↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Medical image segmentation struggles with the heterogeneity of MRI modalities and scarcity of annotated data, hindering the development of robust models for under-annotated modalities. Previous methods like generative models and image translation have limitations in handling this heterogeneity and require paired or annotated data, restricting their applicability. Existing approaches either focus on augmenting existing data or translating images between specific modalities, both of which have limitations in terms of scalability and generalizability.
To overcome these challenges, this paper introduces MRGen, a diffusion-based controllable data engine that synthesizes training data for unannotated modalities. MRGen uses a two-stage approach, first pretraining on image-text data and then fine-tuning on mask-annotated data. This allows for controllable generation, guided by text prompts and masks. The extensive experiments across various MRI modalities demonstrate that MRGen can effectively synthesize training data, significantly enhancing the performance of segmentation models on under-annotated modalities.
Key Takeaways#
Why does it matter?#
This paper is important because it addresses the critical challenge of limited annotated data in medical image segmentation, a major bottleneck in the field. By introducing a novel diffusion-based data engine (MRGen), the research offers a practical solution for generating synthetic training data for diverse MRI modalities without requiring paired data. This significantly expands the potential for training accurate and robust segmentation models, improving healthcare applications.
Visual Insights#

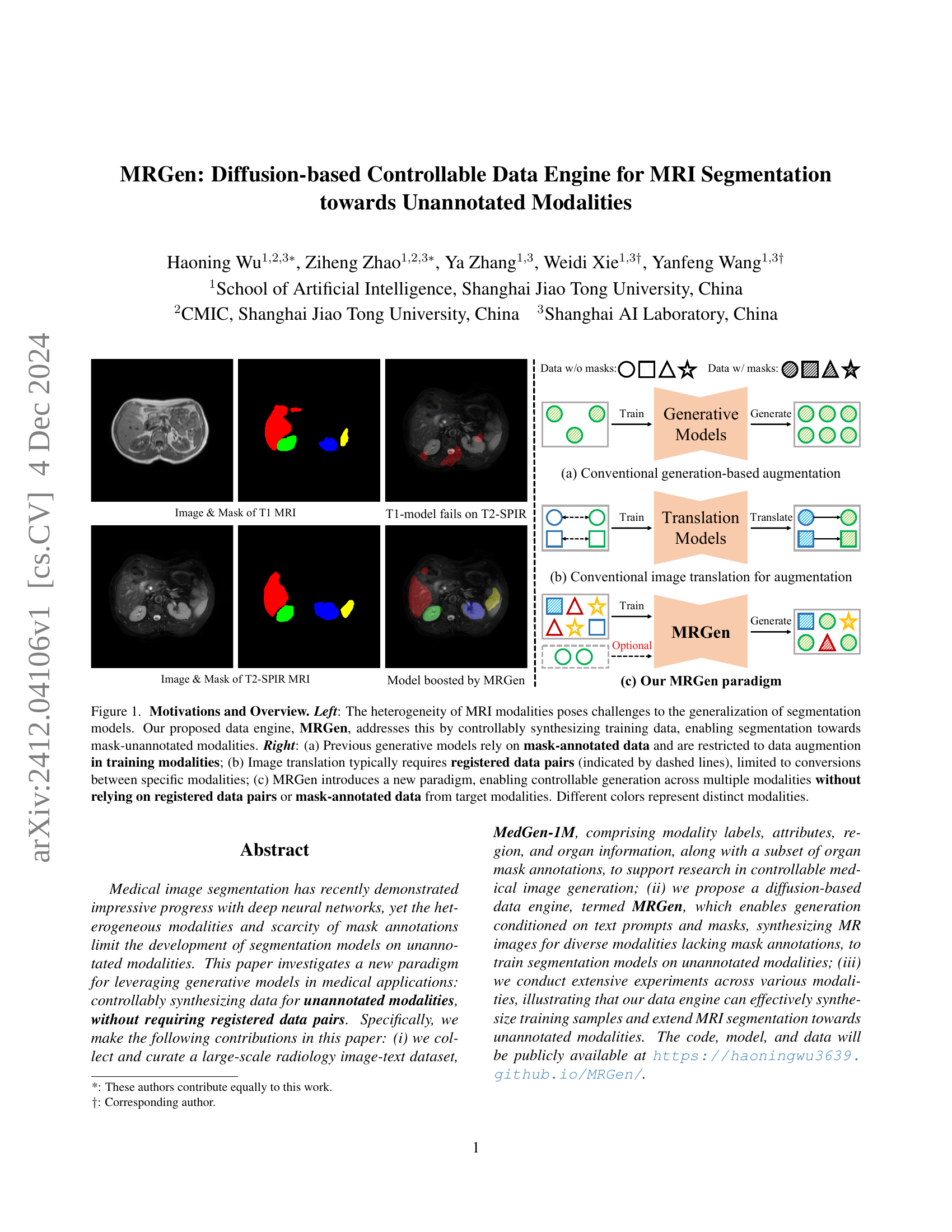
🔼 Figure 1 illustrates the core idea and motivation behind the MRGen model. The left panel shows the challenge of MRI modality heterogeneity for generalizable segmentation models. The right panel compares MRGen to prior approaches. (a) shows conventional generation-based augmentation methods limited to training data with masks. (b) shows conventional image translation limited to registered data pairs. (c) shows MRGen, which generates data across modalities without requiring registered data or mask annotations. Different colors represent different MRI modalities.
read the caption
Figure 1: Motivations and Overview. Left: The heterogeneity of MRI modalities poses challenges to the generalization of segmentation models. Our proposed data engine, MRGen, addresses this by controllably synthesizing training data, enabling segmentation towards mask-unannotated modalities. Right: (a) Previous generative models rely on mask-annotated data and are restricted to data augmention in training modalities; (b) Image translation typically requires registered data pairs (indicated by dashed lines), limited to conversions between specific modalities; (c) MRGen introduces a new paradigm, enabling controllable generation across multiple modalities without relying on registered data pairs or mask-annotated data from target modalities. Different colors represent distinct modalities.
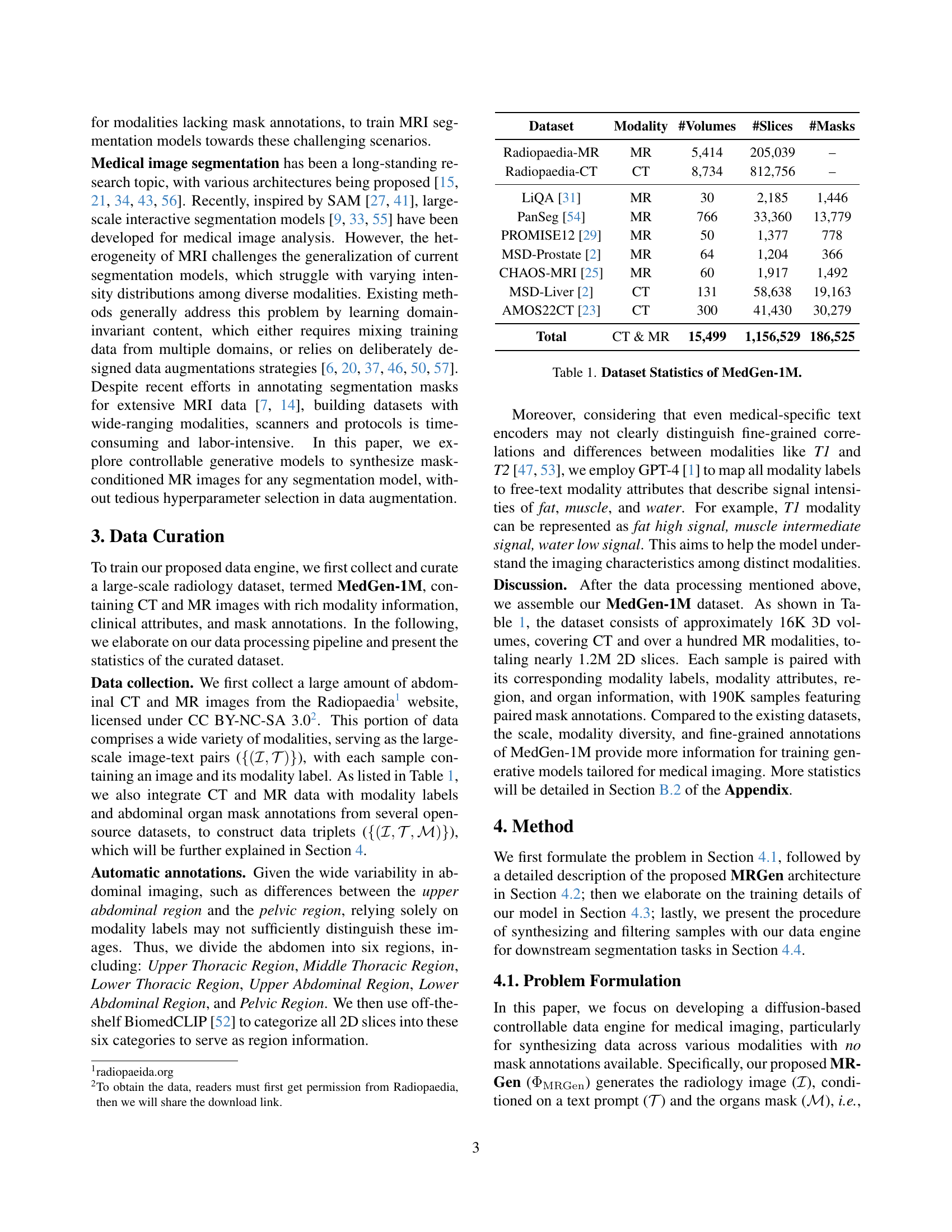
| Dataset | Modality | #Volumes | #Slices | #Masks |
|---|---|---|---|---|
| Radiopaedia-MR | MR | 5,414 | 205,039 | – |
| Radiopaedia-CT | CT | 8,734 | 812,756 | – |
| LiQA [31] | MR | 30 | 2,185 | 1,446 |
| PanSeg [54] | MR | 766 | 33,360 | 13,779 |
| PROMISE12 [29] | MR | 50 | 1,377 | 778 |
| MSD-Prostate [2] | MR | 64 | 1,204 | 366 |
| CHAOS-MRI [25] | MR | 60 | 1,917 | 1,492 |
| MSD-Liver [2] | CT | 131 | 58,638 | 19,163 |
| AMOS22CT [23] | CT | 300 | 41,430 | 30,279 |
| Total | CT & MR | 15,499 | 1,156,529 | 186,525 |
🔼 This table presents a summary of the MedGen-1M dataset, a large-scale radiology image-text dataset used in the paper. It details the number of volumes, slices, and masks for both CT and MR images across several different datasets. The inclusion of mask counts highlights the availability of annotations for training controlled data generation.
