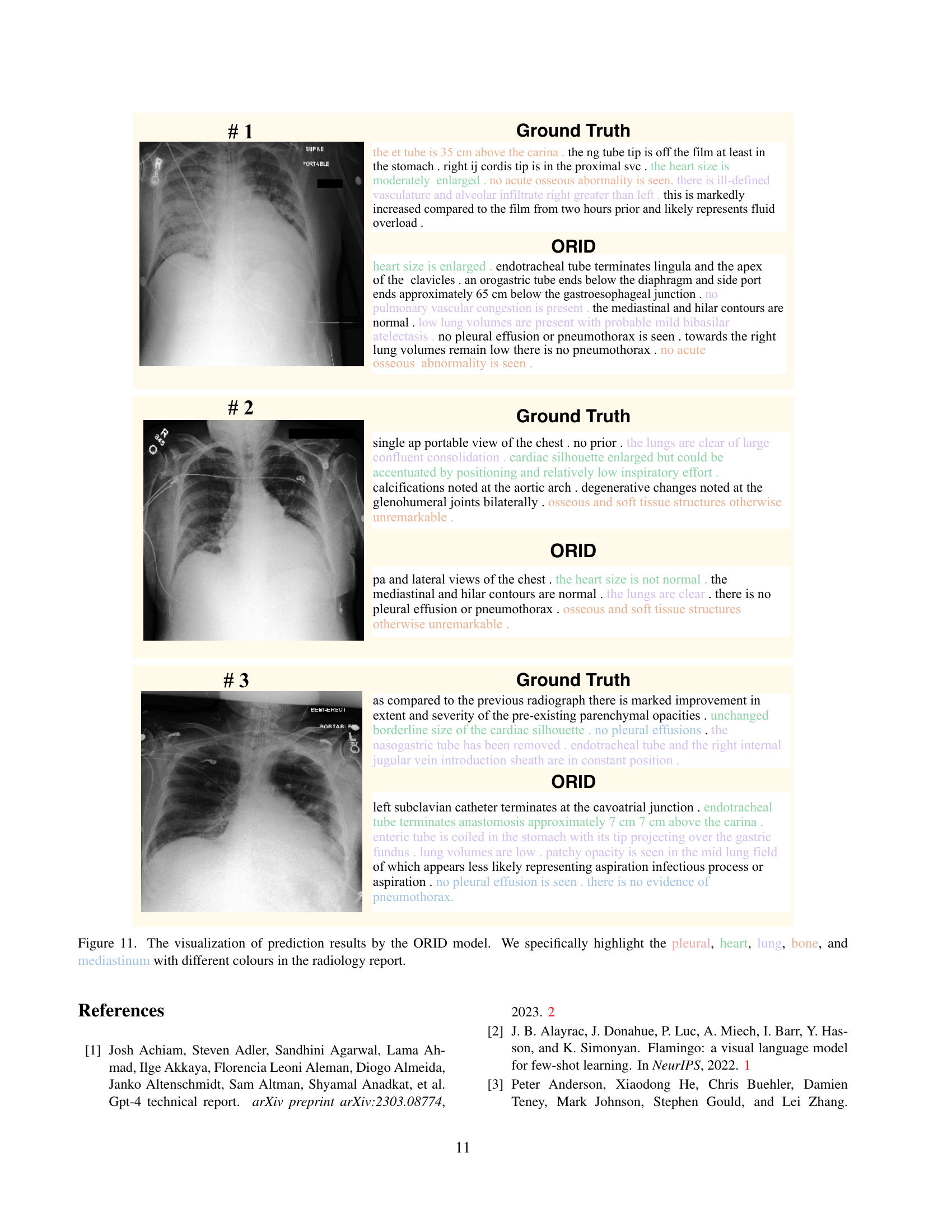
🔼 Figure 7 displays a qualitative comparison of radiology reports generated using different configurations of the ORID framework. It showcases the impact of individual components like the Organ-based Cross-modal Fusion (OCF) module and the Organ Importance Coefficient Analysis (OICA) module. The figure highlights that integrating both modules leads to more comprehensive and accurate reports by emphasizing clinically significant regions based on importance scores and incorporating organ-specific details. The ground truth report is included for comparison to the reports generated by each variation of the model.
read the caption
Figure 7: Qualitative examples of generated radiology reports with different modules.

