↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Medical image segmentation is crucial for diagnosis, but creating effective models is challenging due to variations in data (modality, target, size). Current approaches often lack generalizability. This study addresses these issues by investigating transfer learning capabilities of models pre-trained on large, full-body CT datasets.
The research employs STU-Net, a scalable model, evaluated on 87 public datasets. Results show that pre-trained models, fine-tuned on downstream tasks, dramatically improve performance, especially for smaller and larger datasets. Furthermore, effective transfer learning is possible across different imaging modalities and targets (structures and lesions), showcasing the robustness and versatility of the approach.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image segmentation because it provides a large-scale benchmark and a readily available, well-performing baseline model (STU-Net). It addresses the critical issue of transfer learning across diverse datasets and modalities, offering insights into effective training strategies and paving the way for more robust and generalizable medical image analysis models. Its open-source nature further enhances its impact, enabling wider adoption and collaboration within the research community.
Visual Insights#

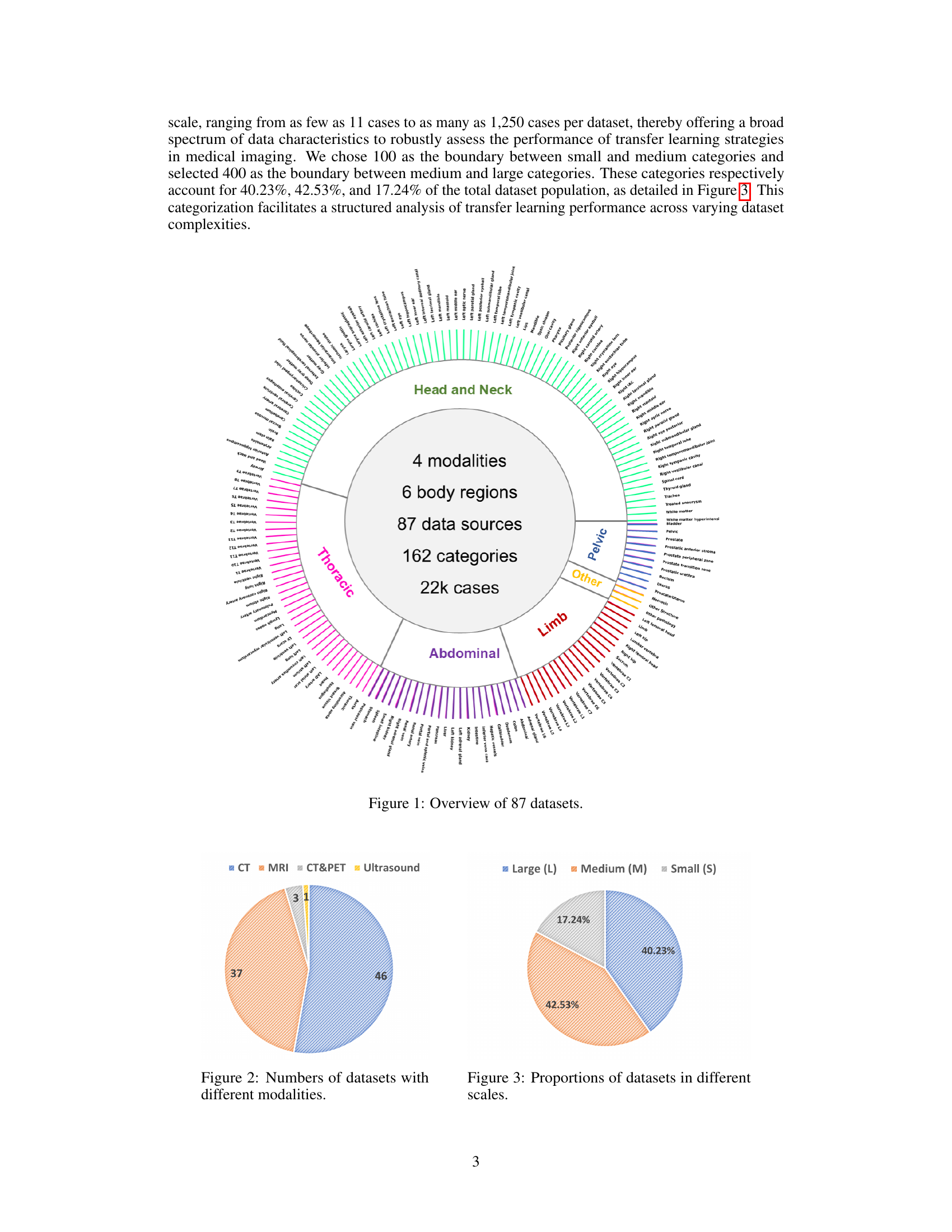
🔼 This figure provides a visual summary of the 87 datasets used in the study. It shows the distribution of datasets across different modalities (CT, MRI, CT&PET, Ultrasound), body regions (Head and Neck, Thoracic, Abdominal, Pelvic, Limb, Other), and dataset sizes (Small, Medium, Large). The pie charts illustrate the proportions of datasets categorized by modality and size, respectively. The circular graphic displays the different datasets, grouping them by the body region they cover, and uses color-coding to represent the imaging modality. This visualization helps to understand the diversity and scale of the datasets used in the benchmark.
read the caption
Figure 1: Overview of 87 datasets.
| Modality | CT | CT | CT | CT | MRI | MRI | MRI | MRI | CT&PET | Ultrasound | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target | Lesion | Seen Sturctute | Unseen Sturctute | Lesion & Sturcture | Lesion | Seen Sturctute | Unseen Sturctute | Lesion & Sturcture | Lesion | Lesion | ||
| datsets | 11 | 17 | 9 | 9 | 12 | 9 | 9 | 7 | 3 | 1 | 87 |
🔼 This table summarizes the distribution of 87 publicly available datasets used in the study. It categorizes these datasets based on imaging modality (CT, MRI, CT&PET, Ultrasound), target (lesions, structures, or a combination of both), and whether the target structures were seen during training or were unseen (new). The numbers in the table represent the count of datasets that fall into each specific combination of these categories.
read the caption
Table 1: Summary of number of datasets across targets and modalities.