read the caption
Table 1: Dataset Statistics of MedGen-1M.
In-depth insights#
MRI Modality Gap#
The MRI modality gap highlights a critical challenge in medical image analysis: the significant variability in image appearance across different MRI sequences. This variability stems from differing pulse sequences, acquisition parameters, and even scanner hardware, leading to substantial differences in image contrast, resolution, and tissue characteristics. This heterogeneity makes it difficult to train robust deep learning models that can generalize well to unseen modalities. A model trained on one MRI sequence (e.g., T1-weighted) may perform poorly on another (e.g., T2-weighted) because it hasn’t learned to disentangle the modality-specific artifacts from the underlying anatomical information. Addressing this gap requires developing techniques that can either make the data more homogenous across modalities (e.g., through data augmentation or domain adaptation), or train models that are inherently robust to these modality variations. This might involve using techniques like multi-modal training, adversarial learning, or specialized architectures that explicitly model modality differences.
MRGen Engine#
The MRGen engine, as described in the research paper, is a diffusion-based controllable data engine designed to address the challenges of medical image segmentation in MRI. Its core innovation lies in the controllable synthesis of training data for modalities lacking mask annotations. This is particularly significant for MRI, where data heterogeneity and annotation costs are major obstacles. The engine leverages a two-stage training process. First, a text-guided generative model is pre-trained on a large-scale, multi-modal image-text dataset. Second, a mask-conditioned finetuning step enables controllable generation guided by both text and mask inputs. This allows the engine to synthesize training samples for downstream tasks and importantly, extend segmentation capabilities to under-annotated modalities. The efficacy of this approach is demonstrated through experiments showcasing improved segmentation performance on unseen, mask-unannotated datasets when compared to baselines using traditional augmentation or image translation methods. The MedGen-1M dataset is a significant component, providing the foundation for training and evaluation, showing a large-scale, rich radiology data resource with diverse modalities and annotations that further proves its effectiveness. Overall, the MRGen engine presents a novel paradigm in medical image generation, facilitating training for segmentation models across various MRI modalities, and potentially other medical imaging domains, where data scarcity remains a critical hurdle.
Diffusion Synthesis#
Diffusion models offer a powerful approach to data synthesis, particularly in scenarios with limited labeled data. By iteratively adding noise to an image and then learning to reverse this process, they can generate new samples that resemble the original training data. This is particularly relevant for medical imaging, where obtaining large, annotated datasets is often expensive and time-consuming. In the context of MRI segmentation, diffusion synthesis offers a way to augment existing datasets with synthetic images, increasing the diversity and size of the training set and potentially improving the generalization capabilities of segmentation models. A key advantage of this technique is the ability to control the generation process by conditioning on various factors, such as text descriptions or segmentation masks, allowing for targeted synthesis and improved control over the generated images. However, challenges remain, such as dealing with the high dimensionality of medical images, ensuring the fidelity and realism of generated samples, and avoiding overfitting to the training data. Further research in developing more efficient and robust diffusion models, and in exploring novel conditioning strategies, is crucial to unlock the full potential of this powerful technique for medical image analysis.
Segmentation Gains#
A hypothetical section titled “Segmentation Gains” in a medical image segmentation research paper would likely detail the improvements achieved by the proposed method compared to existing techniques. This would involve a quantitative analysis showing increased Dice Similarity Coefficients (DSC) or Intersection over Union (IoU) scores across different anatomical structures and imaging modalities. The analysis might be broken down by modality (e.g., T1-weighted MRI, T2-weighted MRI, CT) or organ, highlighting where the greatest gains were observed. A discussion of these results would be crucial, explaining the reasons for improvement. This might involve factors like the effectiveness of the data augmentation strategy, the model’s ability to generalize to unseen data, or the impact of using a novel architecture or training paradigm. The analysis might also delve into the impact of the proposed method on clinically relevant metrics, such as reducing false positives or improving the detection of subtle lesions, which would have greater clinical significance.
Future Scope#
The future scope of research in this area is promising. Improving the handling of extremely small organ masks is crucial for generating higher-quality images, perhaps through data augmentation techniques or architectural modifications to the model itself. Addressing false-negative generation, where organs are produced that weren’t in the input mask, is also vital. This might involve refining the model’s training process or implementing more sophisticated data filtering. Furthermore, exploring more diverse modalities and datasets will improve generalization. Expanding the dataset to include more varied CT data and collecting more MRI data will enrich the training data and potentially improve the model’s handling of varied organ morphologies. Developing a more robust and comprehensive data filtering pipeline is necessary to ensure that high-quality samples meet downstream task requirements. Finally, investigating advanced architectural modifications could improve generation efficiency and accuracy, potentially utilizing more efficient deep learning architectures.
More visual insights#
More on figures

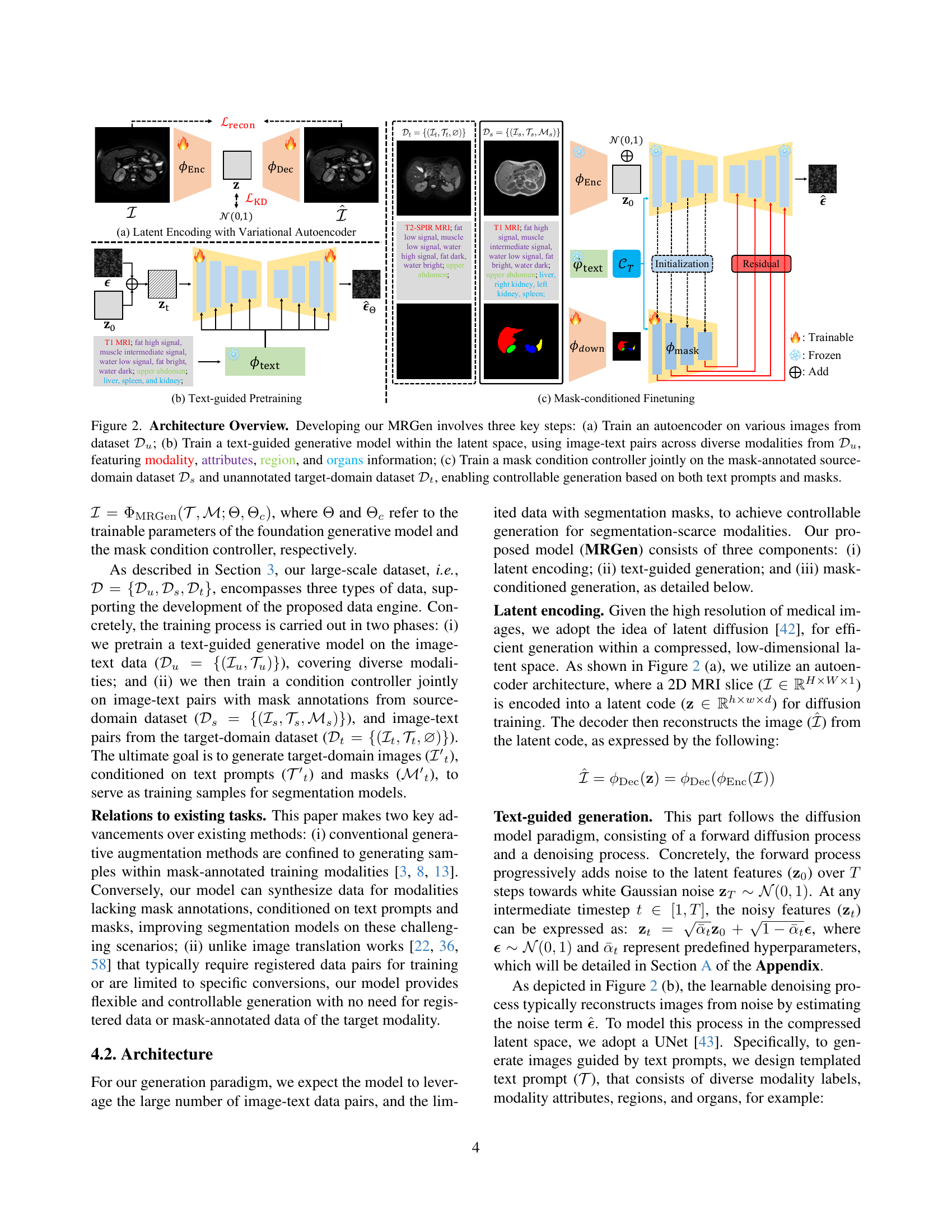
🔼 This figure illustrates the architecture of MRGen, a diffusion-based controllable data engine. It details the three key steps involved in developing MRGen: 1) training an autoencoder on a variety of images from dataset 𝒟u to learn a compressed latent representation; 2) training a text-guided generative model in the latent space using image-text pairs from 𝒟u, incorporating modality, attributes, region, and organ information; 3) jointly training a mask condition controller on mask-annotated source-domain data 𝒟s and unannotated target-domain data 𝒟t to enable controllable generation based on text prompts and masks. This architecture allows MRGen to generate high-quality medical images for modalities lacking annotations.