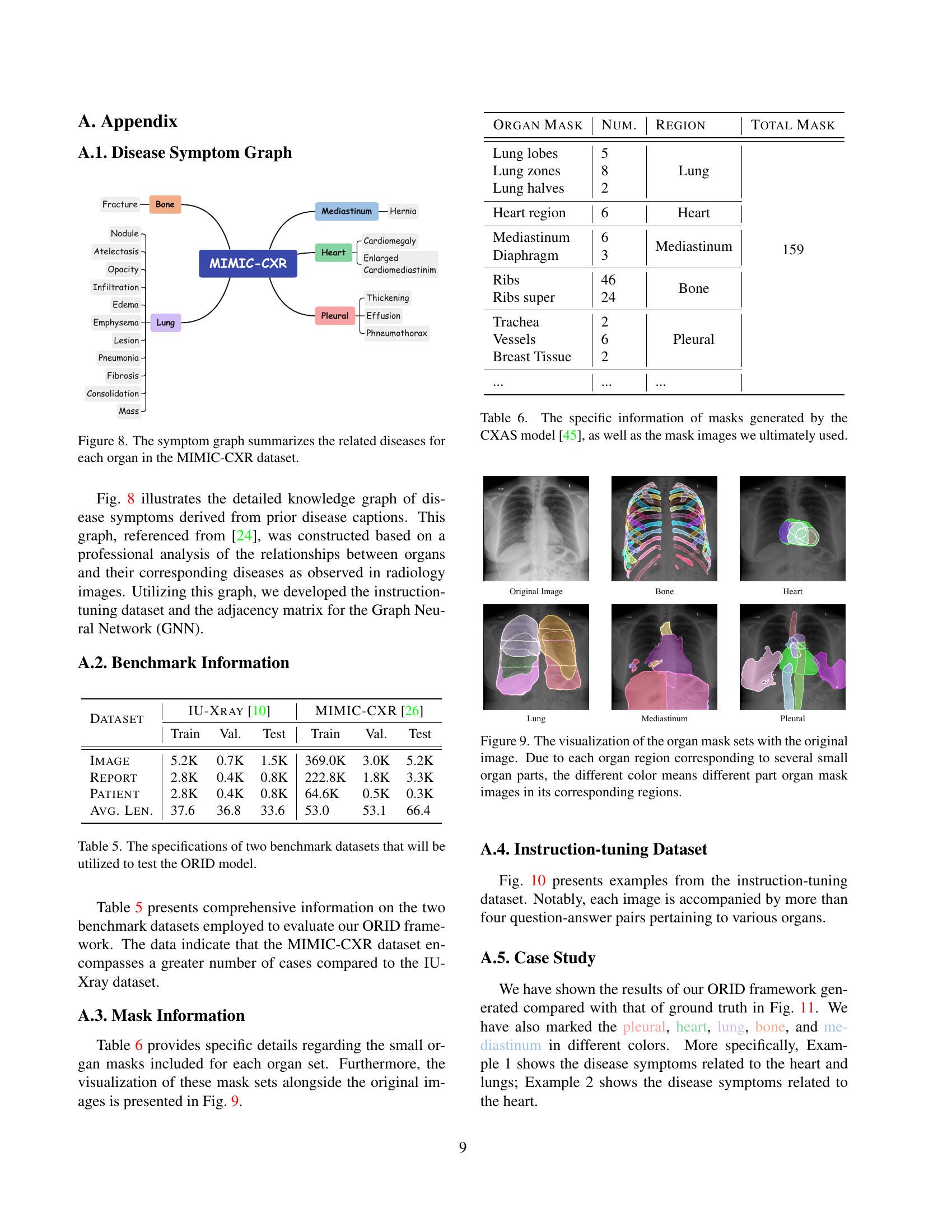
🔼 This figure visualizes the relationships between organs (lung, heart, bone, pleura, mediastinum) and their associated diseases, as derived from analyzing MIMIC-CXR dataset captions. The graph shows how various diseases manifest in specific organs. It serves as a knowledge base used in the ORID framework to improve the accuracy and relevance of generated radiology reports.
read the caption
Figure 8: The symptom graph summarizes the related diseases for each organ in the MIMIC-CXR dataset.

🔼 Figure 9 visualizes organ masks overlaid on an original chest X-ray image. Each organ (lung, heart, etc.) is segmented into multiple sub-regions. The different colors represent these sub-regions within a given organ, highlighting the detailed segmentation performed to isolate the specific areas of interest for analysis. This detailed segmentation is a key component of the proposed ORID framework, providing more granular information for the model during radiology report generation.
read the caption
Figure 9: The visualization of the organ mask sets with the original image. Due to each organ region corresponding to several small organ parts, the different color means different part organ mask images in its corresponding regions.
More on tables
| Method | Precision | Recall | F1-Score |
|---|---|---|---|
| R2Gen [7] | 0.333 | 0.273 | 0.276 |
| CMMRL [43] | 0.342 | 0.294 | 0.292 |
| R2GenCMN [6] | 0.334 | 0.275 | 0.278 |
| METransformer [56] | 0.364 | 0.309 | 0.311 |
| ORID(Ours) | 0.435 | 0.295 | 0.352 |
🔼 This table presents a comparison of clinical efficacy metrics for different radiology report generation models using the MIMIC-CXR dataset. The metrics evaluated assess the precision, recall, and F1-score of the generated reports in identifying clinically significant observations. The best performing model for each metric is highlighted in bold, and the most important metrics are shaded in grey to emphasize their relative importance in evaluating the overall clinical effectiveness of the generated reports. This allows readers to directly compare the performance of various models in terms of their ability to produce clinically relevant and accurate radiology reports.
