↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Current methods for aligning molecules and their textual descriptions often treat molecules as whole entities, neglecting the crucial fine-grained relationships between substructures and descriptive phrases. This limitation hinders accurate and explainable predictions in molecule-related tasks. This paper introduces challenges in aligning molecules and texts using LLMs.
The proposed solution is MolReFlect, a teacher-student framework that tackles these issues. It leverages a larger teacher LLM to label detailed alignments between molecular substructures and textual phrases. A smaller student LLM then refines these alignments using in-context learning and chain-of-thought reasoning. The results demonstrate that MolReFlect significantly outperforms existing methods on benchmark datasets, showcasing its effectiveness in improving the accuracy and explainability of molecule-text alignment.
Key Takeaways#
Why does it matter?#
This paper is important because it presents MolReFlect, a novel framework that significantly improves the accuracy and explainability of molecule-text alignment. This addresses a critical challenge in molecule understanding and generation, paving the way for more accurate and explainable predictions in various applications, including drug discovery and materials science. The innovative teacher-student approach and the introduction of fine-grained alignments offer new avenues for research in LLMs applied to chemistry.
Visual Insights#

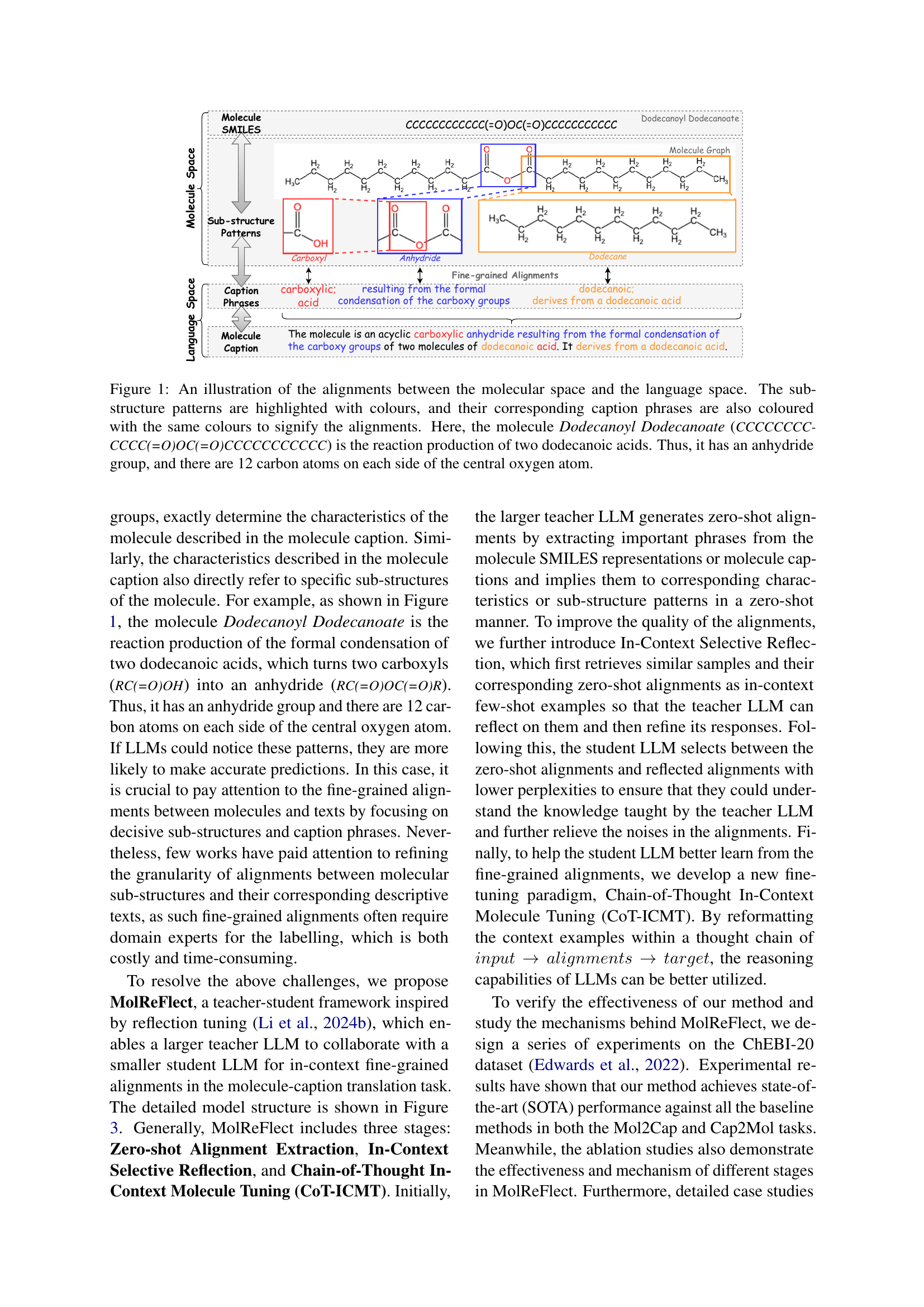
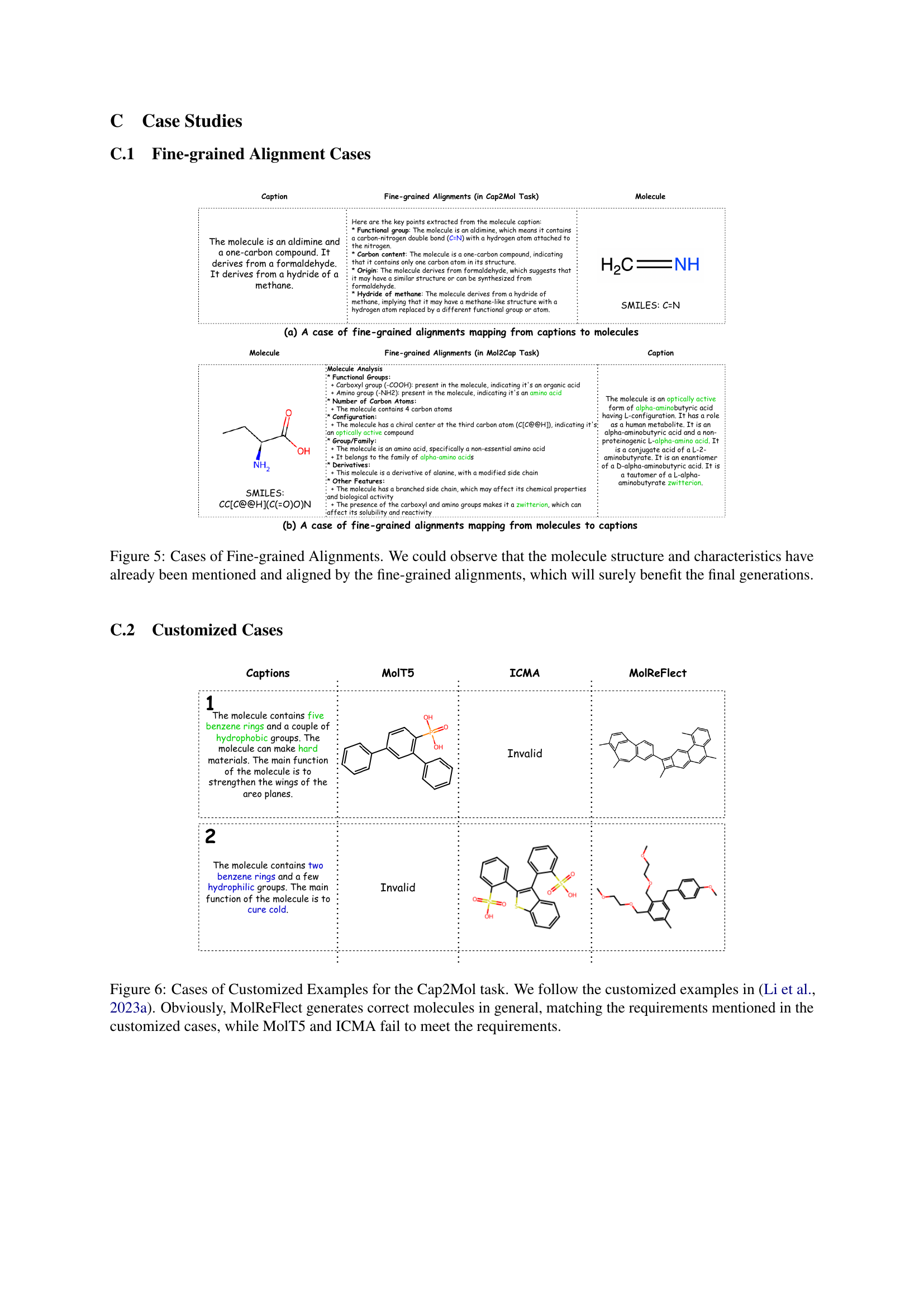
🔼 Figure 1 illustrates the alignment of molecular structures with their textual descriptions. The figure shows Dodecanoyl Dodecanoate, a molecule formed by the condensation of two dodecanoic acid molecules. Its chemical structure is represented, with key sub-structures (like the anhydride group and the dodecyl chains) color-coded and linked to corresponding phrases in its caption. This visual demonstrates the fine-grained alignment between molecular components and their linguistic representations, highlighting the central goal of the MolReFlect framework.
read the caption
Figure 1: An illustration of the alignments between the molecular space and the language space. The sub-structure patterns are highlighted with colours, and their corresponding caption phrases are also coloured with the same colours to signify the alignments. Here, the molecule Dodecanoyl Dodecanoate (CCCCCCCCCCCC(=O)OC(=O)CCCCCCCCCCC) is the reaction production of two dodecanoic acids. Thus, it has an anhydride group, and there are 12 carbon atoms on each side of the central oxygen atom.