read the caption
Figure 2: Architecture Overview. Developing our MRGen involves three key steps: (a) Train an autoencoder on various images from dataset 𝒟usubscript𝒟𝑢\mathcal{D}_{u}caligraphic_D start_POSTSUBSCRIPT italic_u end_POSTSUBSCRIPT; (b) Train a text-guided generative model within the latent space, using image-text pairs across diverse modalities from 𝒟usubscript𝒟𝑢\mathcal{D}_{u}caligraphic_D start_POSTSUBSCRIPT italic_u end_POSTSUBSCRIPT, featuring modality, attributes, region, and organs information; (c) Train a mask condition controller jointly on the mask-annotated source-domain dataset 𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT and unannotated target-domain dataset 𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT, enabling controllable generation based on both text prompts and masks.

🔼 The figure illustrates the process of creating synthetic medical images using the MRGen model. First, text prompts describing the desired image content and segmentation masks are input into the MRGen model. MRGen then generates radiology images based on these conditions. Finally, a pre-trained SAM2 model automatically filters these generated images, selecting only high-quality images that accurately reflect the input text and masks. This ensures that the synthetic images used for training segmentation models meet quality standards.
read the caption
Figure 3: Synthetic Data Construction Pipeline. MRGen takes text prompt and mask as conditions for controllably generating radiology images and employs a pretrained SAM2 model for automatic filtering to guarantee the quality of generated samples.

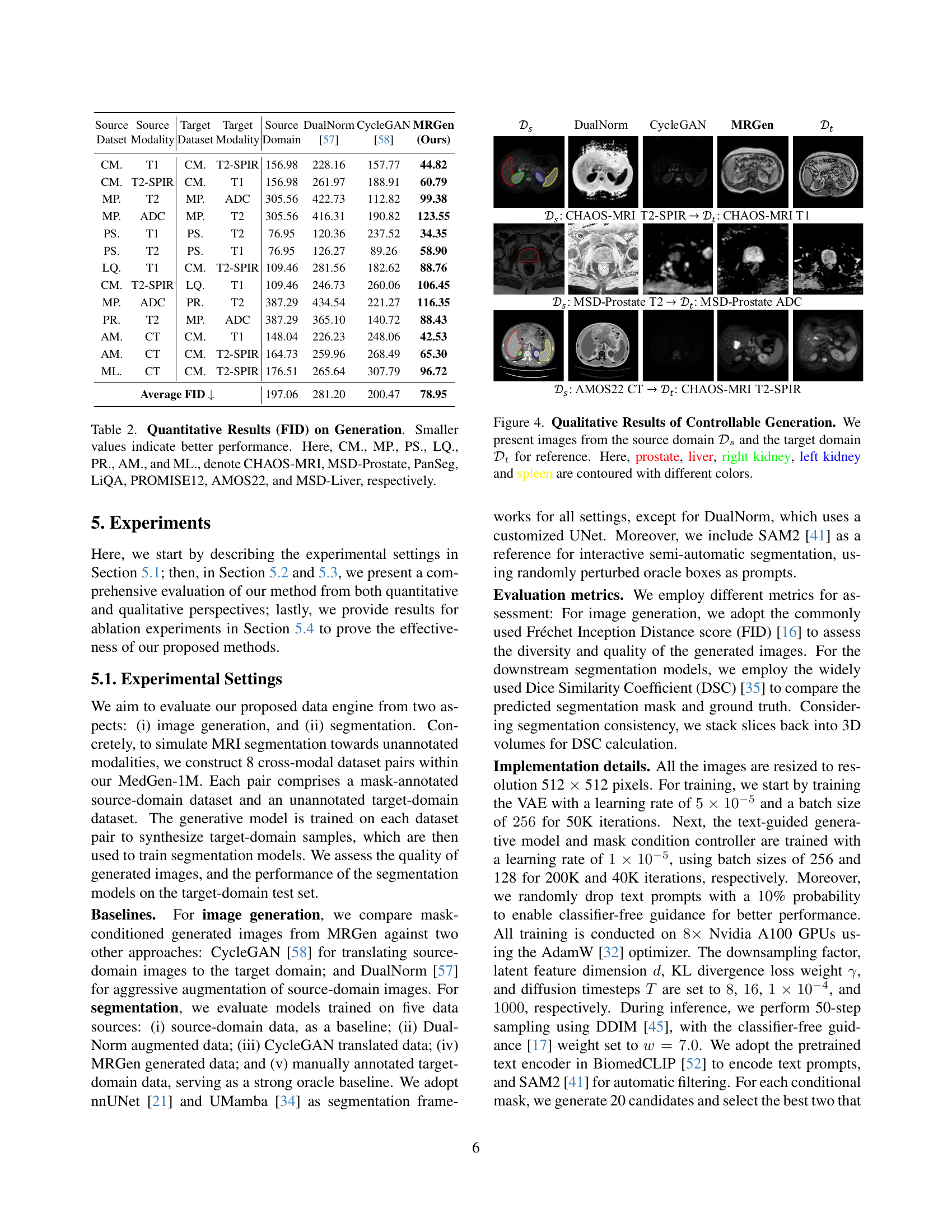
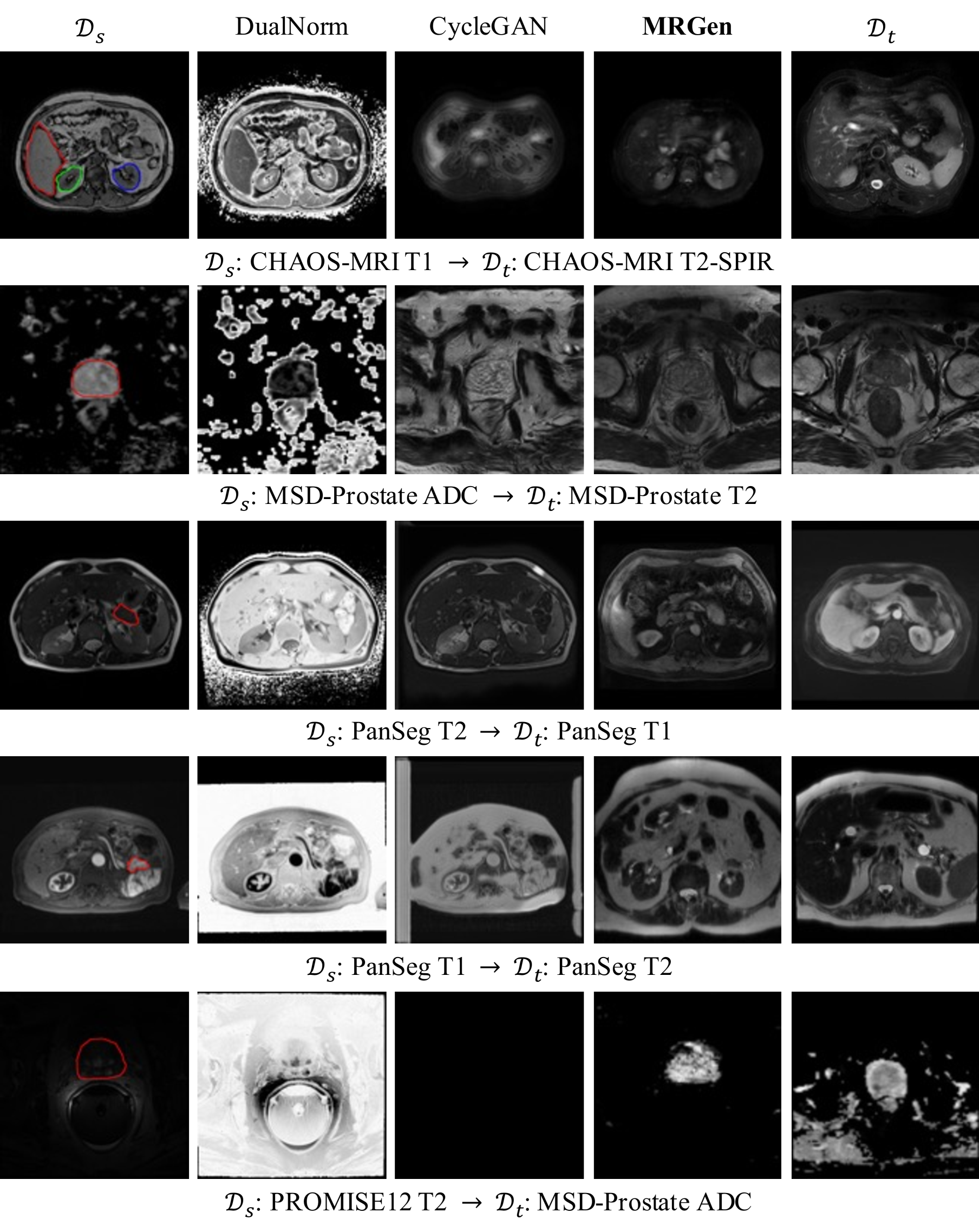
🔼 Figure 4 showcases the effectiveness of the MRGen model in generating high-quality medical images for various modalities. It presents pairs of images; one from a source domain (with existing mask annotations) and the other from a target domain (lacking annotations). MRGen successfully synthesizes images in the target domain (unannotated MRI modalities) that are conditioned upon text descriptions and organ masks. The figure highlights several key organs (prostate, liver, right kidney, left kidney, and spleen) in different colors for easy identification and comparison between the source and target images.