In-depth insights#
CT Transfer Learning#
The study investigates the effectiveness of transfer learning using Computed Tomography (CT) data for medical image segmentation. Full-body CT scans offer a rich source of anatomical information, allowing pre-trained models to generalize well to other imaging modalities (e.g., MRI) and diverse target structures (e.g., organs, lesions). The research explores the impact of various factors on transfer learning success, including the size of the training dataset and the model’s architecture. A key finding is the non-linear relationship between dataset size and performance improvements, suggesting a potential bottleneck effect where increasing the dataset beyond a certain size may not yield proportional performance gains. The ability of CT-pretrained models to effectively transfer to other modalities highlights their potential for broader applicability in clinical settings. Overall, this work provides valuable insights into effective strategies for leveraging CT data in transfer learning for volumetric medical image segmentation.
Dataset Scale Effects#
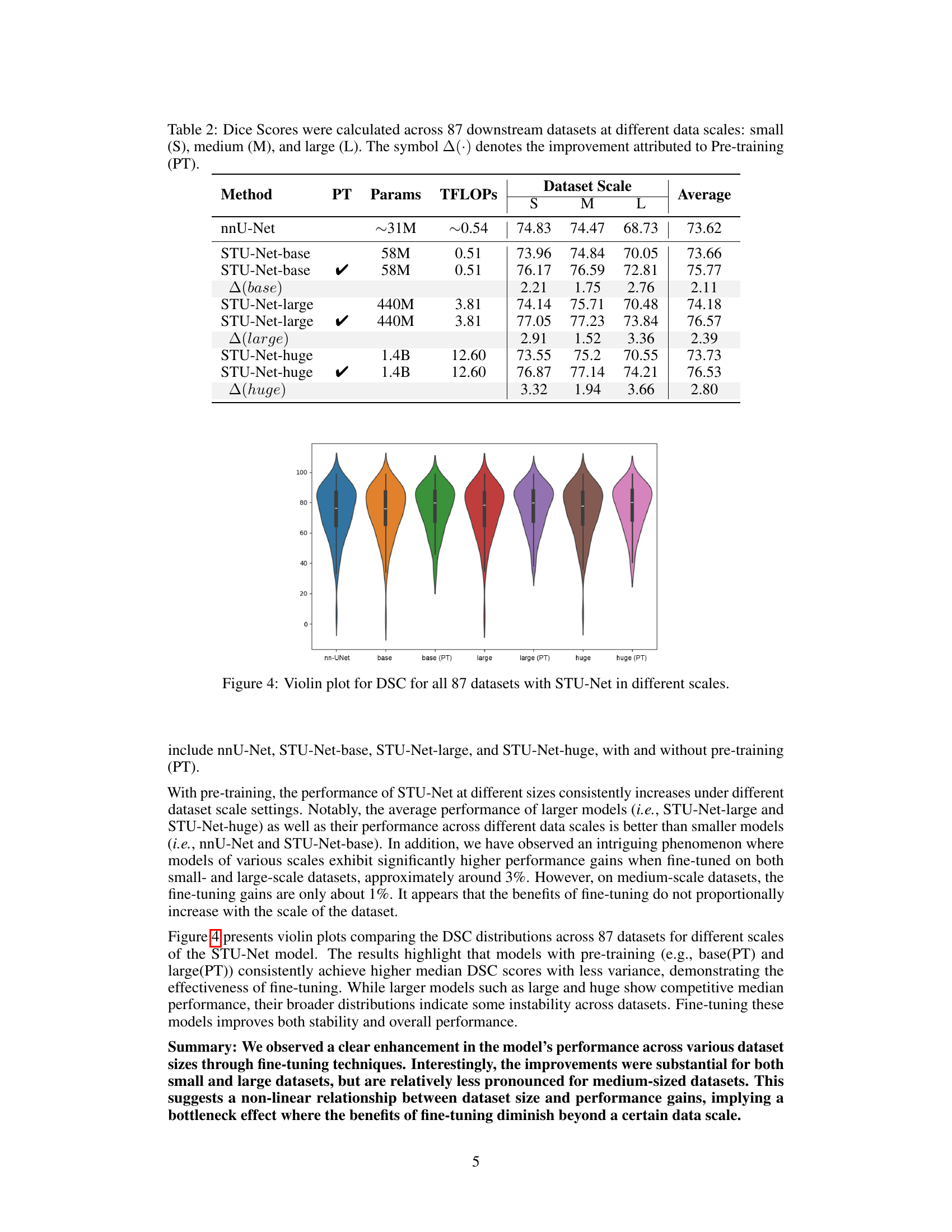
Analyzing the influence of dataset size on model performance reveals a non-linear relationship. The study shows significant improvements in model performance when fine-tuning on both small and large datasets, suggesting that sufficient data, regardless of scale, is crucial for effective model adaptation. However, a bottleneck effect was observed, meaning improvements were less pronounced in medium-sized datasets. This suggests that the benefit of fine-tuning diminishes beyond a certain data scale, indicating an optimal dataset size exists for maximizing transfer learning effectiveness. Further investigation is needed to pinpoint this optimal size and to understand the underlying mechanisms driving this non-linear relationship. This finding has important implications for resource allocation in medical image segmentation, guiding researchers towards more efficient data collection and model training strategies.