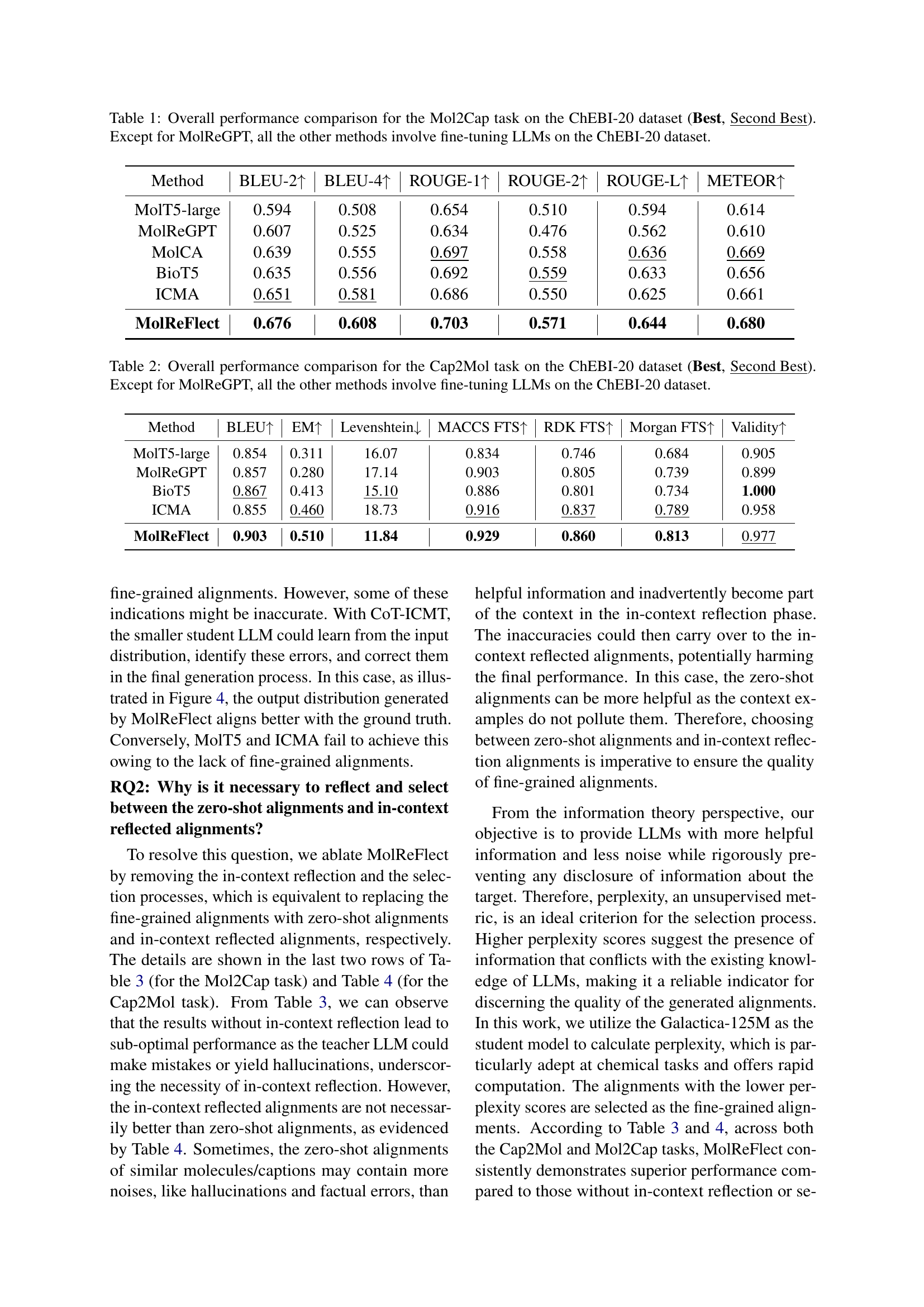
| Method | BLEU-2 ↑ | BLEU-4 ↑ | ROUGE-1 ↑ | ROUGE-2 ↑ | ROUGE-L ↑ | METEOR ↑ |
|---|---|---|---|---|---|---|
| MolT5-large | 0.594 | 0.508 | 0.654 | 0.510 | 0.594 | 0.614 |
| MolReGPT | 0.607 | 0.525 | 0.634 | 0.476 | 0.562 | 0.610 |
| MolCA | 0.639 | 0.555 | 0.697 | 0.558 | 0.636 | 0.669 |
| BioT5 | 0.635 | 0.556 | 0.692 | 0.559 | 0.633 | 0.656 |
| ICMA | 0.651 | 0.581 | 0.686 | 0.550 | 0.625 | 0.661 |
| MolReFlect | 0.676 | 0.608 | 0.703 | 0.571 | 0.644 | 0.680 |
🔼 This table presents a comprehensive comparison of the performance of various models on the molecule captioning task using the ChEBI-20 dataset. It specifically evaluates the Mol2Cap (molecule-to-caption) subtask, comparing different methods’ effectiveness in generating accurate and relevant captions for given molecular structures. The models are evaluated based on several standard metrics, including BLEU scores (BLEU-2 and BLEU-4), ROUGE scores (ROUGE-1, ROUGE-2, and ROUGE-L), and METEOR score. The table highlights the best and second-best performing models to facilitate easy comparison and identification of top-performing approaches. Note that most methods involve fine-tuning LLMs on the ChEBI-20 dataset, except for MolReGPT.
read the caption
Table 1: Overall performance comparison for the Mol2Cap task on the ChEBI-20 dataset (Best, Second Best). Except for MolReGPT, all the other methods involve fine-tuning LLMs on the ChEBI-20 dataset.
In-depth insights#
Fine-grained Alignment#
Fine-grained alignment, in the context of molecule-text alignment, represents a significant advancement. It moves beyond simply mapping a molecule’s SMILES string or graph to a caption. Instead, it focuses on precisely aligning molecular substructures with specific textual descriptions. This granular approach enables a more nuanced understanding of the relationship between molecules and their textual representations. The benefits extend to increased accuracy and explainability in molecule-related tasks, such as caption generation and property prediction. Achieving fine-grained alignment often requires domain expertise, which makes automated methods particularly valuable. The challenge lies in reliably identifying and associating substructures with their corresponding descriptive phrases. This task demands sophisticated algorithms capable of handling both the structural complexity of molecules and the semantic nuances of natural language. Success in fine-grained alignment unlocks opportunities for more accurate, reliable, and interpretable results in various fields, including drug discovery and materials science.
Teacher-Student Model#
A teacher-student model in the context of a research paper likely involves a large language model (LLM) acting as the teacher and a smaller, potentially more efficient LLM as the student. The teacher’s role is to provide high-quality training data, often in the form of fine-grained alignments between molecular structures and their textual descriptions. This might entail identifying key substructures and their corresponding phrases. The student LLM then learns from this refined data, improving its ability to perform molecule-caption translation tasks. This approach is particularly valuable when high-quality, manually labeled training data is scarce. The teacher-student setup mitigates the need for extensive, expensive human annotation, offering a more efficient and scalable training paradigm. By leveraging the strengths of a powerful teacher to generate accurate alignments and the efficiency of the student LLM to absorb the information, this model architecture promises improved performance and explainability.
In-context Learning#
In-context learning, a paradigm shift in machine learning, is explored in the context of molecule-text alignment. It leverages the ability of large language models (LLMs) to learn from examples provided within the input prompt, rather than relying solely on explicit training data. This is crucial for tasks like molecule captioning, where large, annotated datasets are scarce and expensive to obtain. The core idea is to embed relevant molecule-caption pairs in the prompt, enabling the LLM to perform alignment directly from the input molecule or caption. This approach drastically reduces the reliance on extensive fine-tuning and allows for more efficient and adaptable models. A key strength is the capacity for customization and explainability, potentially offering insights into the LLM’s reasoning process by revealing the specific sub-structures and phrases that drive the alignment. However, challenges remain. Hallucinations – instances where the LLM generates incorrect or nonsensical alignments – are a major concern, requiring refinement through methods such as in-context selective reflection to filter and improve results. Ultimately, in-context learning for molecule-text alignment shows tremendous promise in reducing data requirements, enhancing efficiency, and adding transparency to a complex task.
MolReFlect Framework#
The MolReFlect framework is a teacher-student model designed for fine-grained alignment between molecules and their textual descriptions. It leverages a powerful teacher LLM to initially extract zero-shot alignments, identifying key substructures and their corresponding textual phrases. A crucial innovation is the In-Context Selective Reflection, where the teacher LLM refines these alignments using similar examples as context. This is followed by the Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT), where a smaller student LLM learns from the refined alignments within a chain-of-thought framework. This multi-stage approach overcomes limitations of prior methods by focusing on the granular level of alignment, leading to improved performance in molecule-caption translation. The framework’s teacher-student architecture is particularly effective in managing computational costs and achieving a high level of accuracy. The design promotes explainability by explicitly outlining the alignment process, and its overall efficiency suggests applicability across various molecule-related tasks.
Future Directions#
Future research could explore expanding MolReFlect’s capabilities to encompass more diverse molecular representations beyond SMILES and SELFIES, potentially including 3D structures and other advanced formats. Improving the robustness of MolReFlect to noisy or incomplete data is crucial, as real-world datasets are often imperfect. Investigating methods for handling uncertainty and incorporating error correction techniques would significantly enhance the model’s reliability. Developing a more efficient and scalable training framework is another key area for future work, particularly for larger and more complex molecules. This might involve exploring transfer learning or other techniques to reduce training time and computational costs. Finally, integrating MolReFlect with other machine learning models for downstream tasks, such as molecule property prediction or drug discovery, could unlock significant value in practical applications. A focus on explainability and interpretability is also warranted to fully realize the potential of fine-grained alignments, allowing for a deeper understanding of the relationships between molecular structure and textual descriptions.
