↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Accurate brain tumor segmentation from MRI scans is crucial for effective treatment, but manual segmentation is time-consuming and prone to errors. Existing AI models often struggle with variations in MRI quality and patient demographics, leading to unreliable results in diverse populations. This necessitates development of robust AI models for reliable tumor detection across different populations.
The researchers used a method called MedNeXt, a comprehensive model ensembling technique combined with thorough postprocessing. This approach demonstrated excellent performance on the BraTS 2024 SSA (Sub-Saharan Africa) and pediatric datasets, achieving high accuracy in identifying tumor regions. The study’s findings show the effectiveness of their approach in handling data variability and achieving improved segmentation accuracy, suggesting advancements towards reliable and accurate AI solutions for global healthcare needs.
Key Takeaways#
Why does it matter?#
This paper is important because it addresses the critical need for reliable and accurate brain tumor segmentation in diverse populations. Improved segmentation methods are crucial for better diagnosis and treatment planning, especially in underserved regions with limited resources. The study’s findings offer valuable insights for researchers working on brain tumor detection and AI-driven medical image analysis, potentially impacting patient care globally.
Visual Insights#

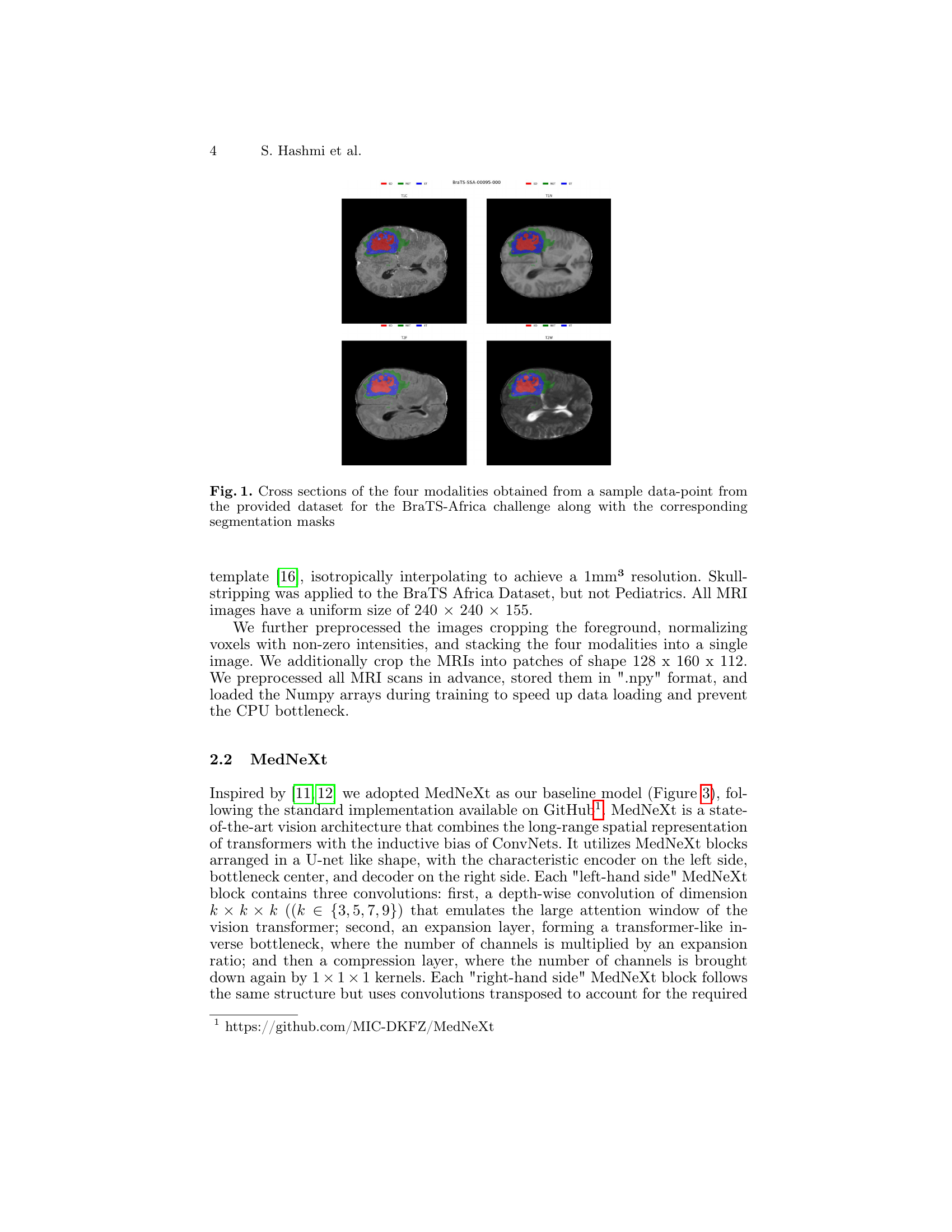
🔼 Figure 1 presents a visual representation of four different MRI modalities (T1, T1Gd, T2, and FLAIR) obtained from a single patient’s brain scan within the BraTS-Africa dataset. Each modality offers a unique perspective on brain tissue properties. The figure includes axial, coronal, and sagittal slices, allowing for a comprehensive 3D visualization of the brain tumor. Crucially, the figure also overlays the corresponding segmentation masks, highlighting the areas identified as tumor by experts. These masks delineate the exact boundaries of the tumor in each slice, serving as the ground truth for evaluating the accuracy of automated brain tumor segmentation algorithms.