Modality Transferability#
The study’s exploration of modality transferability reveals crucial insights into the adaptability of models trained on full-body CT scans to other imaging modalities. The results demonstrate that these models exhibit effective transfer learning, performing well when fine-tuned on datasets with different imaging modalities, such as MRI. This success highlights the power of large-scale, comprehensive pre-training on full-body CT data as a foundation for broader applications. However, the study also reveals a potential bottleneck effect concerning dataset size, with fine-tuning yielding significant improvements on both small and large datasets, yet only moderate gains on medium-sized ones. This suggests a non-linear relationship between data volume and model performance improvements. Further research could explore this bottleneck effect, perhaps by examining the model’s learning dynamics at various dataset sizes to optimize transfer learning efficiency.
Benchmarking Transfer#
Benchmarking transfer learning in medical image segmentation involves a systematic evaluation of pre-trained models’ performance on diverse downstream tasks. The core idea is to assess how effectively knowledge learned from a source domain (e.g., large-scale full-body CT scans) generalizes to various target domains (different modalities, anatomical structures, lesion types, and dataset sizes). This requires a comprehensive benchmark dataset with significant variations in these factors. A key aspect is understanding the impact of dataset size on transfer learning’s effectiveness; a crucial finding might reveal non-linear scaling, with diminishing returns beyond a certain data threshold. Furthermore, analysis of transfer across modalities (e.g., from CT to MRI) and targets (structure vs. lesion segmentation) provides insights into model generalization capabilities and potential limitations. A thorough evaluation should also account for different model architectures and sizes, allowing a comparison of efficiency and accuracy based on the model’s complexity. The ultimate goal is to identify optimal pre-training strategies and establish the conditions under which transfer learning yields significant benefits in medical image analysis. This detailed benchmarking ultimately guides the development of more robust and widely applicable segmentation models.
Future Research#
The paper’s “Future Research” section would benefit from exploring alternative pre-training strategies beyond full-body CT scans. Investigating the effectiveness of pre-training on other modalities, such as MRI or ultrasound, or on specific anatomical regions, could reveal improved transfer learning capabilities. Furthermore, research should focus on optimizing fine-tuning techniques for various dataset sizes to address the observed bottleneck effect. This includes exploring adaptive learning rates and regularization methods tailored to different dataset scales. A deeper investigation into the interaction between model size and dataset size in fine-tuning is crucial, potentially involving exploring more efficient model architectures to mitigate the computational cost of training larger models on larger datasets. Finally, extending the evaluation to more diverse datasets, including those with rarer pathologies or less common imaging modalities, would enhance the generalizability and robustness of the findings, providing valuable insights for future medical image segmentation applications.
More visual insights#
More on figures

🔼 This figure shows a pie chart that presents the number of datasets used in the study, broken down by imaging modality. The modalities included are CT, MRI, CT&PET (combined CT and PET scans), and Ultrasound. The sizes of the slices in the pie chart are proportional to the number of datasets using each modality.
read the caption
Figure 2: Numbers of datasets with different modalities.

🔼 This figure shows the distribution of the 87 datasets used in the study across three different size categories: small, medium, and large. The proportions are presented as percentages, illustrating the relative abundance of datasets in each size category. This breakdown is important for understanding the impact of dataset size on the performance of transfer learning models in medical image segmentation.
read the caption
Figure 3: Proportions of datasets in different scales.

🔼 This violin plot visualizes the Dice Similarity Coefficient (DSC) scores achieved by the STU-Net model across 87 different medical image datasets. The DSC, a common metric for evaluating segmentation performance, is shown separately for various scales (small, base, large, and huge) of the STU-Net model. The violin plot’s shape illustrates the distribution of DSC scores for each model size and helps to reveal trends in performance variation across the datasets and model sizes. It aids in comparing the performance with and without pre-training across different model scales.
read the caption
Figure 4: Violin plot for DSC for all 87 datasets with STU-Net in different scales.