More visual insights#
More on figures

🔼 This figure compares four different fine-tuning methods for language models, specifically focusing on their application in molecule-caption translation. (a) shows the basic Naive Supervised Fine-tuning approach, where the model directly maps input to output without explicit instructions or contextual information. (b) illustrates Instruction Tuning, which adds task instructions to guide the model’s learning. (c) depicts In-Context Molecule Tuning, incorporating similar examples as context for improved performance. Finally, (d) presents the authors’ proposed Chain-of-Thought In-Context Molecule Tuning, which combines the benefits of instruction tuning and in-context learning by structuring the reasoning process in a chain-of-thought format, leading to more explainable and accurate results.
read the caption
Figure 2: Comparisons of four different fine-tuning paradigms, including (a) Naive Supervised Fine-tuning (naive-SFT), (b) Instruction Tuning (Wei et al., 2021), (c) In-Context Molecule Tuning (ICMT) (Li et al., 2024a), and (d) our proposed Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT).

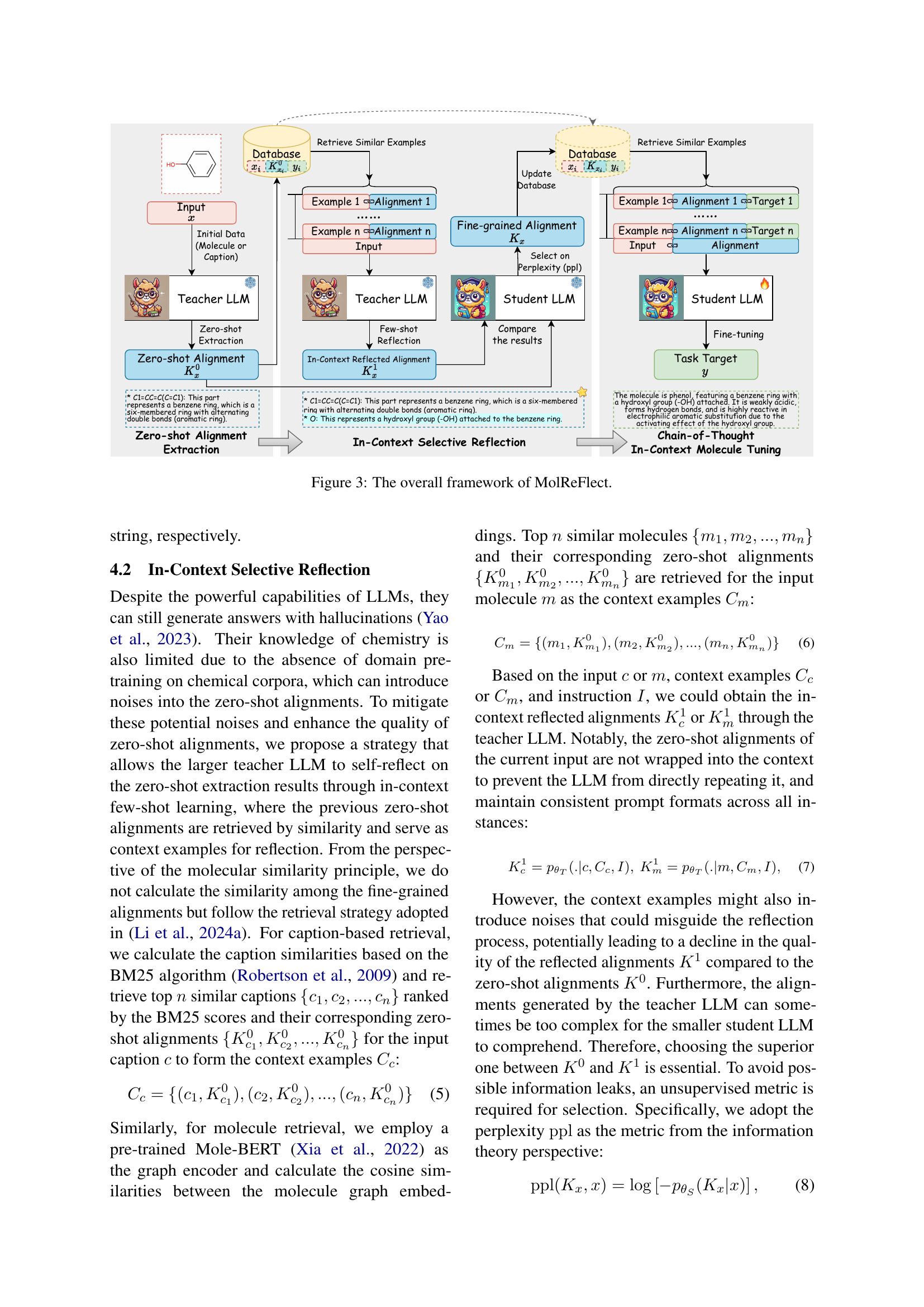
🔼 The figure illustrates the three main stages of the MolReFlect model: Zero-shot Alignment Extraction, In-Context Selective Reflection, and Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT). Zero-shot Alignment Extraction uses a large language model (LLM) to initially extract fine-grained alignments between molecules and their corresponding text descriptions. In-Context Selective Reflection refines these alignments by retrieving similar examples and utilizing the teacher LLM to reflect on these examples. Finally, CoT-ICMT enhances learning by incorporating the fine-grained alignments and reasoning processes in a chain-of-thought format, helping a smaller student LLM refine its molecule-text alignment capabilities. The diagram visually represents the data flow and interaction between the teacher and student LLMs throughout this process.
read the caption
Figure 3: The overall framework of MolReFlect.

🔼 This figure visualizes the embedding distributions of molecules and their corresponding captions using a dimensionality reduction technique like t-SNE or UMAP. The goal is to show how the embeddings of molecules (represented as SMILES or graph structures) and their textual descriptions cluster together in a low-dimensional space. The proximity of molecules and captions in this space indicates the quality of the alignment between the molecular representation and the text. Similar molecules and their related captions should be clustered close together, demonstrating the effectiveness of the model in generating accurate and semantically meaningful captions for molecules, and vice versa.
read the caption
Figure 4: Embedding distributions of molecules and captions.
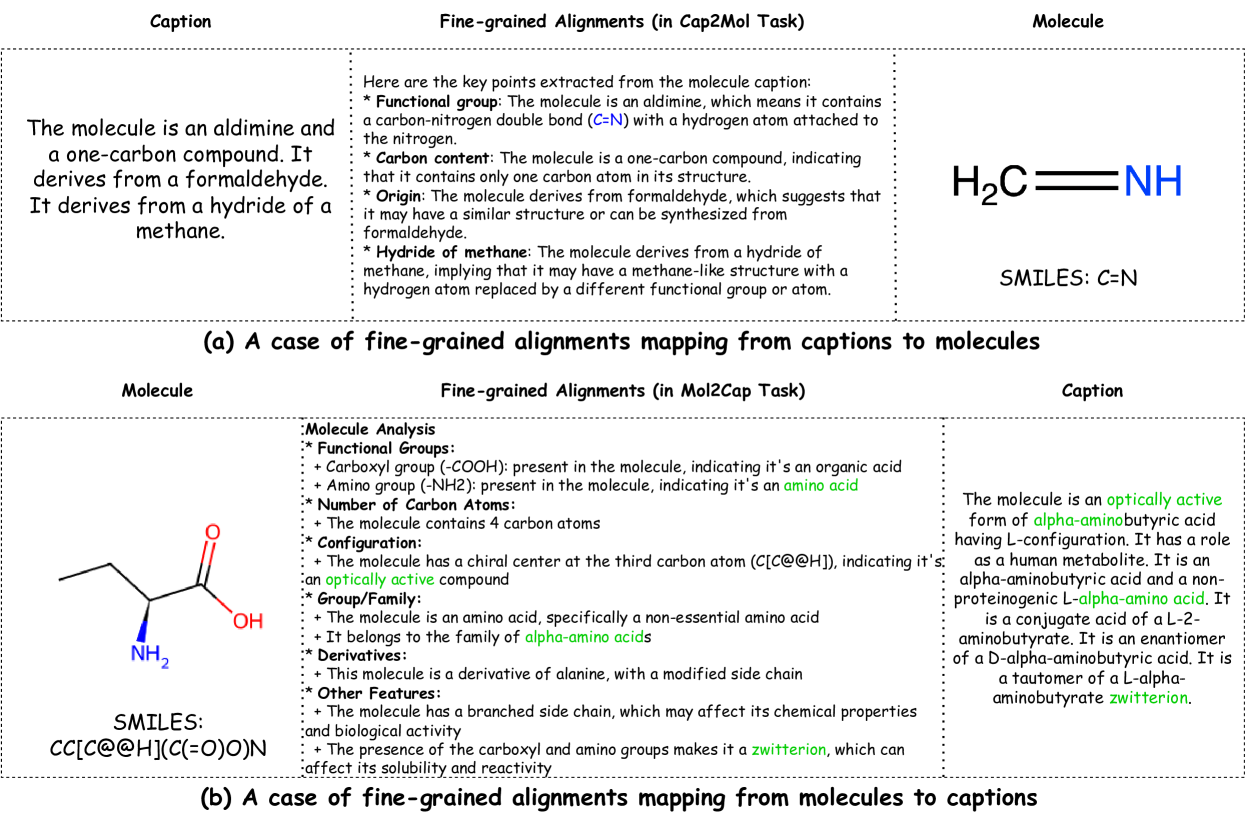
🔼 Figure 5 presents two examples illustrating the concept of fine-grained alignments in the MolReFlect model. The top panel shows how the model extracts key features and characteristics from a molecule caption (e.g., ‘The molecule is an aldimine and a one-carbon compound’) and aligns them to the corresponding substructures of the molecule’s SMILES representation. This alignment process helps the model link textual descriptions to specific molecular components. The bottom panel reverses this process, showing how the model identifies key substructures and characteristics within the molecule’s SMILES and links these to specific descriptive phrases in the caption (e.g., ‘The molecule is an optically active form of alpha-aminobutyric acid having L-configuration.’). These aligned features are crucial for accurate and explainable molecule-caption generation.
read the caption
Figure 5: Cases of Fine-grained Alignments. We could observe that the molecule structure and characteristics have already been mentioned and aligned by the fine-grained alignments, which will surely benefit the final generations.