read the caption
Figure 4: Qualitative Results of Controllable Generation. We present images from the source domain 𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT and the target domain 𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT for reference. Here, prostate, liver, right kidney, left kidney and spleen are contoured with different colors.

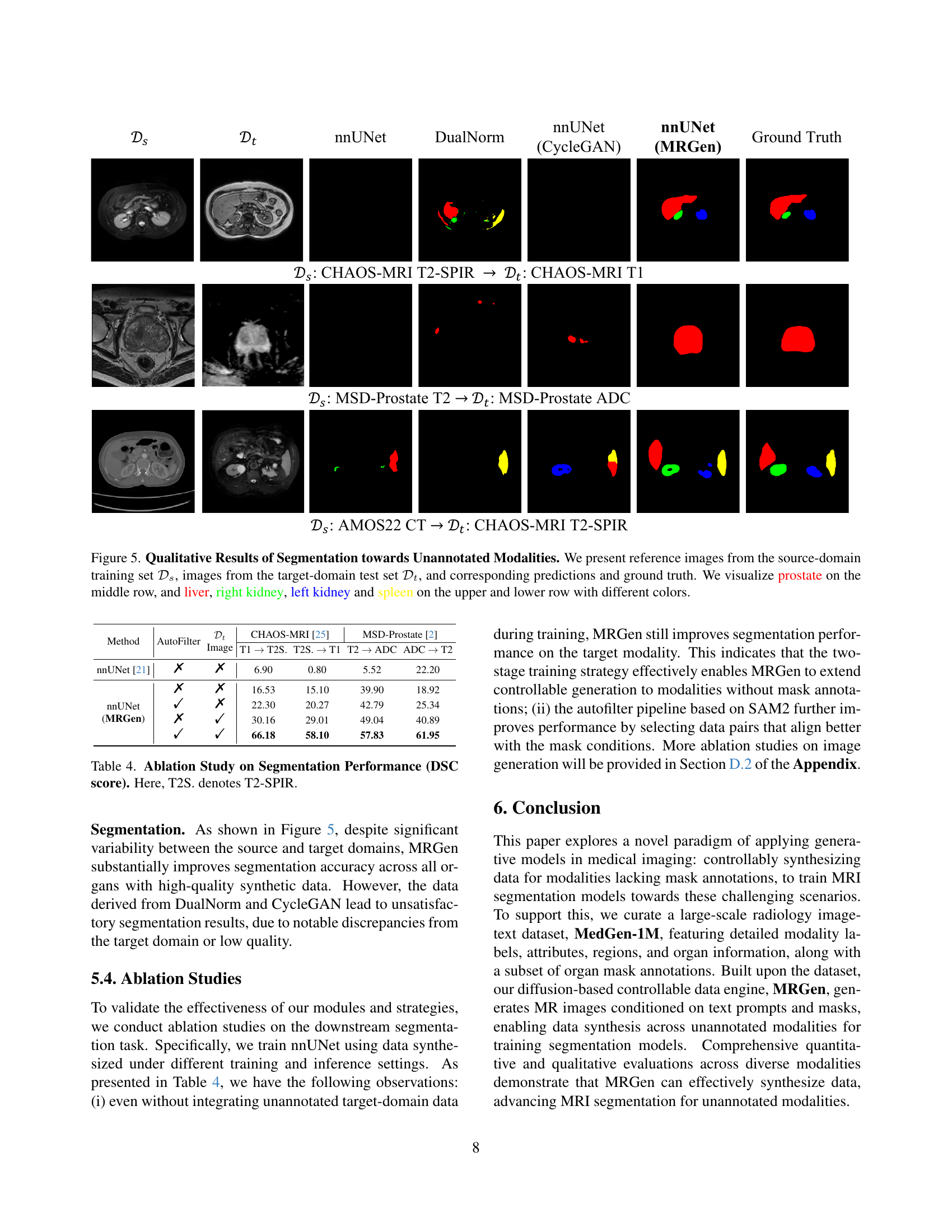
🔼 This figure shows a qualitative comparison of the segmentation results on unseen MRI modalities. The top row displays the source domain training images (from dataset Ds), the middle row contains the target domain test images (from dataset Dt) and the bottom row shows the same target domain images. Each column represents a different segmentation method: nnU-Net, DualNorm, CycleGAN, MRGen and Ground Truth. Different organs (prostate, liver, right kidney, left kidney, and spleen) are color-coded in each image for easy comparison. The results demonstrate the performance of each segmentation method when trained using synthetic data generated by MRGen, compared to other methods and the ground truth.
read the caption
Figure 5: Qualitative Results of Segmentation towards Unannotated Modalities. We present reference images from the source-domain training set 𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT, images from the target-domain test set 𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT, and corresponding predictions and ground truth. We visualize prostate on the middle row, and liver, right kidney, left kidney and spleen on the upper and lower row with different colors.
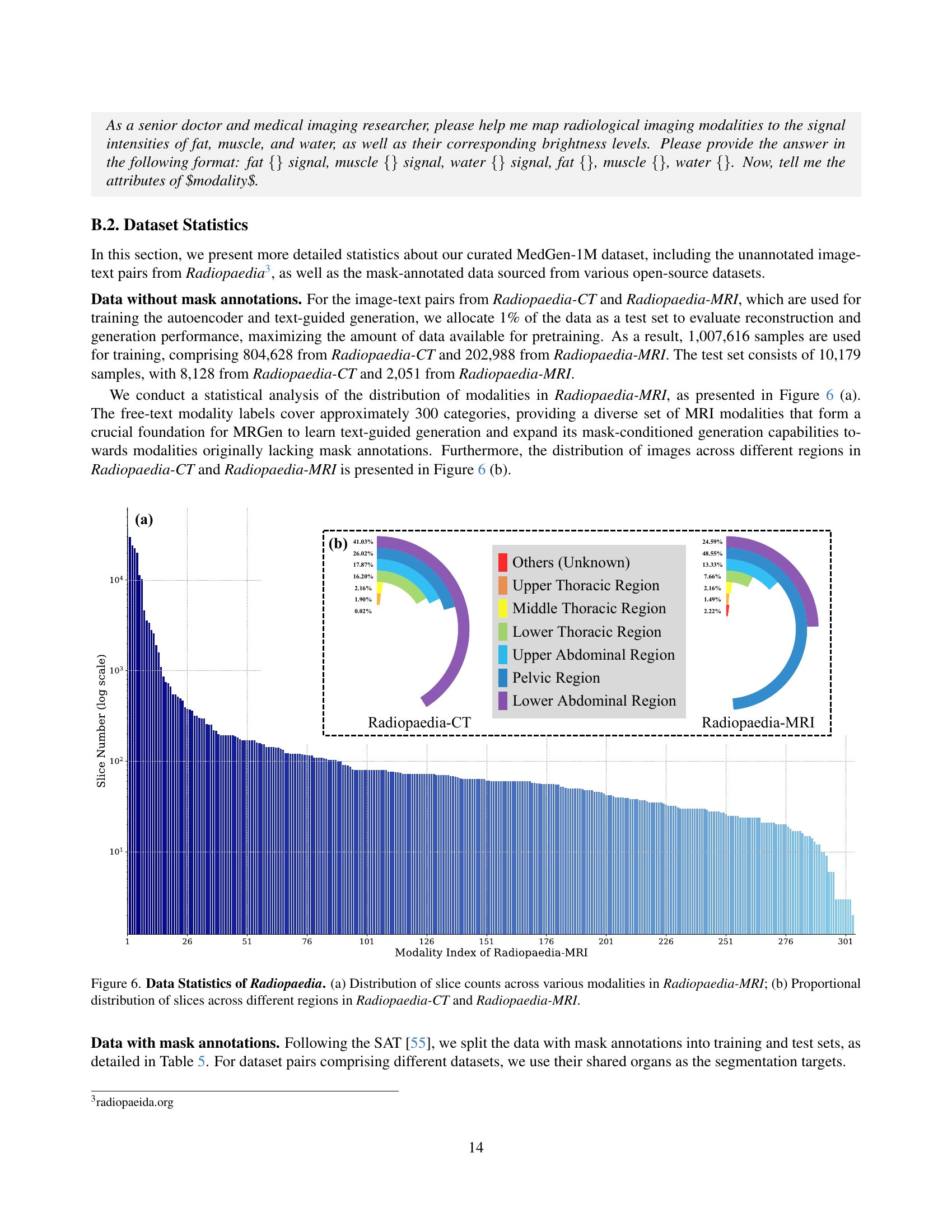
🔼 This figure presents a statistical overview of the Radiopaedia dataset used in the study. Subfigure (a) shows the distribution of the number of slices across different MRI modalities within the Radiopaedia-MRI subset. This illustrates the variety of MRI scans present in the dataset. Subfigure (b) displays the proportional distribution of slices across six anatomical regions (Upper Thoracic, Middle Thoracic, Lower Thoracic, Upper Abdominal, Lower Abdominal, and Pelvic) in both the Radiopaedia-CT and Radiopaedia-MRI subsets. This visualization helps to understand the regional coverage and balance within the dataset.