More on tables
| Method | PT | Params | TFLOPs | Dataset Scale | Average | ||
|---|---|---|---|---|---|---|---|
| nnU-Net | ~31M | ~0.54 | 74.83 | 74.47 | 68.73 | 73.62 | |
| STU-Net-base | 58M | 0.51 | 73.96 | 74.84 | 70.05 | 73.66 | |
| STU-Net-base | ✔ | 58M | 0.51 | 76.17 | 76.59 | 72.81 | 75.77 |
| Δ(base) | 2.21 | 1.75 | 2.76 | 2.11 | |||
| STU-Net-large | 440M | 3.81 | 74.14 | 75.71 | 70.48 | 74.18 | |
| STU-Net-large | ✔ | 440M | 3.81 | 77.05 | 77.23 | 73.84 | 76.57 |
| Δ(large) | 2.91 | 1.52 | 3.36 | 2.39 | |||
| STU-Net-huge | 1.4B | 12.60 | 73.55 | 75.2 | 70.55 | 73.73 | |
| STU-Net-huge | ✔ | 1.4B | 12.60 | 76.87 | 77.14 | 74.21 | 76.53 |
| Δ(huge) | 3.32 | 1.94 | 3.66 | 2.80 |
🔼 Table 2 presents a comprehensive evaluation of transfer learning using the STU-Net model across 87 public datasets. The datasets are categorized into three size groups (small, medium, and large) to analyze the impact of dataset size on performance. The table shows Dice Similarity Coefficients (DSC) achieved with and without pre-training (PT) for three different scales of the STU-Net model (base, large, huge). The difference in DSC scores (Δ(⋅)) between models with and without pre-training is also provided to demonstrate the effectiveness of pre-training on different scale datasets and the impact of model size.
read the caption
Table 2: Dice Scores were calculated across 87 downstream datasets at different data scales: small (S), medium (M), and large (L). The symbol Δ(⋅)Δ⋅\Delta(\cdot)roman_Δ ( ⋅ ) denotes the improvement attributed to Pre-training (PT).
| Method | PT | CT Seen Structure | CT Unseen Structure | CT Lesion | MRI Seen Structure | MRI Unseen Structure | MRI Lesion | US Lesion | CT&PET Lesion |
|---|---|---|---|---|---|---|---|---|---|
| nnU-Net | 82.28 | 69.59 | 58.86 | 87.16 | 83.27 | 68.50 | 49.66 | 58.88 | |
| STU-Net-base | 82.79 | 70.06 | 58.88 | 86.67 | 82.20 | 68.15 | 53.70 | 62.44 | |
| STU-Net-base | ✔ | 85.00 | 73.85 | 63.14 | 87.47 | 82.62 | 69.14 | 54.54 | 66.45 |
| Δ(base) | 2.21 | 3.79 | 4.26 | 0.80 | 0.42 | 0.99 | 0.84 | 4.01 | |
| STU-Net-large | 83.60 | 70.77 | 59.27 | 87.03 | 81.86 | 68.43 | 50.18 | 63.09 | |
| STU-Net-large | ✔ | 85.87 | 75.81 | 63.83 | 87.58 | 82.89 | 69.70 | 52.65 | 68.33 |
| Δ(large) | 2.27 | 5.04 | 4.56 | 0.55 | 1.03 | 1.27 | 2.47 | 5.24 | |
| STU-Net-huge | 82.73 | 70.67 | 57.35 | 86.94 | 82.11 | 68.54 | 49.38 | 62.83 | |
| STU-Net-huge | ✔ | 85.90 | 75.15 | 63.61 | 87.59 | 83.49 | 69.45 | 52.78 | 68.74 |
| Δ(huge) | 3.17 | 4.48 | 6.26 | 0.65 | 1.38 | 0.91 | 3.40 | 5.91 |
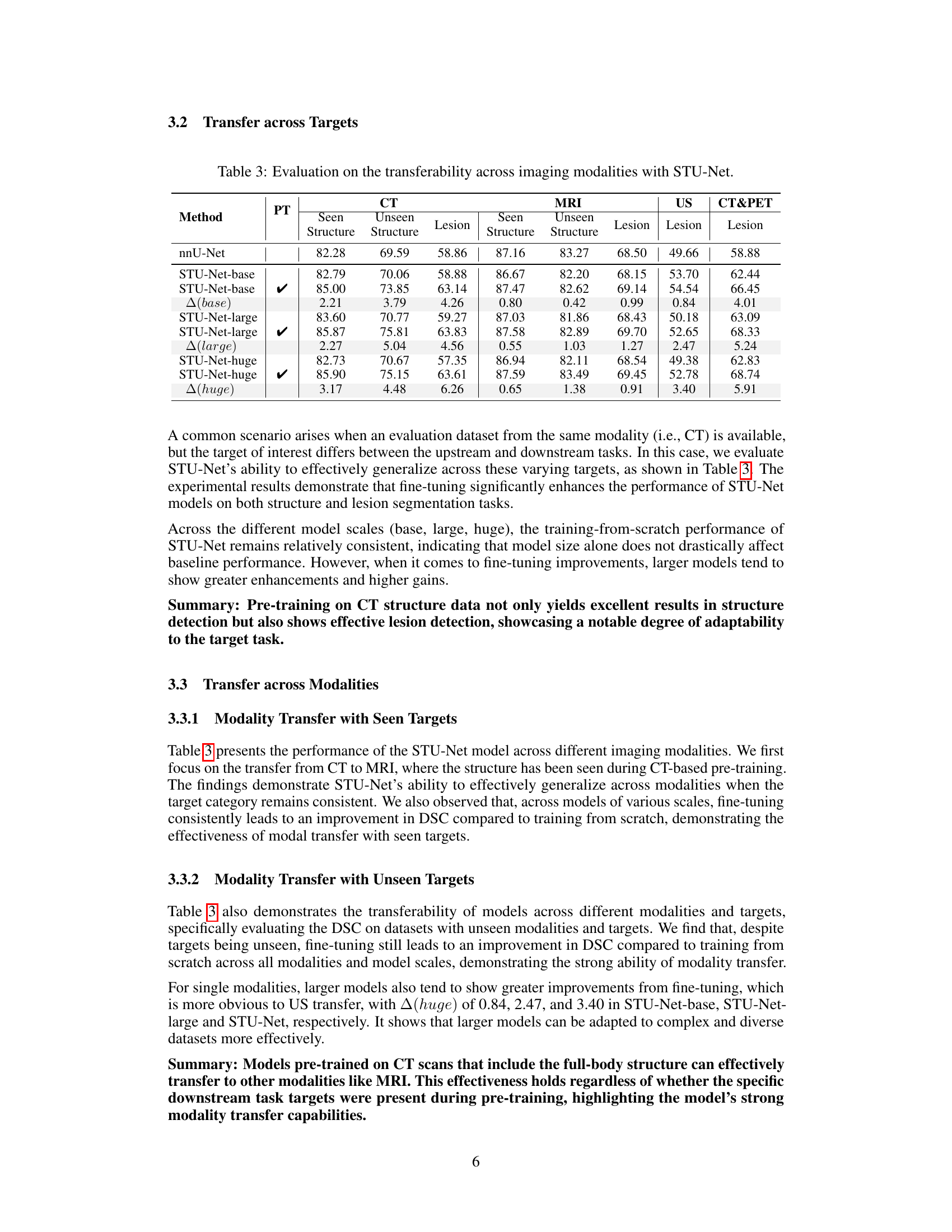