🔼 Figure 6 showcases customized examples for the Cap2Mol task, a molecule generation task from text descriptions. The examples are designed to test the models’ ability to generate molecules that precisely match specific descriptions. The figure directly compares the outputs of three different models: MolReFlect, MolT5, and ICMA. MolReFlect successfully produces molecules that align with the input descriptions in each case. In contrast, MolT5 and ICMA fail to generate correct molecules, highlighting MolReFlect’s superior performance in handling customized, complex descriptions.
read the caption
Figure 6: Cases of Customized Examples for the Cap2Mol task. We follow the customized examples in Li et al. (2023a). Obviously, MolReFlect generates correct molecules in general, matching the requirements mentioned in the customized cases, while MolT5 and ICMA fail to meet the requirements.

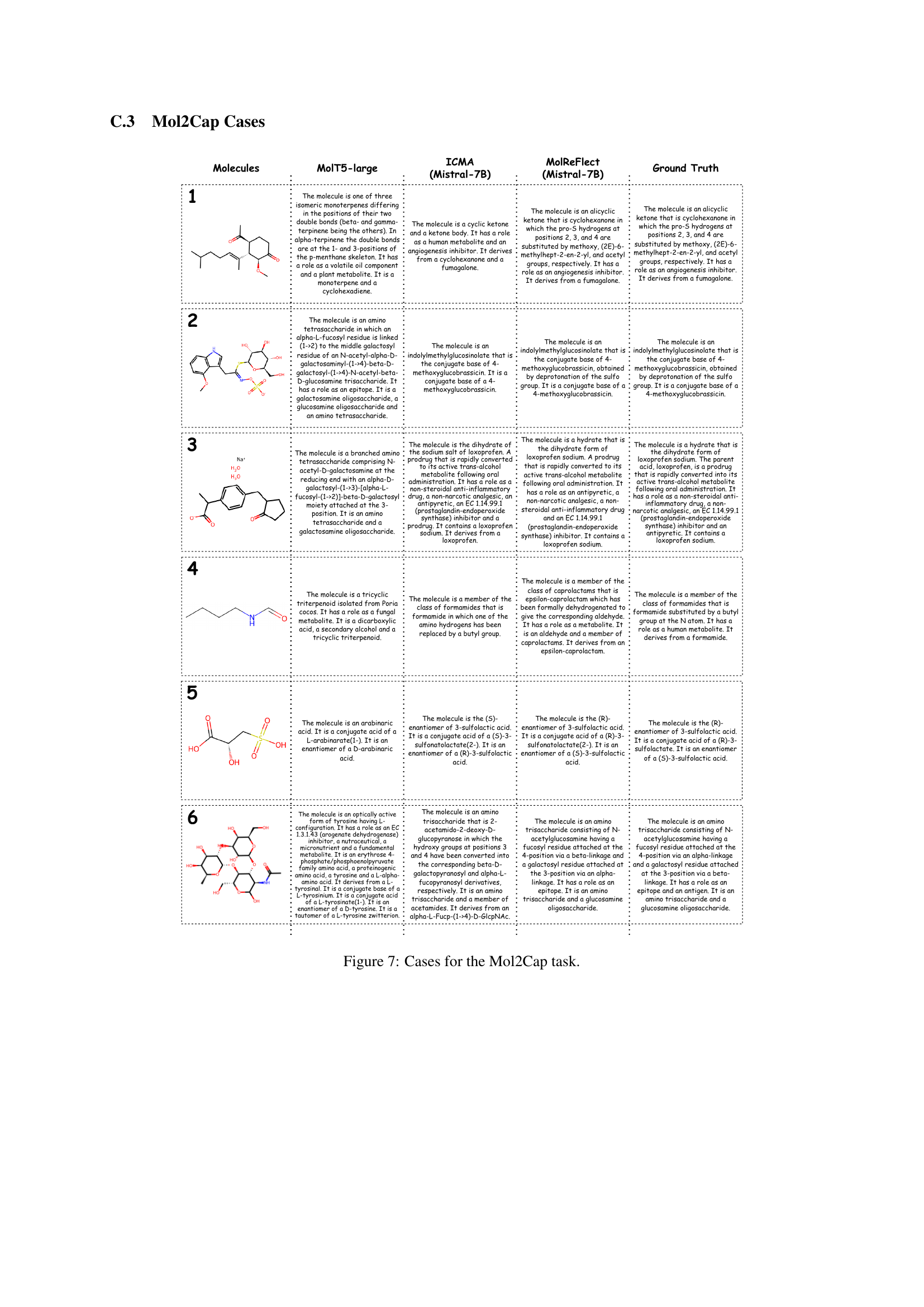
🔼 This figure showcases six examples of the Mol2Cap task, which involves generating molecule captions from SMILES strings. For each example, it shows the SMILES string representation of a molecule, followed by the captions generated by three different models: MolT5-large, ICMA (using Mistral-7B), and MolReFlect (also using Mistral-7B). Finally, the ground truth caption for each molecule is provided. This allows for a direct comparison of the different models’ performance in accurately and comprehensively describing molecules using natural language.
read the caption
Figure 7: Cases for the Mol2Cap task.

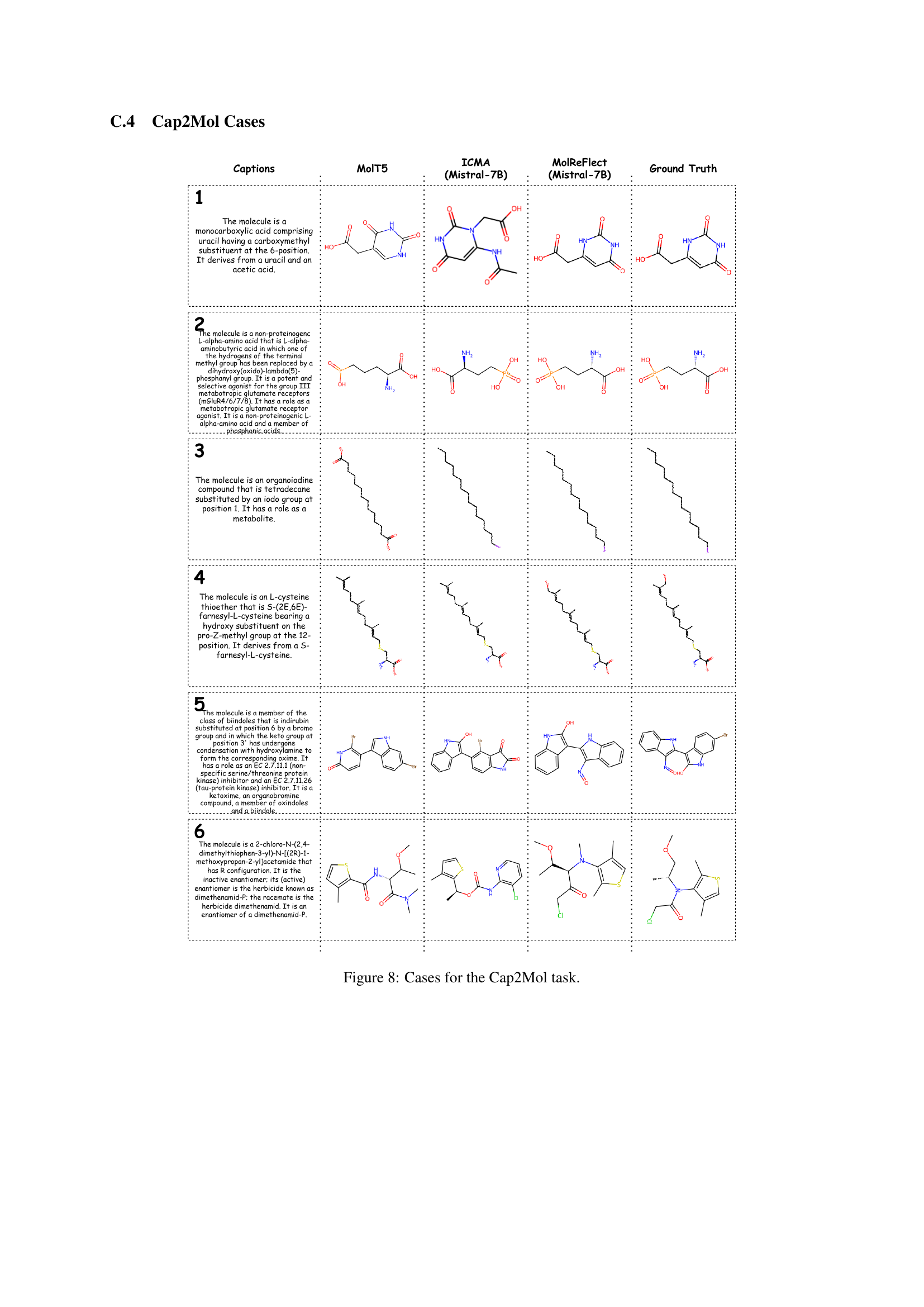
🔼 This figure showcases six examples of the Cap2Mol task, demonstrating the model’s ability to generate molecular structures from textual descriptions. Each example includes the input caption describing the molecule’s properties, the corresponding molecule structures generated by MolT5, ICMA (Mistral-7B), and MolReFlect (Mistral-7B), and the ground truth structure. This visualization helps to compare the performance of different models on this task and highlight MolReFlect’s superior accuracy in generating correct molecular structures based on the given captions.
read the caption
Figure 8: Cases for the Cap2Mol task.

🔼 This figure displays the prompt templates utilized in the Zero-shot Alignment Extraction stage of the MolReFlect model. It shows the specific instructions given to the Large Language Model (LLM) to extract fine-grained alignments from molecule structures (Mol2Cap) and molecule captions (Cap2Mol). The prompts guide the LLM to identify key structural features and descriptive phrases to establish a correspondence between the molecular and textual representations.
read the caption
Figure 9: Prompt templates for Zero-shot Alignment Extraction.

🔼 This figure presents the prompt templates utilized in the In-Context Selective Reflection stage of the MolReFlect framework. It shows how the model prompts the language model (LLM) to refine its zero-shot alignments by providing it with relevant example molecule-alignment pairs. Two templates are shown, one for the Mol2Cap task (molecule to caption) and another for the Cap2Mol task (caption to molecule). The templates leverage a chat-based interaction format (System, User, Assistant roles) to guide the LLM through the refinement process.
read the caption
Figure 10: Prompt templates for In-Context Selective Reflection.