read the caption
Figure 6: Data Statistics of Radiopaedia. (a) Distribution of slice counts across various modalities in Radiopaedia-MRI; (b) Proportional distribution of slices across different regions in Radiopaedia-CT and Radiopaedia-MRI.

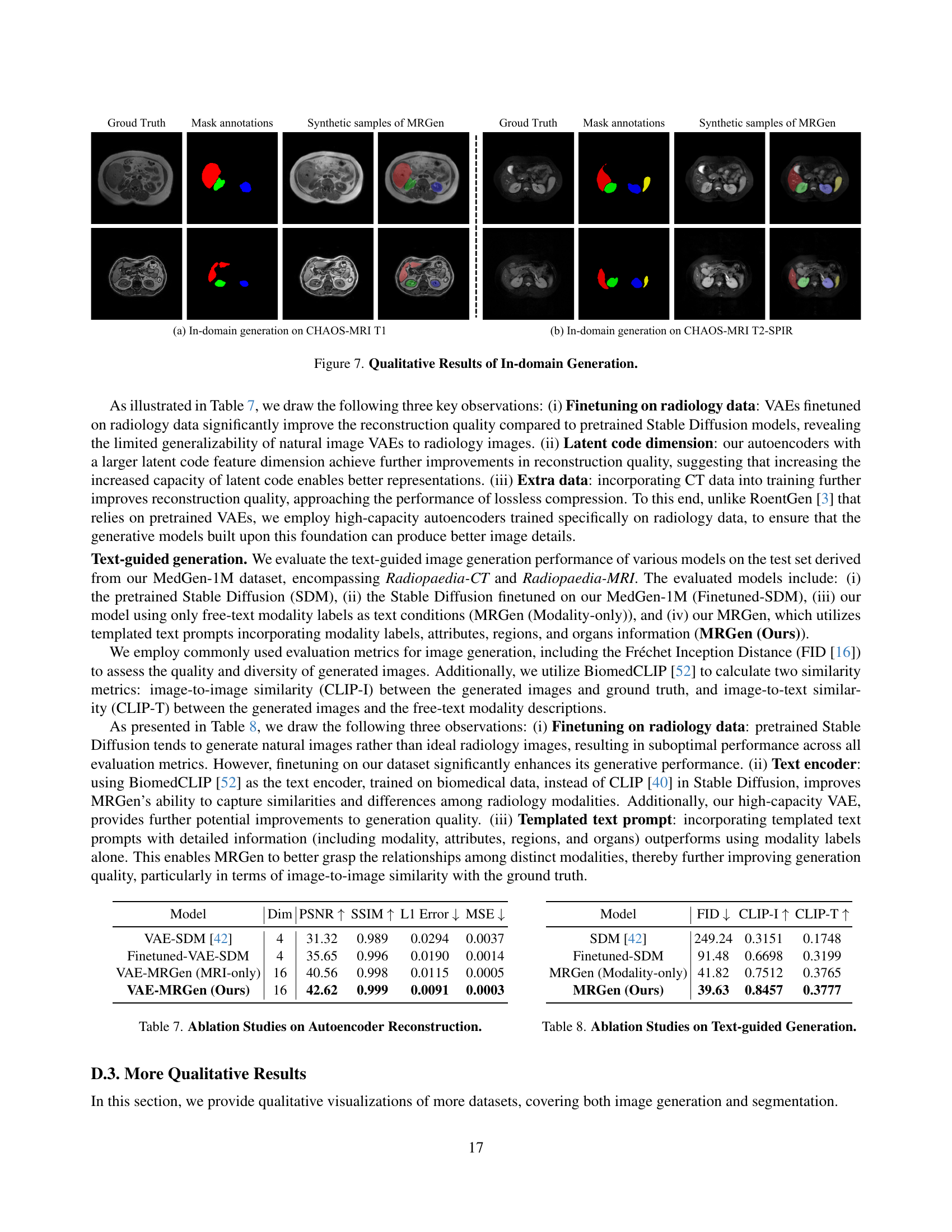
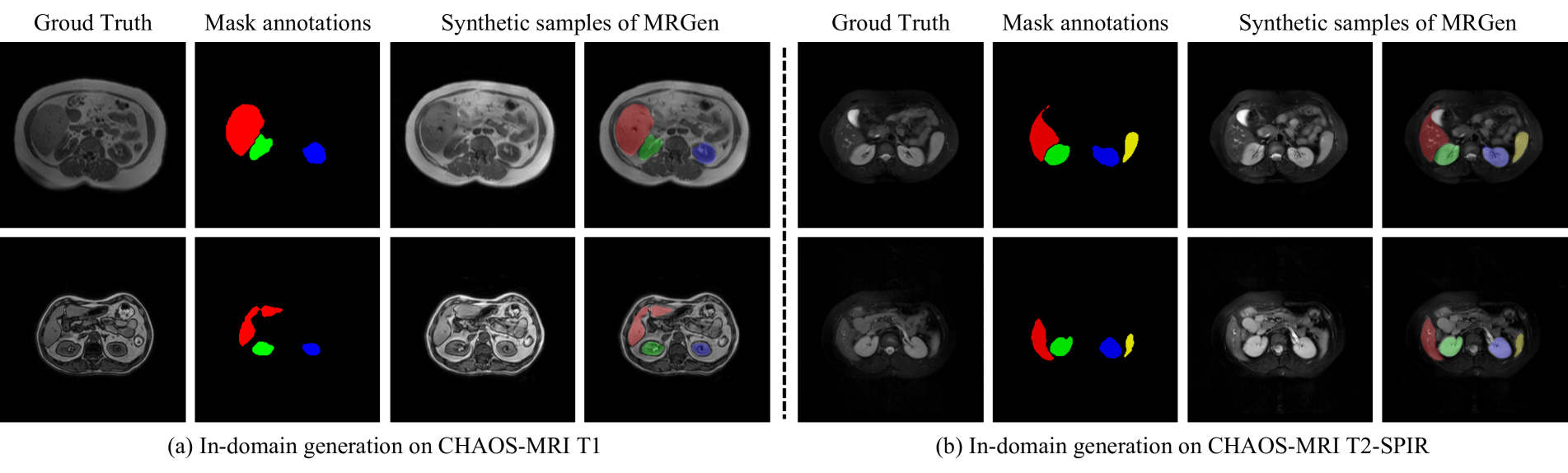
🔼 This figure visualizes the results of in-domain image generation, comparing the ground truth images to those generated by MRGen and other baselines (DualNorm and CycleGAN). The top row shows results for T1-weighted MRI images from the CHAOS-MRI dataset, while the bottom row presents results for T2-SPIR images from the same dataset. It demonstrates MRGen’s capability to generate high-quality images within the same modality as the training data, showcasing its superior performance compared to baselines.
read the caption
Figure 7: Qualitative Results of In-domain Generation.
🔼 This table presents results from ablation studies conducted on the autoencoder part of the MRGen model. The goal was to assess different configurations and their impact on reconstruction quality. Key variations studied include the dimension of the latent space and the datasets used for training. Metrics like PSNR, SSIM, L1 error, and MSE were used to evaluate the reconstruction performance.
read the caption
Table 7: Ablation Studies on Autoencoder Reconstruction.

🔼 This table presents the results of ablation studies performed on the text-guided image generation model. It compares the performance of different model variations, evaluating their FID (Fréchet Inception Distance), CLIP-I (image-to-image similarity using CLIP), and CLIP-T (image-to-text similarity using CLIP) scores. The variations tested include a pretrained Stable Diffusion model, a model fine-tuned on the MedGen-1M dataset, a model using only free-text modality labels, and the full MRGen model utilizing templated text prompts with detailed information. The results show the impact of different training data, text encoding strategies, and text prompt detail levels on the overall performance of the text-guided generation.
read the caption
Table 8: Ablation Studies on Text-guided Generation.