read the caption
Table 2: Comparison of clinical efficacy metrics for the MIMIC-CXR dataset. The best result is presented in bold. The critical metrics have been shaded in grey.
| Diagnosis Model | B@1 | B@4 | MTR. | RGL. |
|---|---|---|---|---|
| LLaVA-Med [32] | 0.441 | 0.158 | 0.179 | 0.378 |
| LLaVA-Med-RRG | 0.501 | 0.198 | 0.211 | 0.400 |
🔼 This table presents a quantitative comparison of the performance of two models: LLaVA-Med-RRG (the model proposed by the authors) and LLaVA-Med (a baseline model) on the task of radiology report generation. The results are presented in terms of four standard metrics used to evaluate natural language generation: BLEU, METEOR, ROUGE-L, and B@4. The best score for each metric is highlighted in bold, and the most important metric (which is indicated as ROUGE-L in the original caption) is shown in gray. The table provides a concise overview of the comparative performance of the two models and is intended to demonstrate the improvement in the report generation quality achieved by the authors’ proposed model.
read the caption
Table 3: Experiment comparison between LLaVA-Med-RRG and LLaVA-Med. The best result is presented in bold. The most important metric is marked in grey.
| # | BL. | Mask | OCF F | OCF C | OICA | Dataset: IU-Xray [10] B@1 | Dataset: IU-Xray [10] B@4 | Dataset: IU-Xray [10] MTR. | Dataset: IU-Xray [10] RGL. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ✓ | 0.475 | 0.165 | 0.187 | 0.371 | ||||
| 2 | ✓ | ✓ | 0.498 | 0.159 | 0.187 | 0.374 | |||
| 3 | ✓ | ✓ | ✓ | 0.501 | 0.170 | 0.206 | 0.360 | ||
| 4 | ✓ | ✓ | ✓ | ✓ | 0.503 | 0.172 | 0.211 | 0.354 | |
| 5 | ✓ | ✓ | ✓ | ✓ | ✓ | 0.501 | 0.198 | 0.211 | 0.400 |
🔼 This ablation study analyzes the impact of different components within the Organ-Regional Information Driven (ORID) framework on the performance of radiology report generation. It compares the baseline model against variations that include or exclude specific modules: the organ mask, organ-based cross-modal fusion (OCF), fine-grained analysis (F), coarse-grained analysis (C), and the organ importance coefficient analysis (OICA). The results are evaluated using four metrics: BLEU@1, BLEU@4, METEOR, and ROUGE-L, with the best-performing metric (ROUGE-L) highlighted in gray. The table demonstrates how each component contributes to the model’s overall performance, illustrating their individual effects and the synergistic benefits when combined.
read the caption
Table 4: Ablation study on different modules of ORID. The best result is presented in bold. The most important metric is marked in grey.
| Dataset | IU-Xray [10] | MIMIC-CXR [26] | ||||
|---|---|---|---|---|---|---|
| Train | Val. | Test | Train | Val. | Test | |
| Image | 5.2K | 0.7K | 1.5K | 369.0K | 3.0K | 5.2K |
| Report | 2.8K | 0.4K | 0.8K | 222.8K | 1.8K | 3.3K |
| Patient | 2.8K | 0.4K | 0.8K | 64.6K | 0.5K | 0.3K |
| Avg. Len. | 37.6 | 36.8 | 33.6 | 53.0 | 53.1 | 66.4 |
🔼 This table presents a detailed comparison of two benchmark datasets: IU-Xray and MIMIC-CXR, used to evaluate the performance of the ORID model for radiology report generation. It shows the number of images, reports, and patients in the training, validation, and testing sets for each dataset. Additionally, it provides the average length of radiology reports in each dataset.
read the caption
Table 5: The specifications of two benchmark datasets that will be utilized to test the ORID model.
| Organ Mask | Num. | Region | Total Mask |
|---|---|---|---|
| Lung lobes | 5 | Lung | 159 |
| Lung zones | 8 | Lung | |
| Lung halves | 2 | Lung | |
| Heart region | 6 | Heart | |
| Mediastinum | 6 | Mediastinum | |
| Diaphragm | 3 | Mediastinum | |
| Ribs | 46 | Bone | |
| Ribs super | 24 | Bone | |
| Trachea | 2 | Pleural | |
| Vessels | 6 | Pleural | |
| Breast Tissue | 2 | Pleural | |
| … | … | … |
🔼 Table 6 provides a detailed breakdown of the organ masks generated using the CXAS model [45]. It lists the number of regions identified for each organ (lung, heart, mediastinum, bone, and pleura), and shows the total number of masks used in the study after combining these regions. This table is essential for understanding the data used in the Organ Importance Coefficient Analysis Module and how the organ-specific masks are used in the cross-modal fusion of visual and textual features. This detailed description of mask generation is important for reproducibility of the results and understanding the framework’s data processing pipeline.
read the caption
Table 6: The specific information of masks generated by the CXAS model [45], as well as the mask images we ultimately used.
Full paper#