🔼 This figure displays the prompt templates utilized in the MolReFlect model when fine-grained alignments are not employed. It shows the structure of the prompts used for both the Mol2Cap (molecule-to-caption) and Cap2Mol (caption-to-molecule) tasks. The prompts are formatted for a chat interface with roles for the system (instructions), user (input), and assistant (model response) to guide the language model in generating the desired outputs.
read the caption
Figure 11: Prompt templates for MolReFlect (w/o Fine-grained Alignments).

🔼 This figure displays the prompt templates utilized in the Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT) phase of the MolReFlect framework. It details the structure of the prompts used to guide the Large Language Model (LLM) in generating fine-grained alignments between molecules and their corresponding text descriptions. The prompts incorporate example molecule-alignment pairs to improve the LLM’s understanding and performance in the task. The templates are designed for the chat interface of LLMs, showing system, user, and assistant roles.
read the caption
Figure 12: Prompt templates for Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT).
More on tables
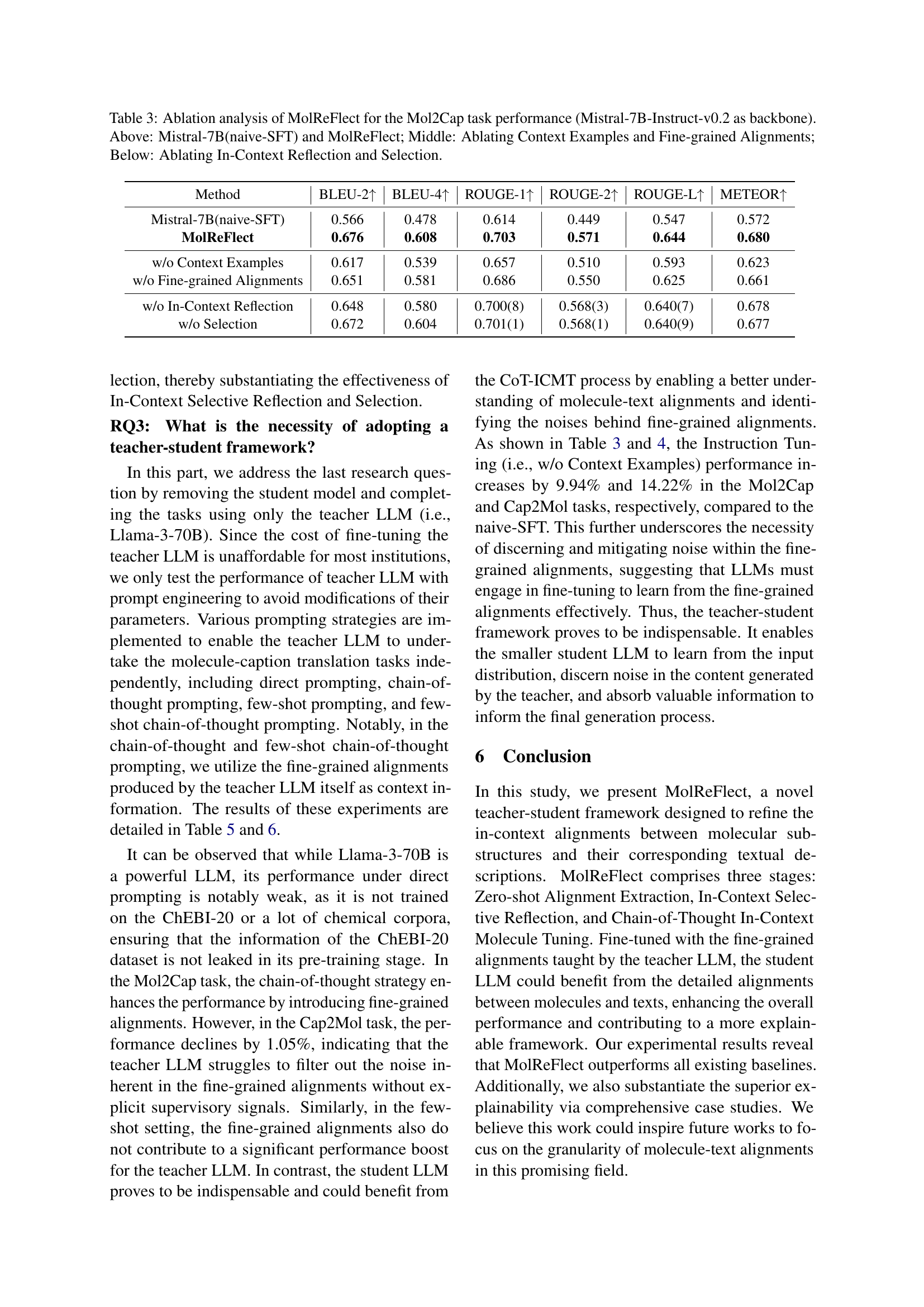
| Method | BLEU↑ | EM↑ | Levenshtein↓ | MACCS FTS↑ | RDK FTS↑ | Morgan FTS↑ | Validity↑ |
|---|---|---|---|---|---|---|---|
| MolT5-large | 0.854 | 0.311 | 16.07 | 0.834 | 0.746 | 0.684 | 0.905 |
| MolReGPT | 0.857 | 0.280 | 17.14 | 0.903 | 0.805 | 0.739 | 0.899 |
| BioT5 | 0.867 | 0.413 | 15.10 | 0.886 | 0.801 | 0.734 | 1.000 |
| ICMA | 0.855 | 0.460 | 18.73 | 0.916 | 0.837 | 0.789 | 0.958 |
| MolReFlect | 0.903 | 0.510 | 11.84 | 0.929 | 0.860 | 0.813 | 0.977 |
🔼 This table presents a comprehensive comparison of various models’ performance on the Cap2Mol task within the ChEBI-20 dataset. The Cap2Mol task involves generating molecule structures (SMILES strings) from given captions describing molecules. The table compares different models using several metrics, including BLEU score (measuring translation quality), Exact Match (the percentage of perfectly generated molecules), Levenshtein distance (measuring the edit distance between generated and ground truth SMILES), and three different types of molecular fingerprint scores (MACCS, RDK, Morgan), assessing the similarity of generated and reference molecule structures. Finally, a validity score is included indicating the proportion of valid SMILES strings generated by each model. The models are compared to establish which ones perform best and the extent to which different approaches succeed in the task. MolReFlect is shown to outperform other models across multiple metrics, showcasing the effectiveness of its approach.
read the caption
Table 2: Overall performance comparison for the Cap2Mol task on the ChEBI-20 dataset (Best, Second Best). Except for MolReGPT, all the other methods involve fine-tuning LLMs on the ChEBI-20 dataset.
| Method | BLEU-2↑ | BLEU-4↑ | ROUGE-1↑ | ROUGE-2↑ | ROUGE-L↑ | METEOR↑ |
|---|---|---|---|---|---|---|
| Mistral-7B(naive-SFT) | 0.566 | 0.478 | 0.614 | 0.449 | 0.547 | 0.572 |
| MolReFlect | 0.676 | 0.608 | 0.703 | 0.571 | 0.644 | 0.680 |
| w/o Context Examples | 0.617 | 0.539 | 0.657 | 0.510 | 0.593 | 0.623 |
| w/o Fine-grained Alignments | 0.651 | 0.581 | 0.686 | 0.550 | 0.625 | 0.661 |
| w/o In-Context Reflection | 0.648 | 0.580 | 0.700(8) | 0.568(3) | 0.640(7) | 0.678 |
| w/o Selection | 0.672 | 0.604 | 0.701(1) | 0.568(1) | 0.640(9) | 0.677 |
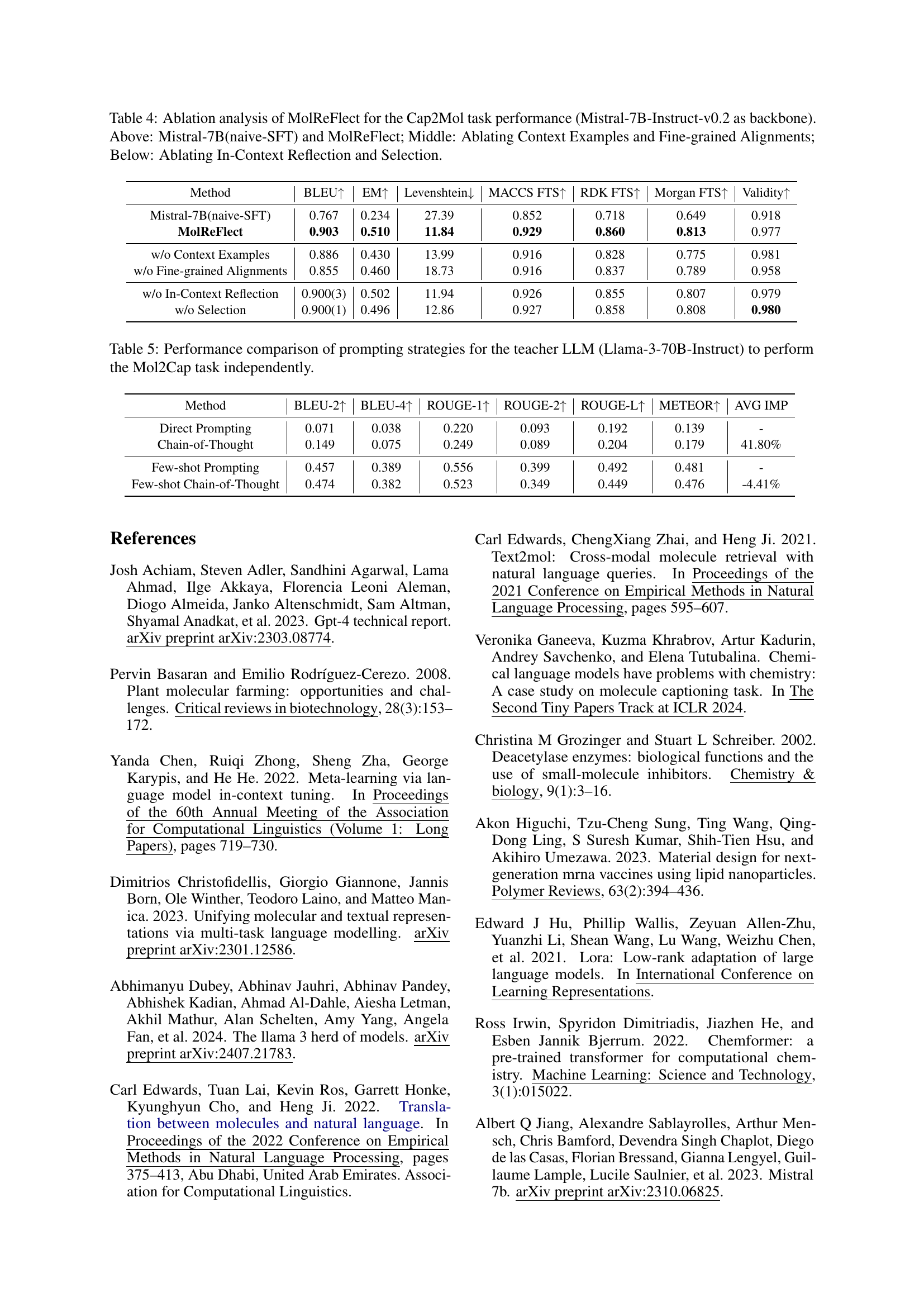
🔼 This table presents an ablation study on the MolReFlect model for the molecule-to-caption (Mol2Cap) task. It shows the impact of different components of the model on performance, using Mistral-7B-Instruct-v0.2 as the base model. The top section compares the baseline naive supervised fine-tuning (naive-SFT) with the full MolReFlect model. The middle section demonstrates the effect of removing context examples and fine-grained alignments separately. The bottom section investigates the impact of removing the in-context reflection and selection stages of MolReFlect. The results are evaluated using BLEU, ROUGE, and METEOR metrics.