🔼 Figure 8 demonstrates limitations of the MRGen model. Subfigure (a) shows that MRGen struggles to generate images with high fidelity when conditioned on extremely small organ masks. Subfigure (b) illustrates that MRGen occasionally produces false negatives, generating organs (e.g. kidneys) not present in the input mask.
read the caption
Figure 8: Failure Cases Analysis. Our proposed MRGen is not without limitations: (a) it may struggle to handle extremely small organ masks; (b) it occasionally produces false-negative samples, such as the unexpected synthesis of kidneys in the given example.

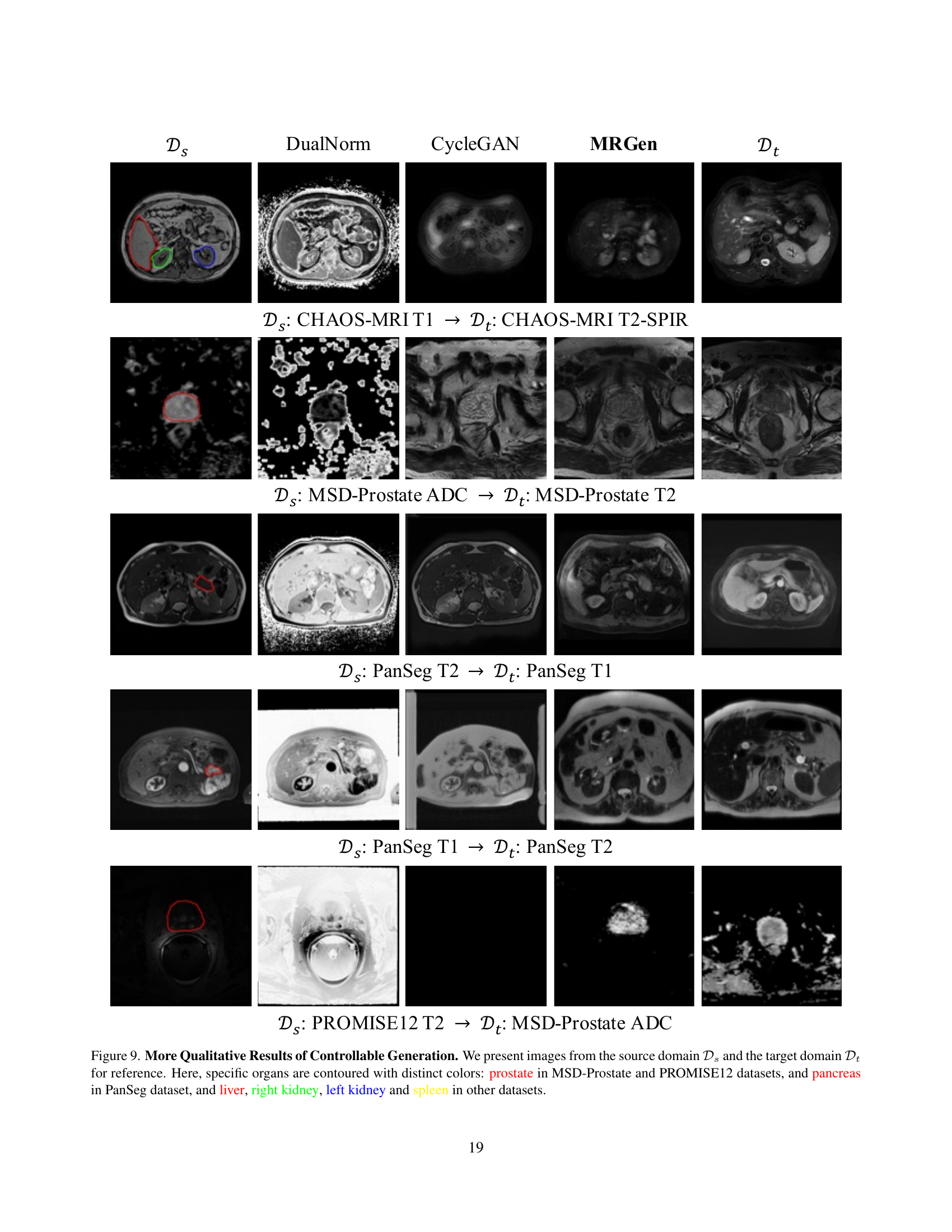
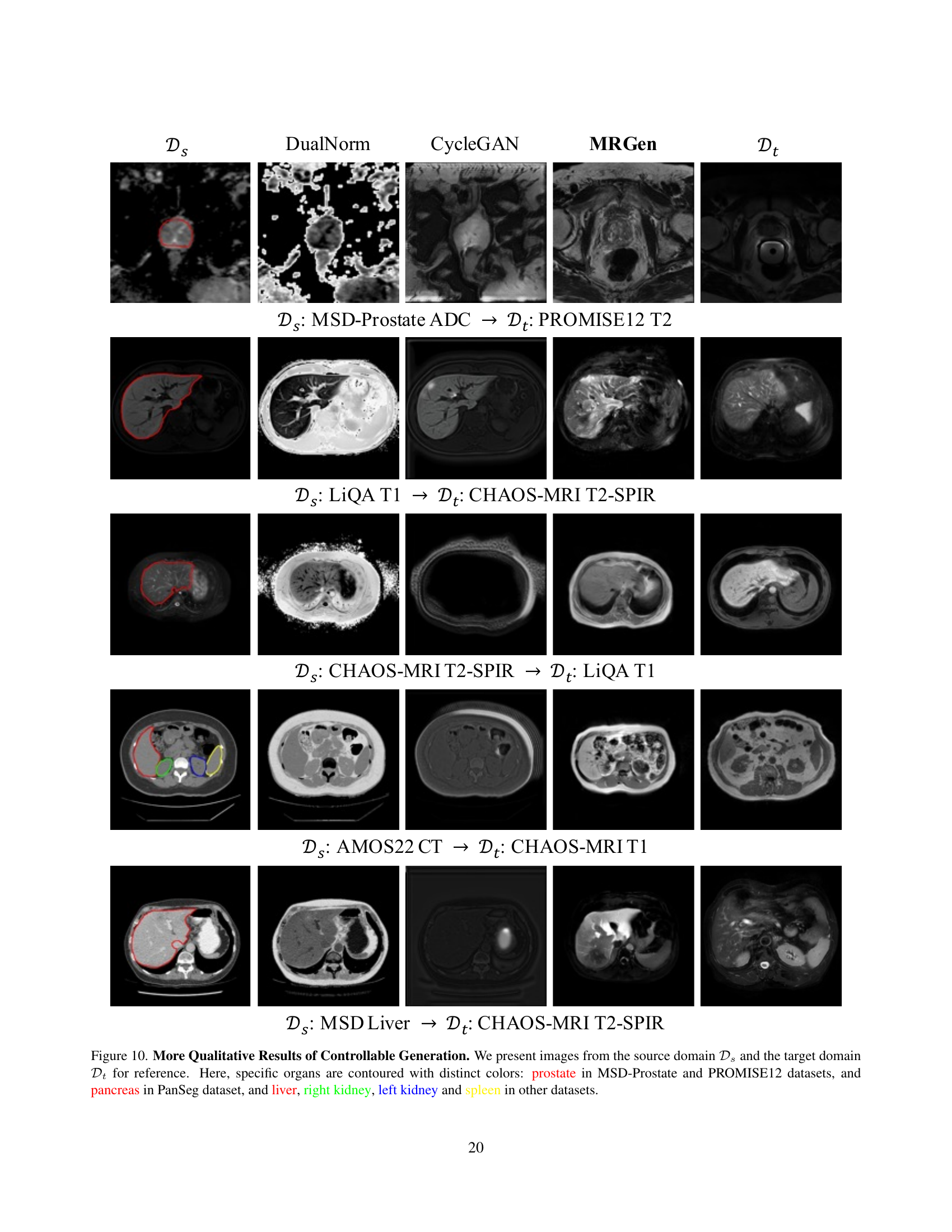
🔼 Figure 9 presents a qualitative comparison of images generated by DualNorm, CycleGAN, and the proposed MRGen model. The figure displays pairs of images, where each pair consists of an image from a source domain (with ground truth annotations) and a corresponding image from a target domain (lacking annotations). The source and target domains are different MRI modalities. Specific organs (prostate, pancreas, liver, right kidney, left kidney, spleen) are highlighted with distinct colors in each image to aid visual comparison and analysis. The purpose is to visually demonstrate the model’s ability to generate realistic and accurate images of various organs across different MRI modalities.
read the caption
Figure 9: More Qualitative Results of Controllable Generation. We present images from the source domain 𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT and the target domain 𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT for reference. Here, specific organs are contoured with distinct colors: prostate in MSD-Prostate and PROMISE12 datasets, and pancreas in PanSeg dataset, and liver, right kidney, left kidney and spleen in other datasets.