read the caption
Figure 1: Cross sections of the four modalities obtained from a sample data-point from the provided dataset for the BraTS-Africa challenge along with the corresponding segmentation masks
| S.No | Model Name | Dice Score (ET) | Dice Score (TC) | Dice Score (WT) | Dice Score (Avg) | HD 95 (ET) | HD 95 (TC) | HD 95 (WT) | HD 95 (Avg) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Baseline | 0.8229 | 0.8151 | 0.8812 | 0.8397 | 39.96 | 46.48 | 25.04 | 37.16 |
| 2 | (+) Change patch size to: 128 × 160 × 112 | 0.852 | 0.839 | 0.912 | 0.8677 | 12.406 | 26.09 | 10.431 | 16.31 |
| 3 | (+) Train on SSA and Adult Gli. | 0.867 | 0.869 | 0.932 | 0.8893 | 15.578 | 22.145 | 8.833 | 15.52 |
| 4 | (+) Finetune the last 10 decoder layers on SSA | 0.874 | 0.870 | 0.933 | 0.8923 | 15.320 | 22.039 | 8.805 | 15.39 |
| 5 | (+) Reinitialize and finetune the last 10 decoder layers on SSA | 0.876 | 0.870 | 0.933 | 0.8930 | 15.328 | 22.040 | 8.746 | 15.37 |
| 6 | (+) Change the binarization threshold 0.7 for ET and TC ,0.5 for WT | 0.883 | 0.873 | 0.933 | 0.8963 | 14.248 | 21.028 | 8.770 | 14.68 |
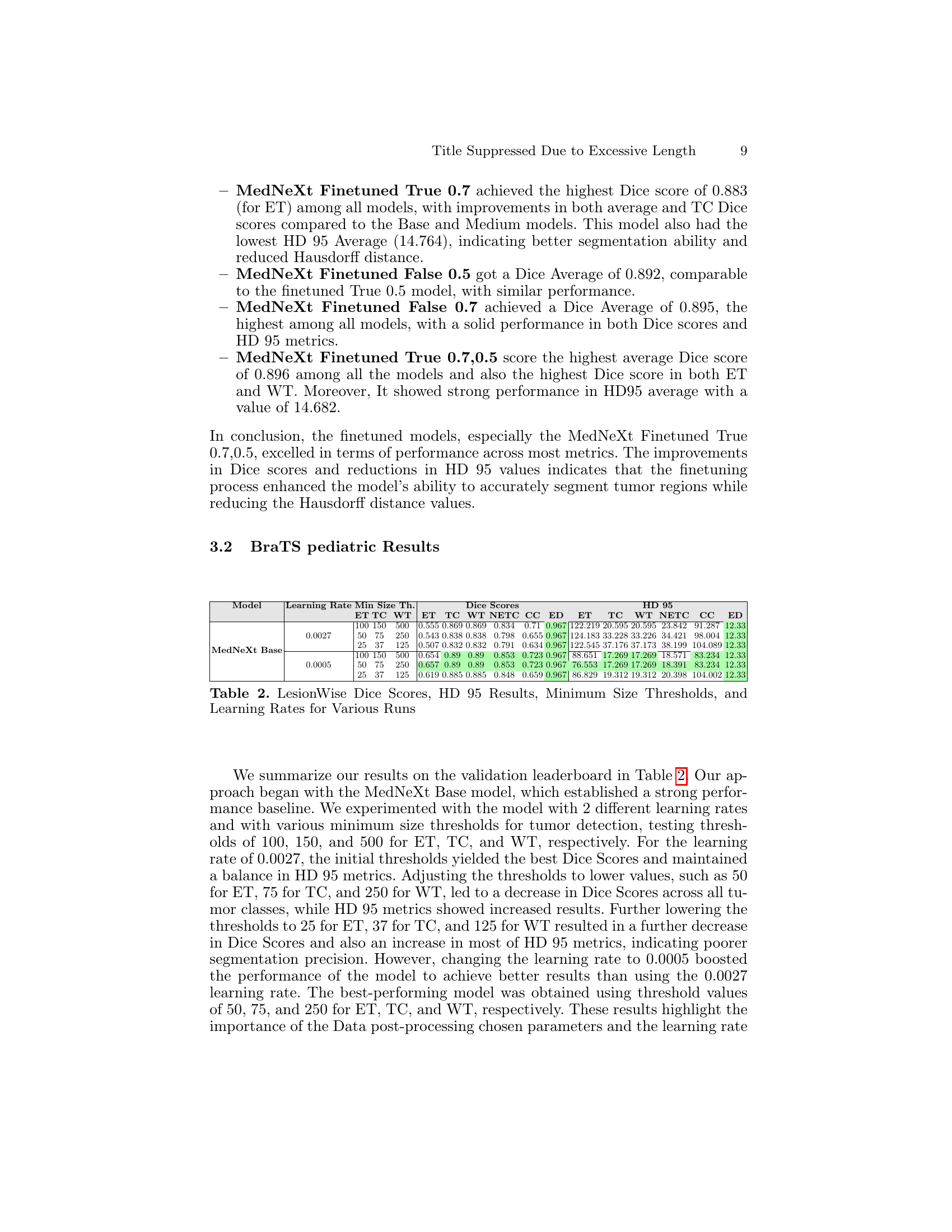
🔼 This table compares the performance of several MedNeXt Base models on the BraTS SSA 2024 dataset. Variations in the models include using different training datasets (SSA only vs. SSA + Adult Glioma), different patch sizes for input images, and different fine-tuning strategies (finetuning last 10 decoder layers with or without re-initializing weights). The comparison is based on the Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD95) metrics for three tumor sub-regions: Enhancing Tumor (ET), Tumor Core (TC), and Whole Tumor (WT). The table shows how these variations affect the accuracy and robustness of the model’s segmentation.
read the caption
Table 1: Comparison of MedNeXt Base Models with Different Training Data, Patch Sizes, and Fine-tuning Strategies
In-depth insights#
Glioma Segmentation#
Glioma segmentation, the process of identifying and delineating gliomas in medical images, is crucial for effective diagnosis and treatment planning. Accurate segmentation is challenging due to the inherent variability in glioma appearance, the complexity of tumor boundaries, and the presence of artifacts. Traditional manual segmentation is laborious and subjective; therefore, automated methods using machine learning, particularly deep learning models like U-Net and its variants, have gained significant traction. The BraTS challenge series has been instrumental in advancing the field by providing standardized datasets and evaluation metrics, while also highlighting the need to address distribution shifts across diverse populations and imaging modalities. This research demonstrates the effectiveness of an ensemble approach, specifically the MedNeXt architecture, to mitigate these challenges and improve segmentation accuracy across different patient cohorts, emphasizing the value of incorporating techniques to improve robustness and reliability in real-world applications.