read the caption
Table 3: Ablation analysis of MolReFlect for the Mol2Cap task performance (Mistral-7B-Instruct-v0.2 as backbone). Above: Mistral-7B(naive-SFT) and MolReFlect; Middle: Ablating Context Examples and Fine-grained Alignments; Below: Ablating In-Context Reflection and Selection.
| Method | BLEU↑ | EM↑ | Levenshtein↓ | MACCS FTS↑ | RDK FTS↑ | Morgan FTS↑ | Validity↑ |
|---|---|---|---|---|---|---|---|
| Mistral-7B(naive-SFT) | 0.767 | 0.234 | 27.39 | 0.852 | 0.718 | 0.649 | 0.918 |
| MolReFlect | 0.903 | 0.510 | 11.84 | 0.929 | 0.860 | 0.813 | 0.977 |
| w/o Context Examples | 0.886 | 0.430 | 13.99 | 0.916 | 0.828 | 0.775 | 0.981 |
| w/o Fine-grained Alignments | 0.855 | 0.460 | 18.73 | 0.916 | 0.837 | 0.789 | 0.958 |
| w/o In-Context Reflection | 0.900(3) | 0.502 | 11.94 | 0.926 | 0.855 | 0.807 | 0.979 |
| w/o Selection | 0.900(1) | 0.496 | 12.86 | 0.927 | 0.858 | 0.808 | 0.980 |
🔼 This table presents an ablation study on the MolReFlect model for the Cap2Mol task, which involves generating molecules from captions. It compares the performance of Mistral-7B with naive supervised fine-tuning against MolReFlect’s full model and several ablated versions. The ablations systematically remove components of MolReFlect, such as context examples, fine-grained alignments, in-context reflection, and selection, to isolate the impact of each component on the overall performance. The metrics used include BLEU, Exact Match, Levenshtein, MACCS, RDK, Morgan fingerprint scores, and Validity.
read the caption
Table 4: Ablation analysis of MolReFlect for the Cap2Mol task performance (Mistral-7B-Instruct-v0.2 as backbone). Above: Mistral-7B(naive-SFT) and MolReFlect; Middle: Ablating Context Examples and Fine-grained Alignments; Below: Ablating In-Context Reflection and Selection.
| Method | BLEU-2↑ | BLEU-4↑ | ROUGE-1↑ | ROUGE-2↑ | ROUGE-L↑ | METEOR↑ | AVG IMP |
|---|---|---|---|---|---|---|---|
| Direct Prompting | 0.071 | 0.038 | 0.220 | 0.093 | 0.192 | 0.139 | - |
| Chain-of-Thought | 0.149 | 0.075 | 0.249 | 0.089 | 0.204 | 0.179 | 41.80% |
| Few-shot Prompting | 0.457 | 0.389 | 0.556 | 0.399 | 0.492 | 0.481 | - |
| Few-shot Chain-of-Thought | 0.474 | 0.382 | 0.523 | 0.349 | 0.449 | 0.476 | -4.41% |
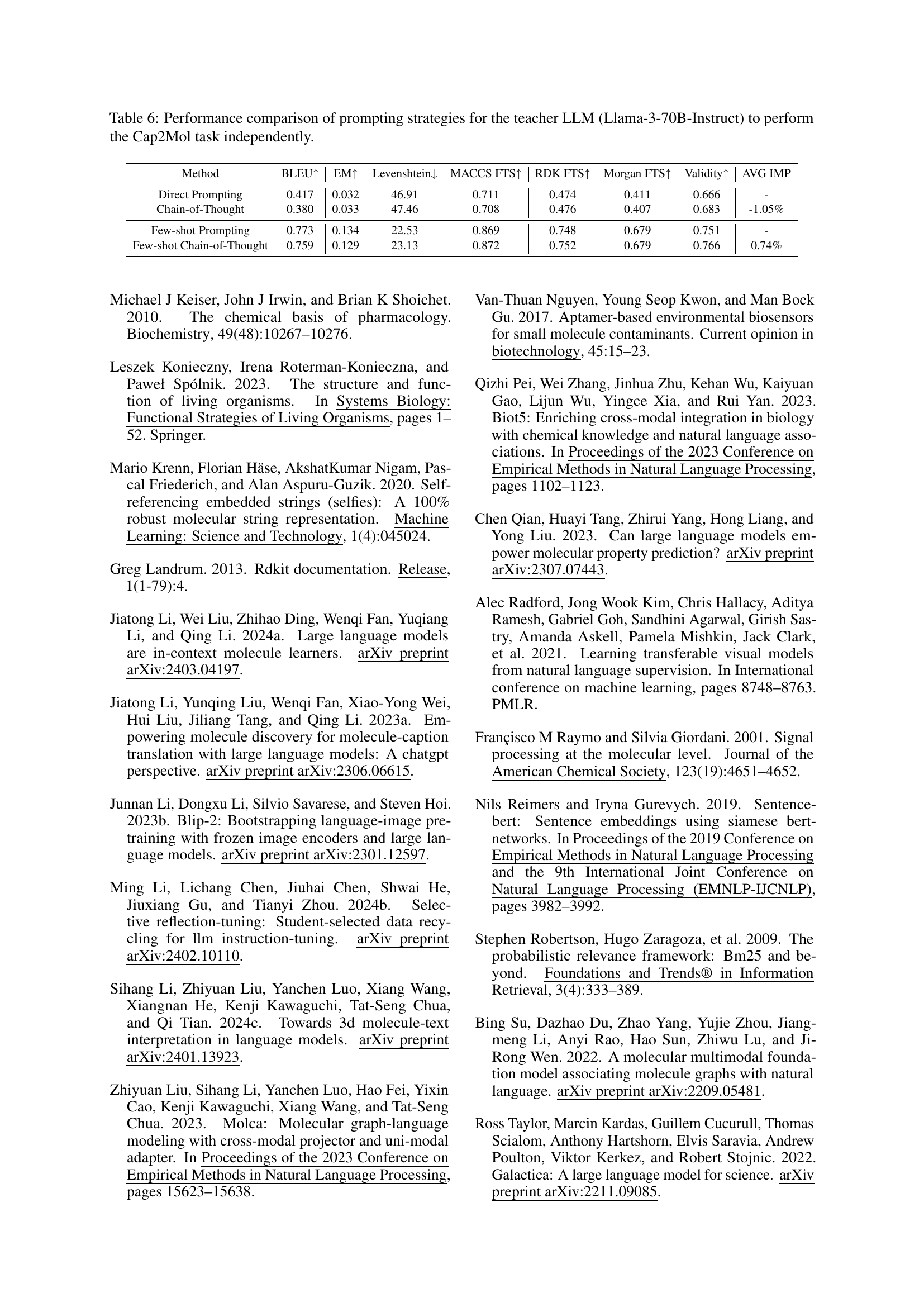
🔼 This table compares the performance of different prompting strategies when using the Llama-3-70B-Instruct large language model (LLM) to perform the molecule-to-caption (Mol2Cap) task without any fine-tuning. It shows how various prompting techniques—direct prompting, chain-of-thought prompting, few-shot prompting, and few-shot chain-of-thought prompting—affect the model’s ability to generate accurate captions for molecules. The results are evaluated using several metrics including BLEU-2, BLEU-4, ROUGE-1, ROUGE-2, ROUGE-L, and METEOR scores, providing a comprehensive assessment of the generated captions’ quality.
read the caption
Table 5: Performance comparison of prompting strategies for the teacher LLM (Llama-3-70B-Instruct) to perform the Mol2Cap task independently.