More on tables
| Source Dataset | Source Modality | Target Dataset | Target Modality | Source Domain | DualNorm [57] | CycleGAN [58] | MRGen (Ours) |
|---|---|---|---|---|---|---|---|
| CM. | T1 | CM. | T2-SPIR | 156.98 | 228.16 | 157.77 | 44.82 |
| CM. | T2-SPIR | CM. | T1 | 156.98 | 261.97 | 188.91 | 60.79 |
| MP. | T2 | MP. | ADC | 305.56 | 422.73 | 112.82 | 99.38 |
| MP. | ADC | MP. | T2 | 305.56 | 416.31 | 190.82 | 123.55 |
| PS. | T1 | PS. | T2 | 76.95 | 120.36 | 237.52 | 34.35 |
| PS. | T2 | PS. | T1 | 76.95 | 126.27 | 89.26 | 58.90 |
| LQ. | T1 | CM. | T2-SPIR | 109.46 | 281.56 | 182.62 | 88.76 |
| CM. | T2-SPIR | LQ. | T1 | 109.46 | 246.73 | 260.06 | 106.45 |
| MP. | ADC | PR. | T2 | 387.29 | 434.54 | 221.27 | 116.35 |
| PR. | T2 | MP. | ADC | 387.29 | 365.10 | 140.72 | 88.43 |
| AM. | CT | CM. | T1 | 148.04 | 226.23 | 248.06 | 42.53 |
| AM. | CT | CM. | T2-SPIR | 164.73 | 259.96 | 268.49 | 65.30 |
| ML. | CT | CM. | T2-SPIR | 176.51 | 265.64 | 307.79 | 96.72 |
| Average FID ↓ | 197.06 | 281.20 | 200.47 | 78.95 |
🔼 This table presents a quantitative comparison of the image generation quality using different methods. The metric used is the Fréchet Inception Distance (FID), where lower scores indicate better performance. The table compares the FID scores achieved by three methods: MRGen (the proposed method), CycleGAN, and DualNorm. The evaluation is performed across multiple cross-modal scenarios, using different source and target MRI modalities. Acronyms for datasets (CHAOS-MRI, MSD-Prostate, PanSeg, LiQA, PROMISE12, AMOS22, and MSD-Liver) are defined for better readability.
read the caption
Table 2: Quantitative Results (FID) on Generation. Smaller values indicate better performance. Here, CM., MP., PS., LQ., PR., AM., and ML., denote CHAOS-MRI, MSD-Prostate, PanSeg, LiQA, PROMISE12, AMOS22, and MSD-Liver, respectively.
| Source Dataset | Source Modality | Target Dataset | Target Modality | DualNorm [57] | nnUNet [21] | UMamba [34] |
|---|---|---|---|---|---|---|
| 𝒬s | CycleGAN | MRGen | 𝒬s | CycleGAN | MRGen | Oracle Box |
| — | — | — | — | — | — | — |
| Same Datasets, Different Modalities | ||||||
| CHAOS-MRI | T1 | CHAOS-MRI | T2-SPIR | 14.00 | 6.90 | 7.58 |
| CHAOS-MRI | T2-SPIR | CHAOS-MRI | T1 | 12.50 | 0.80 | 1.38 |
| MSD-Prostate | T2 | MSD-Prostate | ADC | 1.43 | 5.52 | 40.92 |
| MSD-Prostate | ADC | MSD-Prostate | T2 | 12.94 | 22.20 | 57.06 |
| PanSeg | T1 | PanSeg | T2 | 0.21 | 0.68 | 2.40 |
| PanSeg | T2 | PanSeg | T1 | 0.11 | 0.30 | 3.59 |
| Different Datasets, Different Modalities | ||||||
| LiQA | T1 | CHAOS-MRI | T2-SPIR | 19.23 | 16.20 | 7.84 |
| CHAOS-MRI | T2-SPIR | LiQA | T1 | 31.09 | 15.80 | 41.02 |
| MSD-Prostate | ADC | PROMISE12 | T2 | 1.43 | 17.19 | 42.13 |
| PROMISE12 | T2 | MSD-Prostate | ADC | 9.84 | 19.20 | 59.95 |
| Different Datasets, Different Modalities, CT - MRI | ||||||
| AMOS22 | CT | CHAOS-MRI | T1 | 19.78 | 0.11 | 6.75 |
| AMOS22 | CT | CHAOS-MRI | T2-SPIR | 16.09 | 8.88 | 52.49 |
| MSD-Liver | CT | CHAOS-MRI | T2-SPIR | 1.58 | 3.12 | 10.14 |
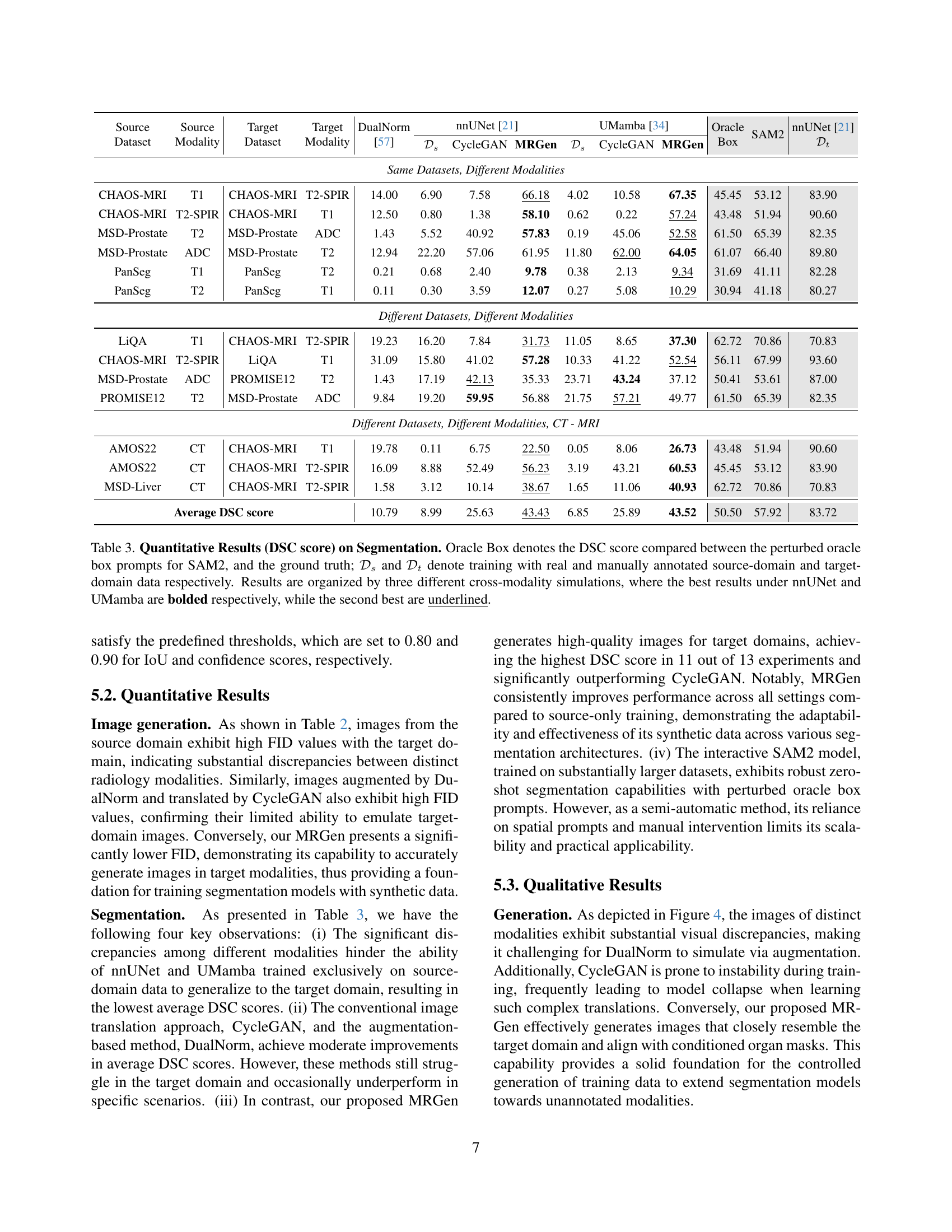
| Average DSC score | 10.79 | 8.99 | 25.63 | 43.43 | 6.85 | 25.89 |
🔼 This table presents the quantitative segmentation results using the Dice Similarity Coefficient (DSC) metric. It compares the performance of different methods: nnUNet and UMamba trained on various data sources. The data sources include: real manually annotated source-domain data (Ds), source-domain data augmented using DualNorm, source-domain data translated to the target domain using CycleGAN, target-domain data generated by the proposed MRGen model, and finally, real manually annotated target-domain data (Dt) acting as an oracle. The table is organized into three sections representing different cross-modality scenarios, highlighting the best and second-best performing models (nnUNet and UMamba) for each scenario. The ‘Oracle Box’ column shows the DSC score of using SAM2 with perturbed oracle box prompts as input.