MedNeXt Model#
The MedNeXt model, a state-of-the-art architecture for medical image segmentation, is the core of this research. Its unique design combines the strengths of convolutional neural networks (CNNs) and transformers, leveraging the inductive bias of CNNs for local feature extraction and the long-range dependencies captured by transformers. This hybrid approach is particularly well-suited for the complex and nuanced task of brain tumor segmentation in diverse datasets, which often exhibit significant variations in image quality and patient demographics. The study explores different MedNeXt configurations (Base and Medium) and training strategies, demonstrating its adaptability and robustness. Key innovations include the use of a schedule-free AdamW optimizer, improving training efficiency and stability, and the implementation of deep supervision to mitigate vanishing gradients. Furthermore, the impact of various pre-processing steps, model ensembling, and post-processing techniques are thoroughly investigated to optimize the final segmentation results. The integration of these elements demonstrates a holistic approach towards achieving high accuracy in brain tumor segmentation. Fine-tuning strategies on specific datasets further enhance the model’s performance, showcasing its capability to adapt to varied imaging characteristics.
Dataset Challenges#
Dataset challenges in brain tumor segmentation are significant. Data scarcity for certain demographics (e.g., pediatric patients, sub-Saharan African populations) limits the generalizability of models trained on predominantly Western datasets. Data heterogeneity stemming from diverse imaging protocols and equipment quality across different centers further compounds the problem. Annotation inconsistencies between different annotators introduce variability and uncertainty in the ground truth labels. Addressing these requires a multi-pronged approach including creating more representative datasets, developing robust training strategies that handle noisy or incomplete data, and employing advanced evaluation metrics that go beyond simple Dice scores to capture the nuances of segmentation performance across varied data distributions.
Ensemble Methods#
Ensemble methods, in the context of brain tumor segmentation, leverage the power of combining multiple models to improve predictive accuracy and robustness. The core idea is that diverse models, trained on the same data or different subsets, will make different errors. By aggregating their predictions, usually through averaging or weighted voting, the ensemble can mitigate individual model weaknesses and achieve a performance superior to any single constituent model. This approach is particularly valuable in medical imaging where data scarcity and inherent variability can limit the reliability of individual models. The success of an ensemble hinges critically on the diversity of its components. This can be achieved through variations in model architectures, training data, or hyperparameters. Careful consideration should be given to the ensemble strategy, such as simple averaging, weighted averaging, or more sophisticated techniques like stacking or boosting. Post-processing steps are often crucial for refining the ensemble predictions, such as smoothing or thresholding, which can further enhance accuracy and reduce noise. The paper likely demonstrates the benefits of ensembling in improving overall segmentation performance, highlighting the value of this approach for robust and reliable clinical applications.
Future Directions#
Future research directions stemming from this work could explore several promising avenues. Expanding the dataset to include a more geographically and demographically diverse patient pool is crucial for enhancing the generalizability of the MedNeXt model. This includes incorporating data from different MRI scanners and acquisition protocols to mitigate biases. Improving the robustness of the model in the face of noisy or low-quality MRI scans is also important. This might involve exploring more sophisticated data augmentation techniques or developing more robust loss functions that are less sensitive to outliers. Furthermore, investigating the incorporation of additional modalities such as PET or diffusion tensor imaging (DTI) into the model could improve its accuracy and provide a more comprehensive understanding of brain tumors. Finally, exploring the use of explainable AI (XAI) techniques would be invaluable for gaining a better understanding of the model’s decision-making process, facilitating better clinical interpretation and trust in automated diagnostic tools. This would allow radiologists to more confidently utilize MedNeXt for tumor segmentation, potentially leading to earlier and more effective intervention.