| Method | BLEU↑ | EM↑ | Levenshtein↓ | MACCS FTS↑ | RDK FTS↑ | Morgan FTS↑ | Validity↑ | AVG IMP |
|---|---|---|---|---|---|---|---|---|
| Direct Prompting | 0.417 | 0.032 | 46.91 | 0.711 | 0.474 | 0.411 | 0.666 | - |
| Chain-of-Thought | 0.380 | 0.033 | 47.46 | 0.708 | 0.476 | 0.407 | 0.683 | -1.05% |
| Few-shot Prompting | 0.773 | 0.134 | 22.53 | 0.869 | 0.748 | 0.679 | 0.751 | - |
| Few-shot Chain-of-Thought | 0.759 | 0.129 | 23.13 | 0.872 | 0.752 | 0.679 | 0.766 | 0.74% |
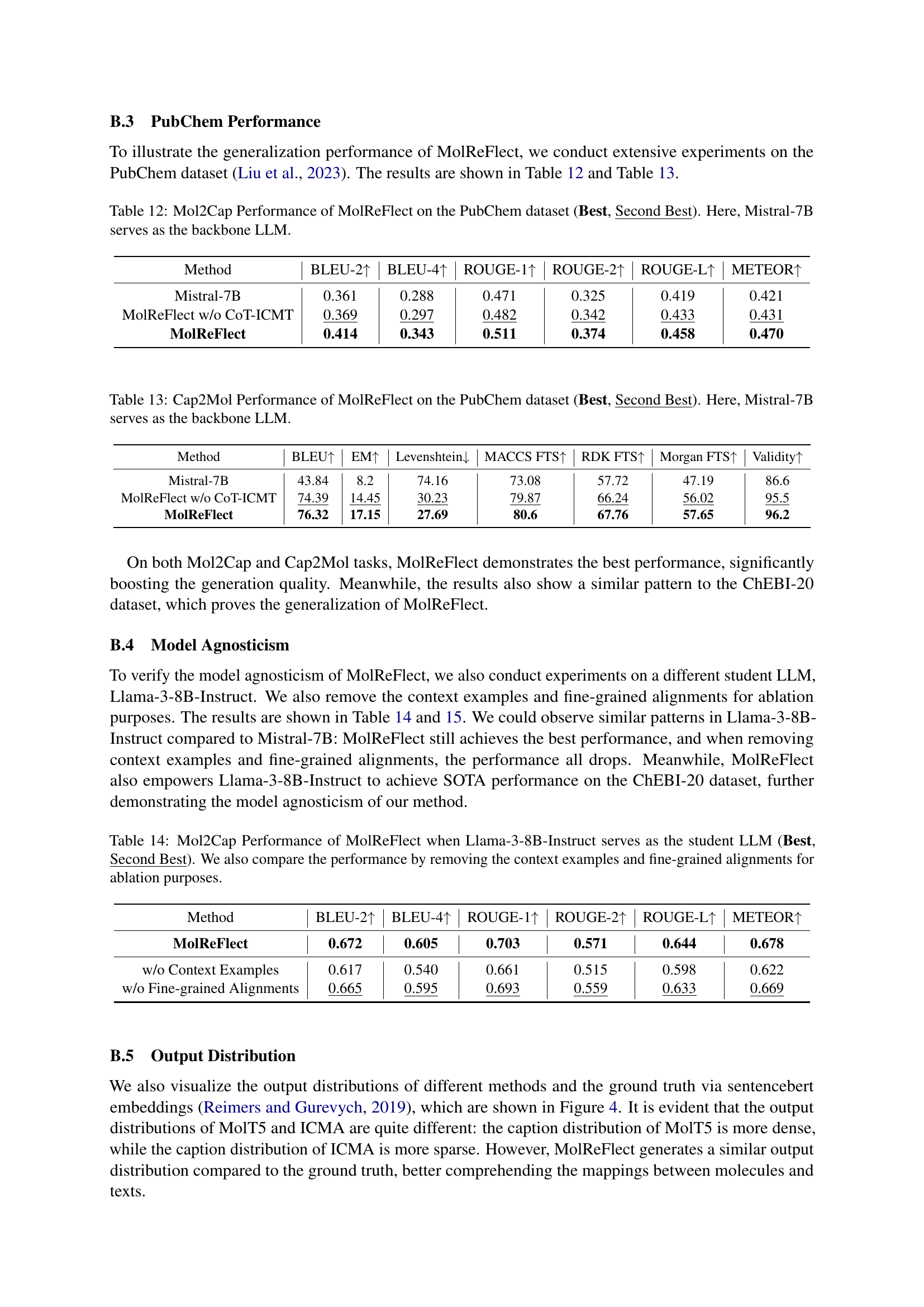
🔼 This table compares the performance of different prompting strategies when using the Llama-3-70B-Instruct large language model (LLM) to perform the Cap2Mol task (generating molecules from captions) independently, without a smaller student LLM. The strategies tested include direct prompting, chain-of-thought prompting, few-shot prompting, and few-shot chain-of-thought prompting. The table shows the performance metrics (BLEU, Exact Match, Levenshtein distance, MACCS, RDK, Morgan fingerprint scores, and Validity) for each prompting strategy to assess which approach is most effective for this task when using only the large LLM.
read the caption
Table 6: Performance comparison of prompting strategies for the teacher LLM (Llama-3-70B-Instruct) to perform the Cap2Mol task independently.
| Item | Value |
|---|---|
| int4 | True |
| temperature | 0.75 |
| top_p | 0.85 |
| top_k | 40 |
| num_return_sequences | 1 |
| max_new_tokens | 512 |
| number-of-examples | 2 |
🔼 This table details the hyperparameters used for the larger teacher Language Model (LLM) in the MolReFlect framework. These settings control aspects of the model’s behavior during the zero-shot alignment extraction and in-context selective reflection stages. Specific parameters include the quantization method (int4), temperature, top_p and top_k values for sampling, the number of return sequences, and the maximum number of new tokens to generate.
read the caption
Table 7: Hyper-parameters for the larger teacher LLM.
| Item | Value |
|---|---|
| macro batch size | 32 |
| micro batch size | 1 |
| steps | 8000 |
| warm-up steps | 1000 |
| cutoff length | 4096 |
| number-of-examples | 2 |
| learning rate | 2e-4 |
| lora_r | 32 |
| lora_alpha | 64 |
| lora_dropout | 0.1 |
| int8 | True |
| fp16 | True |
| temperature | 0.75 |
| top_p | 0.85 |
| top_k | 40 |
| num_return_sequences | 1 |
| max_new_tokens | 512 |
🔼 This table lists the hyperparameters used for fine-tuning the smaller student Large Language Model (LLM) in the MolReFlect framework. It includes settings for batch size (both macro and micro), training steps, learning rate, LoRA (Low-Rank Adaptation) parameters (r, alpha, and dropout), and other settings relevant to the generation process (integer type, precision, temperature, top_p, top_k, and max_new_tokens).
read the caption
Table 8: Hyper-parameters for the smaller student LLM.
| Item | semantic similarity | perplexity |
|---|---|---|
| molecules | 0.2483 | 2.246 |
| zero-shot alignments | 0.4983 | 2.066 |
| in-context reflected alignments | 0.4985 | 2.070 |
| fine-grained alignments | 0.5029 | 1.995 |
🔼 This table presents a quantitative analysis of the quality of different molecular alignments generated for the molecule captioning task (Mol2Cap). It compares the average semantic similarity and perplexity scores of four types of alignments: original molecules, zero-shot alignments, in-context reflected alignments, and fine-grained alignments. The perplexity score measures how well a language model predicts the alignment, while semantic similarity assesses the closeness in meaning between the alignment and the original molecule. This comparison helps evaluate the effectiveness of different alignment methods in representing molecular structures and their associated textual descriptions.
read the caption
Table 9: Average semantic similarity and perplexity scores of different alignments and the original molecules in the training set for the Mol2Cap task.
| Item | semantic similarity | perplexity |
|---|---|---|
| captions | 0.2483 | 2.758 |
| zero-shot alignments | 0.2721 | 2.426 |
| in-context reflected alignments | 0.2377 | 2.351 |
| fine-grained alignments | 0.2524 | 2.230 |
🔼 This table presents a quantitative analysis of the quality of various molecule-caption alignments generated during the Cap2Mol task. It compares the average semantic similarity and perplexity scores for four different alignment types: original molecule captions, zero-shot alignments, in-context reflected alignments, and the final fine-grained alignments. The semantic similarity score measures how well the alignment captures the meaning of the original caption, while the perplexity score reflects the uncertainty or randomness of the alignment. By comparing these scores across the different alignment types, the table helps evaluate the effectiveness of the MolReFlect framework’s different stages in refining the molecule-caption alignments.
read the caption
Table 10: Average semantic similarity and perplexity scores of different alignments and the original molecule captions in the training set for the Cap2Mol task.
| Tasks | BACE | BBBP |
|---|---|---|
| Mistral7B | 0.4926 | 0.4829 |
| ICMA | 0.7995 | 0.6775 |
| MolReFlect | 0.8795 | 0.8925 |
🔼 This table presents the area under the ROC curve (ROC-AUC) scores achieved by the MolReFlect model and several baseline models on two molecular property prediction tasks: the Binding Affinity to the Beta-Secretase 1 (BACE) and Blood-Brain Barrier Permeability (BBBP) tasks. The MoleculeNet dataset (Wu et al., 2018) is used for this evaluation. The scores indicate the performance of each model in predicting whether a molecule exhibits a certain property (e.g., binding affinity or BBBP permeability). Higher scores represent better performance.
read the caption
Table 11: ROC-AUC (%) scores of MolReFlect on the BACE and BBBP task from the MoleculeNet dataset (Wu et al., 2018) (Best, Second Best).