🔼 This table presents the results of an experiment evaluating the transfer learning capabilities of the STU-Net model across different imaging modalities. It shows the Dice Similarity Coefficient (DSC) scores achieved when fine-tuning the model on various datasets, comparing performance with and without pre-training on full-body CT scans. The table breaks down the results by model variant (base, large, and huge), dataset modality (CT, MRI, US, CT&PET), and target type (seen and unseen structures, and lesions). The change in DSC score from using pre-training is also shown (△). This allows for a comparison of the effectiveness of transfer learning based on the size and type of the pre-trained model, target consistency, and data modality.
read the caption
Table 3: Evaluation on the transferability across imaging modalities with STU-Net.
| Method | PT | Head and Neck | Pelvic | Limb | Thoracic | Abdominal | other | Bone | Vessel |
|---|---|---|---|---|---|---|---|---|---|
| nnU-Net | 79.16 | 84.2 | 62.9 | 73.15 | 89.52 | 63.84 | 65.47 | 80.46 | |
| STU-Net-base | 75.85 | 84.11 | 66.12 | 73.48 | 89.61 | 65.98 | 67.10 | 80.19 | |
| STU-Net-base | ✔ | 80.68 | 84.99 | 72.74 | 74.12 | 89.71 | 68.22 | 71.29 | 81.24 |
| Δ(base) | 4.83 | 0.88 | 6.62 | 0.64 | 0.10 | 2.24 | 4.19 | 1.05 | |
| STU-Net-large | 79.75 | 84.52 | 66.31 | 74.00 | 89.93 | 66.59 | 67.75 | 80.86 | |
| STU-Net-large | ✔ | 81.51 | 85.15 | 74.16 | 74.80 | 90.28 | 68.82 | 72.39 | 82.06 |
| Δ(large) | 1.76 | 0.63 | 7.85 | 0.8 | 0.35 | 2.23 | 4.64 | 1.20 | |
| STU-Net-huge | 78.99 | 84.29 | 65.88 | 73.90 | 89.85 | 65.99 | 67.32 | 80.53 | |
| STU-Net-huge | ✔ | 80.37 | 85.99 | 73.97 | 74.92 | 90.37 | 69.86 | 72.33 | 82.00 |
| Δ(huge) | 1.38 | 1.70 | 8.09 | 1.02 | 0.52 | 3.87 | 5.01 | 1.47 |
🔼 This table presents the Dice Similarity Coefficient (DSC) scores achieved by different models (nnU-Net and STU-Net with varying sizes) when performing segmentation tasks across various anatomical structures. It shows the baseline performance of each model when trained from scratch and compares it to the performance after pre-training on full-body CT scans, highlighting the impact of pre-training and model size on the effectiveness of transfer learning across different anatomical structures (Head and Neck, Pelvic, Limb, Thoracic, Abdominal, Other, Bone, Vessel). The results indicate the ability of the models, especially larger ones, to generalize to various downstream tasks, even for complex structures like bones and vessels, and demonstrate the impact of transfer learning from full-body CT on improving segmentation accuracy.
read the caption
Table 4: Evaluation on the transferability across different structures.