More visual insights#
More on figures

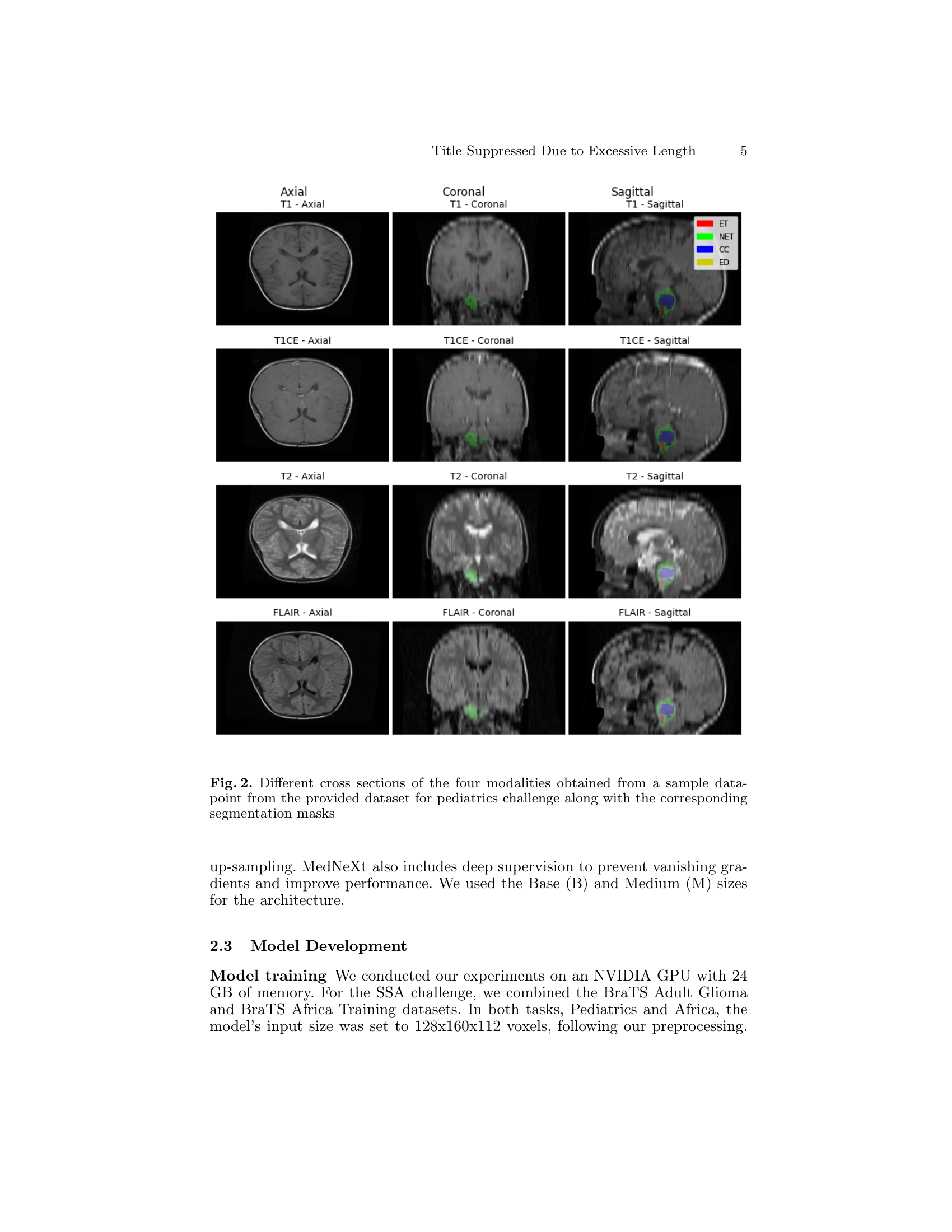
🔼 Figure 2 displays various cross-sections (axial, coronal, and sagittal views) of brain MRI scans from a pediatric patient. Each modality (T1, T1ce, T2, and FLAIR) is shown, illustrating the different tissue contrasts revealed by each imaging sequence. Overlaid on these images are corresponding segmentation masks which delineate different tumor regions of interest (ET: enhancing tumor, NETC: non-enhancing tumor core, CC: cystic component, ED: peritumoral edema). This figure visually demonstrates the complexity of pediatric brain tumor segmentation and the necessity of multi-modal image analysis for accurate results.
read the caption
Figure 2: Different cross sections of the four modalities obtained from a sample data-point from the provided dataset for pediatrics challenge along with the corresponding segmentation masks

🔼 The figure shows the architecture of the MedNeXt model, which is a U-Net-like network that uses MedNeXt blocks. These blocks combine the strengths of convolutional neural networks and transformers. The encoder downsamples the input, the bottleneck processes the features, and the decoder upsamples the output to the original resolution. Deep supervision is used to improve training stability and accuracy. Residual connections add the outputs of each layer to the input of the subsequent layer to prevent vanishing gradients.
read the caption
Figure 3: (a) Architectural design of the MedNeXt. The network has 4 Encoder and Decoder layers each, with a bottleneck layer. MedNeXt blocks are present in Up and Downsampling layers as well. Deep Supervision is used at each decoder layer, with lower loss weights at lower resolutions. All residuals are additive while convolutions are padded to retain tensor sizes. For further details, we refer to [18]
Full paper#