| Method | BLEU-2 ↑ | BLEU-4 ↑ | ROUGE-1 ↑ | ROUGE-2 ↑ | ROUGE-L ↑ | METEOR ↑ |
|---|---|---|---|---|---|---|
| Mistral-7B | 0.361 | 0.288 | 0.471 | 0.325 | 0.419 | 0.421 |
| MolReFlect w/o CoT-ICMT | 0.369 | 0.297 | 0.482 | 0.342 | 0.433 | 0.431 |
| MolReFlect | 0.414 | 0.343 | 0.511 | 0.374 | 0.458 | 0.470 |
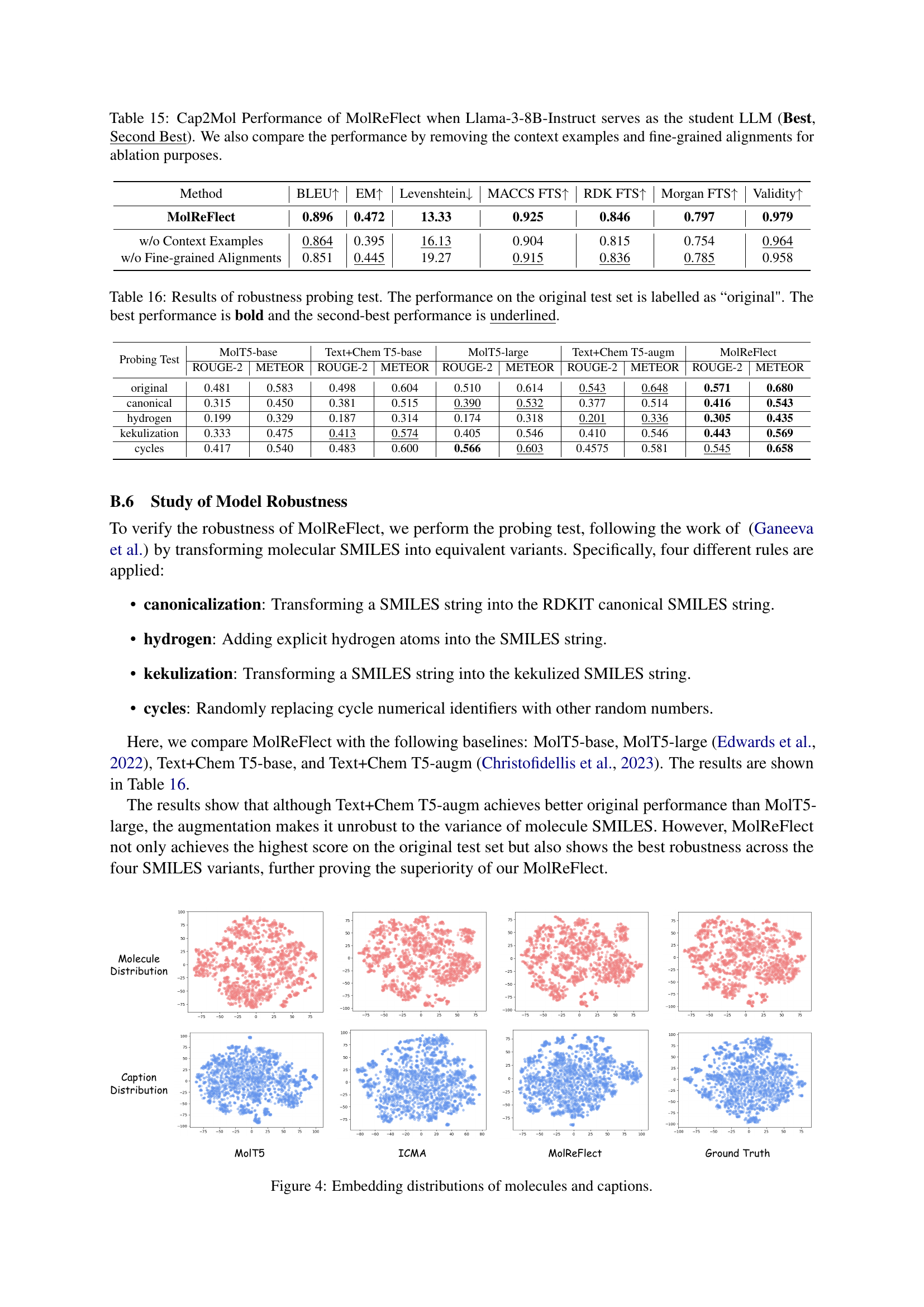
🔼 This table presents a comparison of the performance of different models on the Mol2Cap task using the PubChem dataset. Specifically, it contrasts the performance of the Mistral-7B model (baseline) with MolReFlect, both with and without Chain-of-Thought In-Context Molecule Tuning (CoT-ICMT). The metrics used for evaluation include BLEU scores (BLEU-2, BLEU-4), ROUGE scores (ROUGE-1, ROUGE-2, ROUGE-L), and METEOR score. The results showcase MolReFlect’s performance improvement over the baseline model, highlighting the effectiveness of the fine-tuning method.
read the caption
Table 12: Mol2Cap Performance of MolReFlect on the PubChem dataset (Best, Second Best). Here, Mistral-7B serves as the backbone LLM.
| Method | BLEU↑ | EM↑ | Levenshtein↓ | MACCS FTS↑ | RDK FTS↑ | Morgan FTS↑ | Validity↑ |
|---|---|---|---|---|---|---|---|
| Mistral-7B | 43.84 | 8.2 | 74.16 | 73.08 | 57.72 | 47.19 | 86.6 |
| MolReFlect w/o CoT-ICMT | 74.39 | 14.45 | 30.23 | 79.87 | 66.24 | 56.02 | 95.5 |
| MolReFlect | 76.32 | 17.15 | 27.69 | 80.6 | 67.76 | 57.65 | 96.2 |
🔼 This table presents the results of the Cap2Mol task on the PubChem dataset using MolReFlect, a novel teacher-student framework for molecule-caption alignment. The table compares the performance of MolReFlect against several baseline methods. Metrics used for evaluation include BLEU score, exact match rate, Levenshtein distance, MACCS, RDK, and Morgan fingerprints, and the validity of generated SMILES strings. Mistral-7B served as the base large language model for all methods in this comparison.
read the caption
Table 13: Cap2Mol Performance of MolReFlect on the PubChem dataset (Best, Second Best). Here, Mistral-7B serves as the backbone LLM.
| Method | BLEU-2↑ | BLEU-4↑ | ROUGE-1↑ | ROUGE-2↑ | ROUGE-L↑ | METEOR↑ |
|---|---|---|---|---|---|---|
| MolReFlect | 0.672 | 0.605 | 0.703 | 0.571 | 0.644 | 0.678 |
| w/o Context Examples | 0.617 | 0.540 | 0.661 | 0.515 | 0.598 | 0.622 |
| w/o Fine-grained Alignments | 0.665 | 0.595 | 0.693 | 0.559 | 0.633 | 0.669 |
🔼 This table presents the performance of the MolReFlect model on the Mol2Cap task of the ChEBI-20 dataset using Llama-3-8B-Instruct as the student LLM. It shows the performance metrics (BLEU-2, BLEU-4, ROUGE-1, ROUGE-2, ROUGE-L, METEOR) achieved by MolReFlect under different conditions: with all components intact, without context examples, and without fine-grained alignments. This allows for an analysis of the contribution of each component to the model’s overall performance. The best and second-best results are indicated.
read the caption
Table 14: Mol2Cap Performance of MolReFlect when Llama-3-8B-Instruct serves as the student LLM (Best, Second Best). We also compare the performance by removing the context examples and fine-grained alignments for ablation purposes.
| Method | BLEU↑ | EM↑ | Levenshtein↓ | MACCS FTS↑ | RDK FTS↑ | Morgan FTS↑ | Validity↑ |
|---|---|---|---|---|---|---|---|
| MolReFlect | 0.896 | 0.472 | 13.33 | 0.925 | 0.846 | 0.797 | 0.979 |
| w/o Context Examples | 0.864 | 0.395 | 16.13 | 0.904 | 0.815 | 0.754 | 0.964 |
| w/o Fine-grained Alignments | 0.851 | 0.445 | 19.27 | 0.915 | 0.836 | 0.785 | 0.958 |
🔼 This table presents the results of the Cap2Mol task using MolReFlect with Llama-3-8B-Instruct as the student LLM. It shows the overall performance metrics (BLEU, Exact Match, Levenshtein distance, and various fingerprint scores) comparing MolReFlect’s performance against two ablation studies. The ablation studies remove either context examples or fine-grained alignments to analyze the impact of these components on the model’s ability to generate molecules from captions.
read the caption
Table 15: Cap2Mol Performance of MolReFlect when Llama-3-8B-Instruct serves as the student LLM (Best, Second Best). We also compare the performance by removing the context examples and fine-grained alignments for ablation purposes.
| Probing Test | MolT5-base ROUGE-2 | MolT5-base METEOR | Text+Chem T5-base ROUGE-2 | Text+Chem T5-base METEOR | MolT5-large ROUGE-2 | MolT5-large METEOR | Text+Chem T5-augm ROUGE-2 | Text+Chem T5-augm METEOR | MolReFlect ROUGE-2 | MolReFlect METEOR |
|---|---|---|---|---|---|---|---|---|---|---|
| original | 0.481 | 0.583 | 0.498 | 0.604 | 0.510 | 0.614 | 0.543 | 0.648 | 0.571 | 0.680 |
| canonical | 0.315 | 0.450 | 0.381 | 0.515 | 0.390 | 0.532 | 0.377 | 0.514 | 0.416 | 0.543 |
| hydrogen | 0.199 | 0.329 | 0.187 | 0.314 | 0.174 | 0.318 | 0.201 | 0.336 | 0.305 | 0.435 |
| kekulization | 0.333 | 0.475 | 0.413 | 0.574 | 0.405 | 0.546 | 0.410 | 0.546 | 0.443 | 0.569 |
| cycles | 0.417 | 0.540 | 0.483 | 0.600 | 0.566 | 0.603 | 0.4575 | 0.581 | 0.545 | 0.658 |
🔼 This table presents the results of a robustness probing test conducted on various molecule captioning models. The test evaluates model performance under different transformations of SMILES strings (canonicalization, hydrogen addition, kekulization, and cycle modification). The ‘original’ row shows performance on the original test set without SMILES string transformations. The best performance for each metric (ROUGE-2 and METEOR) under each transformation is shown in bold, with the second-best performance underlined. This allows for a comparison of how well different models maintain accuracy when faced with variations in input data representation.
read the caption
Table 16: Results of robustness probing test. The performance on the original test set is labelled as “original'. The best performance is bold and the second-best performance is underlined.
Full paper#