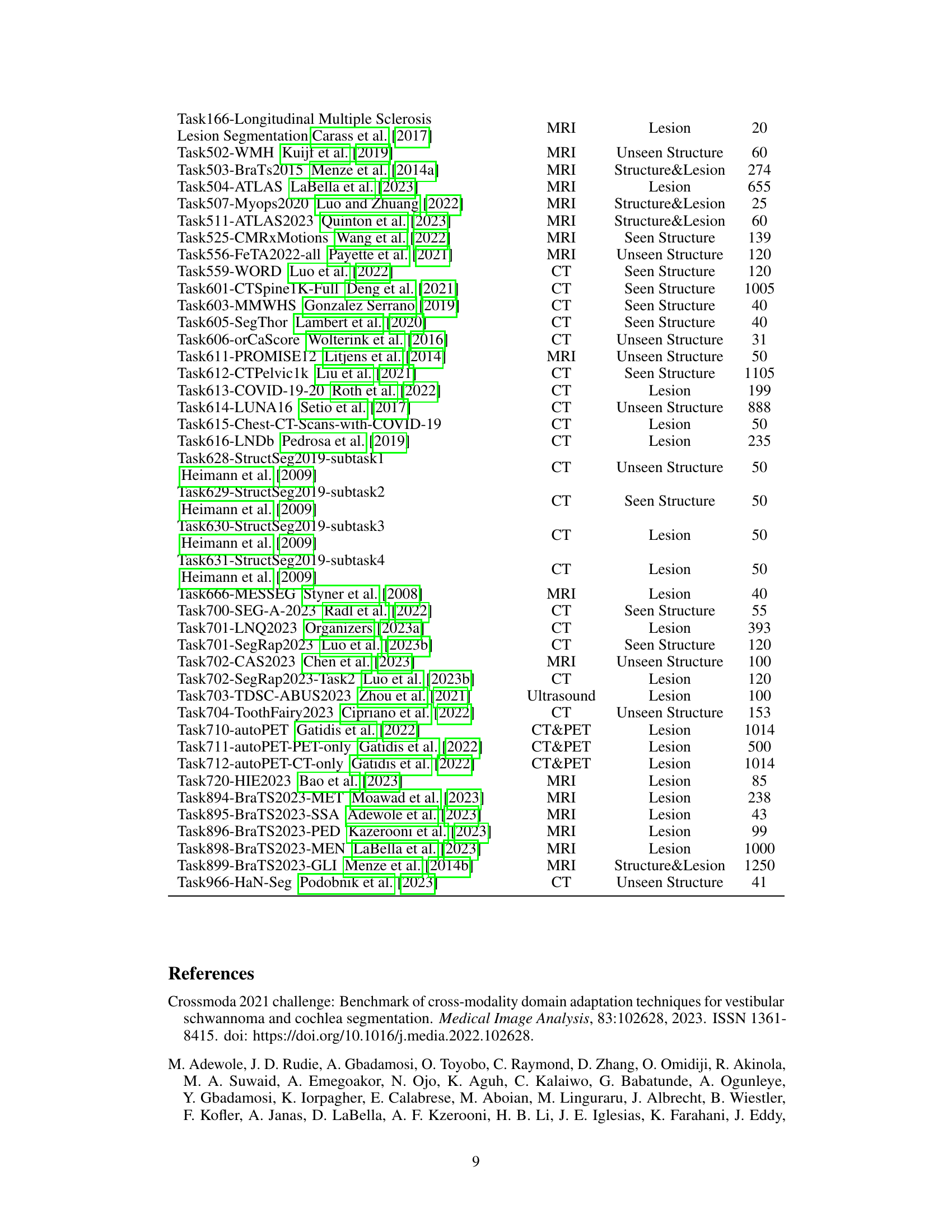
| Dataset | Modality | Target | Case |
|---|---|---|---|
| Task001-BrainTumour Antonelli et al. (2022) | MRI | Lesion | 484 |
| Task002-Heart Antonelli et al. (2022) | MRI | Seen Structure | 20 |
| Task003-Liver Antonelli et al. (2022) | CT | Structure&Lesion | 130 |
| Task004-Hippocampus Antonelli et al. (2022) | MRI | Unseen Structure | 260 |
| Task005-Prostate Antonelli et al. (2022) | MRI | Seen Structure | 31 |
| Task006-Lung Antonelli et al. (2022) | CT | Lesion | 63 |
| Task007-Pancreas Antonelli et al. (2022) | CT | Structure&Lesion | 280 |
| Task008-HepaticVessel Antonelli et al. (2022) | CT | Structure&Lesion | 303 |
| Task009-Spleen Antonelli et al. (2022) | CT | Seen Structure | 40 |
| Task010-Colon Antonelli et al. (2022) | CT | Lesion | 125 |
| Task011-BTCV Landman et al. (2015) | CT | Seen Structure | 30 |
| Task012-BTCV-Cervix Landman et al. (2015) | CT | Seen Structure | 30 |
| Task013-ACDC Bernard et al. (2018) | MRI | Seen Structure | 200 |
| Task019-BraTS21 Baid et al. (2021) | MRI | Lesion | 1250 |
| Task020-AbdomenCT1K Ma et al. (2021) | CT | Seen Structure | 1000 |
| Task021-KiTS2021 Heller et al. (2023) | CT | Structure&Lesion | 300 |
| Task023-FLARE22 Ma et al. (2023) | CT | Seen Structure | 70 |
| Task029-LITS Bilic et al. (2023) | CT | Structure&Lesion | 130 |
| Task034-Instance22 Li et al. (2023) | CT | Unseen Structure | 100 |
| Task036-KiPA22 He et al. (2020) | CT | Structure&Lesion | 70 |
| Task037-CHAOS-Task-3-5-Variant1 Kavur et al. (2021) | MRI | Seen Structure | 40 |
| Task039-Parse22 Luo et al. (2023a) | CT | Seen Structure | 100 |
| Task040-ATM22 Zhang et al. (2023) | CT | Unseen Structure | 300 |
| Task041-ISLES2022 Hernandez Petzsche et al. (2022) | MRI | Lesion | 250 |
| Task044-CrossMoDA23 DOR (2023) | MRI | Structure&Lesion | 226 |
| Task044-KiTS23 Heller et al. (2021) | CT | Structure&Lesion | 489 |
| Task050-LAScarQS22-task1 Li et al. (2022) | MRI | Seen Structure | 60 |
| Task051-AMOS-CT Ji et al. (2022) | CT | Seen Structure | 300 |
| Task051-LAScarQS22-task2 Li et al. (2022) | MRI | Seen Structure | 130 |
| Task052-AMOS-MR Ji et al. (2022) | MRI | Seen Structure | 60 |
| Task053-AMOS-Task2 Ji et al. (2022) | MRI | Seen Structure | 360 |
| Task083-VerSe2020 Sekuboyina et al. (2021) | CT | Seen Structure | 350 |