read the caption
Table 3: Quantitative Results (DSC score) on Segmentation. Oracle Box denotes the DSC score compared between the perturbed oracle box prompts for SAM2, and the ground truth; 𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT and 𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT denote training with real and manually annotated source-domain and target-domain data respectively. Results are organized by three different cross-modality simulations, where the best results under nnUNet and UMamba are bolded respectively, while the second best are underlined.
| Method | AutoFilter | \mathcal{D}_{t} Image | CHAOS-MRI [25] | MSD-Prostate [2] |
|---|---|---|---|---|
| T1 \rightarrow T2S | T2S \rightarrow T1 | T2 \rightarrow ADC | ADC \rightarrow T2 | |
| nnUNet [21] | ✗ | ✗ | 6.90 | 0.80 |
| nnUNet (MRGen) | ✗ | ✗ | 16.53 | 15.10 |
| ✓ | ✗ | 22.30 | 20.27 | |
| ✗ | ✓ | 30.16 | 29.01 | |
| ✓ | ✓ | 66.18 | 58.10 |
🔼 This ablation study analyzes the impact of different components of the MRGen model on the Dice Similarity Coefficient (DSC) scores achieved in MRI segmentation tasks. Specifically, it compares the performance of nnUNet models trained using synthetic data generated by MRGen with various configurations. The configurations include variations in whether or not an automatic filter is used and whether training incorporates both source domain data (with mask annotations) and target domain data (without mask annotations). The results illustrate how these design choices affect the ability of the model to generalize to unseen modalities.
read the caption
Table 4: Ablation Study on Segmentation Performance (DSC score). Here, T2S. denotes T2-SPIR.
| Dataset | Organs | Modality | Train # Vol. | Train # Slc. | Train # Slc. w/ mask | Test # Vol. | Test # Slc. | Test # Slc. w/ mask |
|---|---|---|---|---|---|---|---|---|
| LiQA [31] | Liver | T1-weighted | 24 | 1,718 | 1,148 | 6 | 467 | 298 |
| PanSeg [54] | Pancreas | T1-weighted | 309 | 14,656 | 5,961 | 75 | 3,428 | 1,400 |
| T2-weighted | 305 | 12,294 | 5,106 | 77 | 2,982 | 1,312 | ||
| MSD-Prostate [2] | Prostate | T2-weighted | 26 | 492 | 100 | 6 | 110 | 83 |
| ADC | 26 | 492 | 100 | 6 | 110 | 83 | ||
| PROMISE12 [29] | Prostate | T2-weighed | 40 | 1,137 | 645 | 10 | 110 | 133 |
| CHAOS-MRI [25] | Liver, Right Kidney, | T1-weighted | 32 | 1,018 | 770 | 8 | 276 | 230 |
| Left Kidney, Spleen | T2-SPIR | 16 | 503 | 388 | 4 | 120 | 104 | |
| MSD-Liver [2] | Liver | CT | 105 | 46,695 | 15,260 | 26 | 11943 | 3903 |
| AMOS22CT [23] | Liver, Right Kidney, | CT | 200 | 26,069 | 19,023 | 100 | 15,361 | 11,256 |
| Left Kidney, Spleen, etc. | ||||||||
| Total | / | / | 1,083 | 105,074 | 48,501 | 218 | 34,907 | 18,802 |
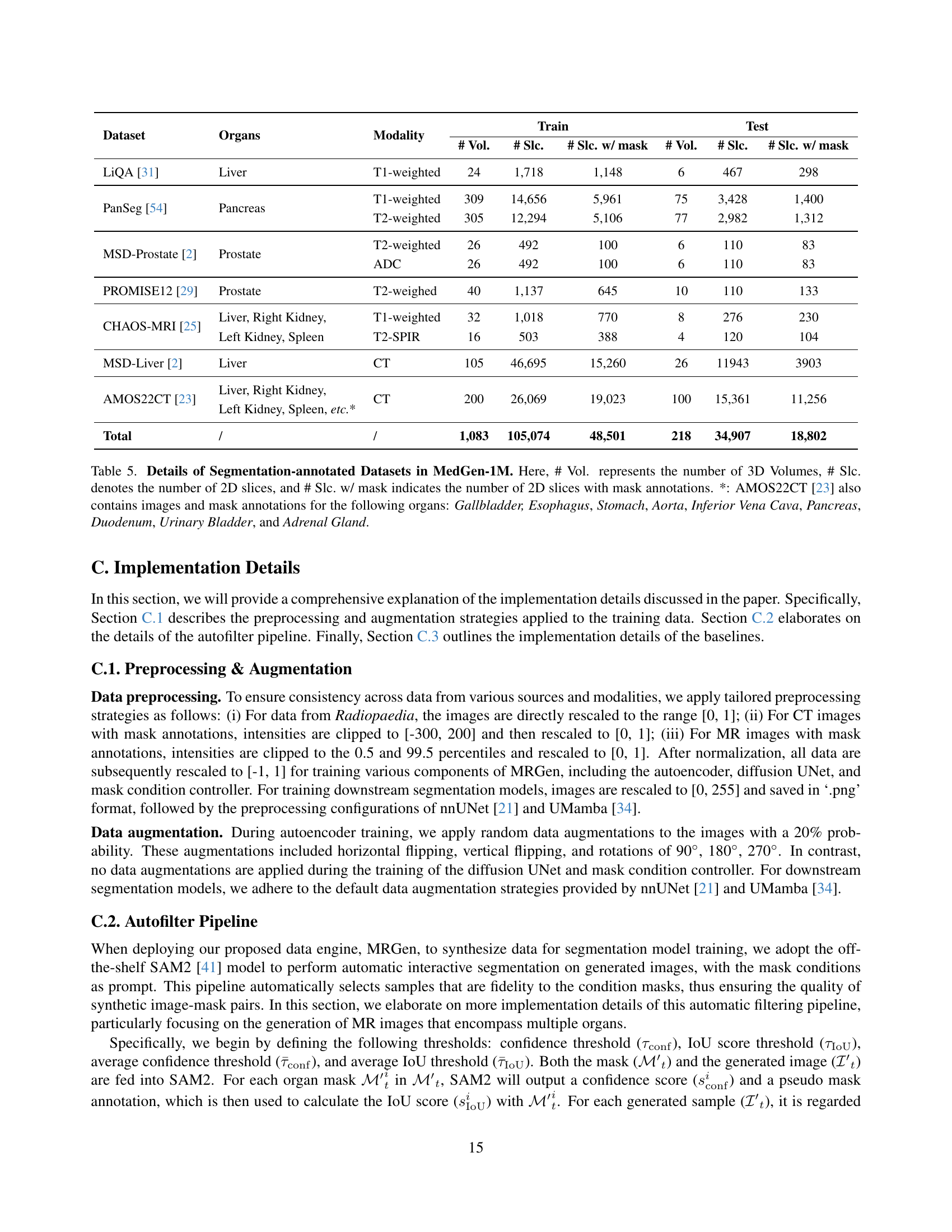
🔼 Table 5 presents details of the segmentation-annotated datasets included in MedGen-1M. It shows the number of 3D volumes, the number of 2D slices, and the number of 2D slices with mask annotations for each dataset. Specifically, it lists the dataset name, the organs annotated, the modality of the images, and the counts for 3D volumes, 2D slices, and 2D slices with masks, both for the training and testing sets. An additional note clarifies that the AMOS22CT dataset contains annotations for additional organs beyond those listed in the main table.
read the caption
Table 5: Details of Segmentation-annotated Datasets in MedGen-1M. Here, # Vol. represents the number of 3D Volumes, # Slc. denotes the number of 2D slices, and # Slc. w/ mask indicates the number of 2D slices with mask annotations. *: AMOS22CT [23] also contains images and mask annotations for the following organs: Gallbladder, Esophagus, Stomach, Aorta, Inferior Vena Cava, Pancreas, Duodenum, Urinary Bladder, and Adrenal Gland.
| Dataset | Source | Modality | Target | Modality | \mathcal{D}_{s} | \mathcal{D}_{t} |
|---|---|---|---|---|---|---|
| \mathcal{D}_{s} | \mathcal{D}_{t} | |||||
| CHAOS-MRI [25] | T1 | T2-SPIR | 90.60 | 88.14 | 4.02 | 67.35 |
| T2-SPIR | T1 | 83.90 | 82.06 | 0.62 | 57.24 |
🔼 Table 6 presents a quantitative comparison of the segmentation performance achieved using the nnUNet model trained on real versus synthetic data. The synthetic data was generated using the MRGen method. The comparison is made for both the source domain (the domain from which the original data for training was obtained) and the target domain (the domain for which the synthetic data was generated for model evaluation). The results are expressed as Dice Similarity Coefficient (DSC) scores, a common metric for evaluating segmentation accuracy.
read the caption
Table 6: More Quantitative Results (DSC score) on Segmentation. We compare the performance of nnUNet [21] trained on real data versus synthetic data generated by our MRGen in both the source domain (𝒟ssubscript𝒟𝑠\mathcal{D}_{s}caligraphic_D start_POSTSUBSCRIPT italic_s end_POSTSUBSCRIPT) and target domain (𝒟tsubscript𝒟𝑡\mathcal{D}_{t}caligraphic_D start_POSTSUBSCRIPT italic_t end_POSTSUBSCRIPT).
Full paper#