| Task103-ADAM2020 Fang et al. (2022) | MRI | Structure&Lesion | 113 |
| Task104-Colorectal-Liver-Metastases Simpson et al. (2024) | CT | Structure&Lesion | 196 |
| Task105-DICOM-LIDC-IDRI-Nodules Fedorov et al. (2018) | CT | Unseen Structure | 1018 |
| Task106-AIIB2023 Nan et al. (2023) | CT | Unseen Structure | 120 |
| Task107-HCC-TACE-Seg Moawad et al. (2021) | CT | Structure&Lesion | 224 |
| Task108-ISBI-MR-Prostate-2013 Bloch et al. (2015) | MRI | Unseen Structure | 79 |
| Task109-SMILE-UHURA2023 Organizers (2023b) | MRI | Unseen Structure | 11 |
| Task110-ISPY1-Tumor-SEG-Radiomics Chitalia et al. (2022) | MRI | Lesion | 160 |
| Task111-LUAD-CT-Survival Goldgof Dmitry et al. (2017) | CT | Lesion | 40 |
| Task112-PROSTATEx-Seg-HiRes Schindele et al. (2020) | MRI | Unseen Structure | 65 |
| Task113-PROSTATEx-Seg-Zones Schindele et al. (2020) | MRI | Unseen Structure | 98 |
| Task114-Prostate-Anatomical-Edge-Cases Thompson et al. (2023) | CT | Seen Structure | 130 |
| Task115-QIBA-VolCT-1B McNitt-Gray et al. (2015) | CT | Lesion | 149 |
| Task116-ISPY1 Chitalia et al. (2022) | MRI | Structure&Lesion | 820 |
| Task166-Longitudinal Multiple Sclerosis Lesion Segmentation Carass et al. (2017) | MRI | Lesion | 20 |
| Task502-WMH Kuijf et al. (2019) | MRI | Unseen Structure | 60 |
| Task503-BraTs2015 Menze et al. (2014a) | MRI | Structure&Lesion | 274 |
| Task504-ATLAS LaBella et al. (2023) | MRI | Lesion | 655 |
| Task507-Myops2020 Luo and Zhuang (2022) | MRI | Structure&Lesion | 25 |
| Task511-ATLAS2023 Quinton et al. (2023) | MRI | Structure&Lesion | 60 |
| Task525-CMRxMotions Wang et al. (2022) | MRI | Seen Structure | 139 |
| Task556-FeTA2022-all Payette et al. (2021) | MRI | Unseen Structure | 120 |
| Task559-WORD Luo et al. (2022) | CT | Seen Structure | 120 |
| Task601-CTSpine1K-Full Deng et al. (2021) | CT | Seen Structure | 1005 |
| Task603-MMWHS Gonzalez Serrano (2019) | CT | Seen Structure | 40 |
| Task605-SegThor Lambert et al. (2020) | CT | Seen Structure | 40 |
| Task606-orCaScore Wolterink et al. (2016) | CT | Unseen Structure | 31 |
| Task611-PROMISE12 Litjens et al. (2014) | MRI | Unseen Structure | 50 |
| Task612-CTPelvic1k Liu et al. (2021) | CT | Seen Structure | 1105 |
| Task613-COVID-19-20 Roth et al. (2022) | CT | Lesion | 199 |
| Task614-LUNA16 Setio et al. (2017) | CT | Unseen Structure | 888 |
| Task615-Chest-CT-Scans-with-COVID-19 | CT | Lesion | 50 |
| Task616-LNDb Pedrosa et al. (2019) | CT | Lesion | 235 |
| Task628-StructSeg2019-subtask1 Heimann et al. (2009) | CT | Unseen Structure | 50 |
| Task629-StructSeg2019-subtask2 Heimann et al. (2009) | CT | Seen Structure | 50 |
| Task630-StructSeg2019-subtask3 Heimann et al. (2009) | CT | Lesion | 50 |
| Task631-StructSeg2019-subtask4 Heimann et al. (2009) | CT | Lesion | 50 |
| Task666-MESSEG Styner et al. (2008) | MRI | Lesion | 40 |
| Task700-SEG-A-2023 Radl et al. (2022) | CT | Seen Structure | 55 |
| Task701-LNQ2023 Organizers (2023a) | CT | Lesion | 393 |
| Task701-SegRap2023 Luo et al. (2023b) | CT | Seen Structure | 120 |
| Task702-CAS2023 Chen et al. (2023) | MRI | Unseen Structure | 100 |
| Task702-SegRap2023-Task2 Luo et al. (2023b) | CT | Lesion | 120 |
| Task703-TDSC-ABUS2023 Zhou et al. (2021) | Ultrasound | Lesion | 100 |
| Task704-ToothFairy2023 Cipriano et al. (2022) | CT | Unseen Structure | 153 |
| Task710-autoPET Gatidis et al. (2022) | CT&PET | Lesion | 1014 |
| Task711-autoPET-PET-only Gatidis et al. (2022) | CT&PET | Lesion | 500 |
| Task712-autoPET-CT-only Gatidis et al. (2022) | CT&PET | Lesion | 1014 |
| Task720-HIE2023 Bao et al. (2023) | MRI | Lesion | 85 |
| Task894-BraTS2023-MET Moawad et al. (2023) | MRI | Lesion | 238 |
| Task895-BraTS2023-SSA Adewole et al. (2023) | MRI | Lesion | 43 |
| Task896-BraTS2023-PED Kazerooni et al. (2023) | MRI | Lesion | 99 |
| Task898-BraTS2023-MEN LaBella et al. (2023) | MRI | Lesion | 1000 |
| Task899-BraTS2023-GLI Menze et al. (2014b) | MRI | Structure&Lesion | 1250 |
| Task966-HaN-Seg Podobnik et al. (2023) | CT | Unseen Structure | 41 |
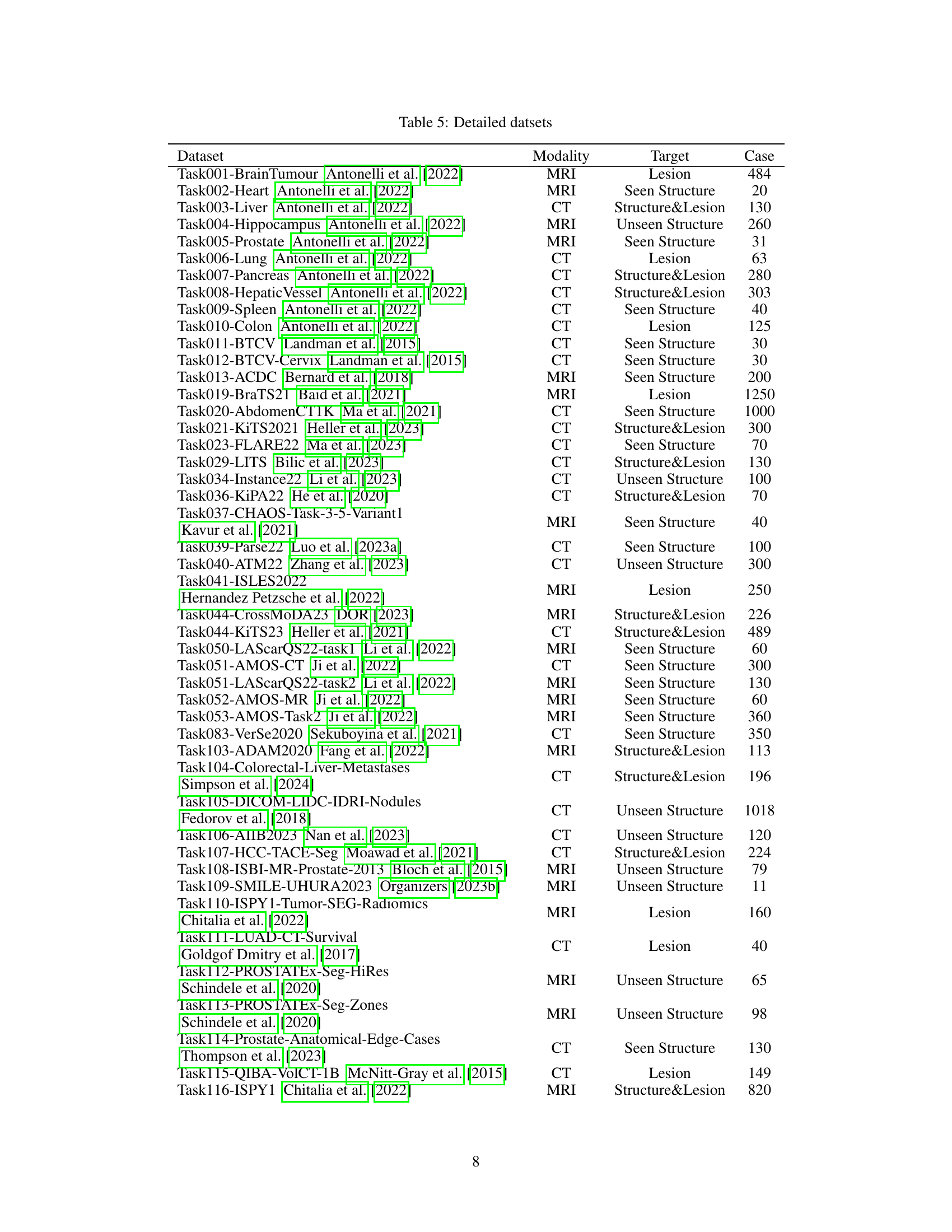
🔼 Table 5 presents detailed information for each of the 87 public datasets used in the study. For each dataset, it lists the dataset name, imaging modality (e.g., CT, MRI, PET), the segmentation target (e.g., structure, lesion, or both), and the number of cases included. This provides a comprehensive overview of the diversity in terms of modality, target, and data size across the datasets, crucial context for understanding the results of the transfer learning experiments.
read the caption
Table 5: Detailed datsets
Full paper#