↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Current medical AI tools, while promising, are mostly English-centric, limiting their use in non-English speaking populations. Existing multilingual models often compromise medical text comprehension when handling image data. This highlights a need for models catering to diverse languages and handling both text and image data effectively. The disparity in language availability creates accessibility challenges, particularly in regions where languages like Arabic are prevalent, hindering progress towards truly global healthcare solutions.
This paper introduces BiMediX2, a bilingual (Arabic-English) Large Multimodal Model (LMM) specializing in medical applications. Built on the Llama3.1 architecture, it excels at understanding medical images while retaining strong text-based medical knowledge. It uses a new 1.6M sample bilingual dataset called BiMed-V, and introduces BiMed-MBench, a bilingual benchmark for LMMs. BiMediX2 outperforms current models in both understanding medical images and text-based medical evaluations, setting a new benchmark in bilingual multimodal medical evaluations and offering a more inclusive approach to healthcare AI.
Key Takeaways#
Why does it matter?#
BiMediX2’s bilingual, multimodal approach is crucial for researchers exploring inclusive healthcare solutions. It offers a robust framework for developing similar models in other languages, bridging healthcare access gaps. The extensive BiMed-V dataset provides valuable resources for multimodal medical research. Finally, BiMed-MBench sets a new standard for evaluating bilingual medical LMMs, fostering further advancements in the field.
Visual Insights#
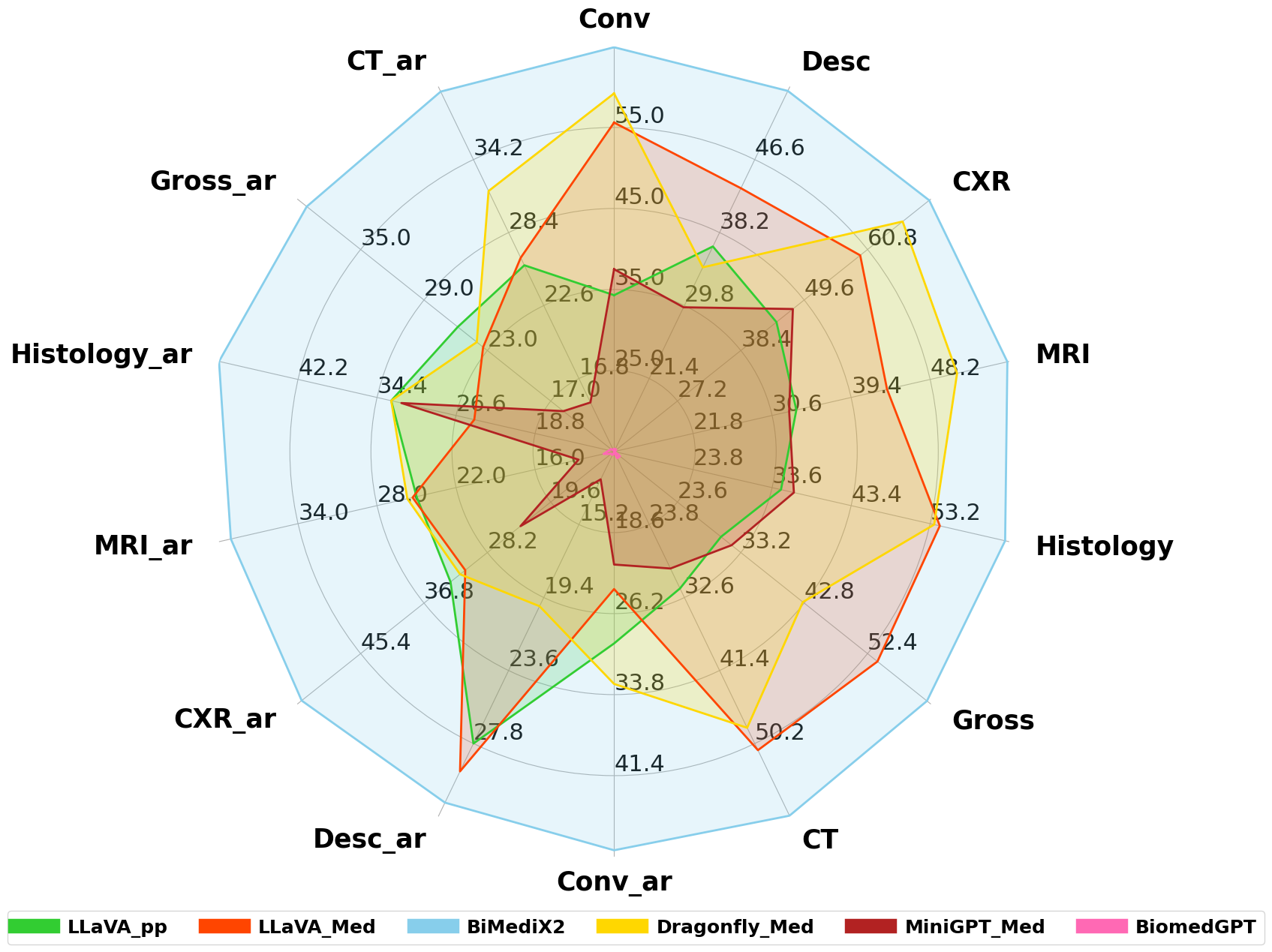
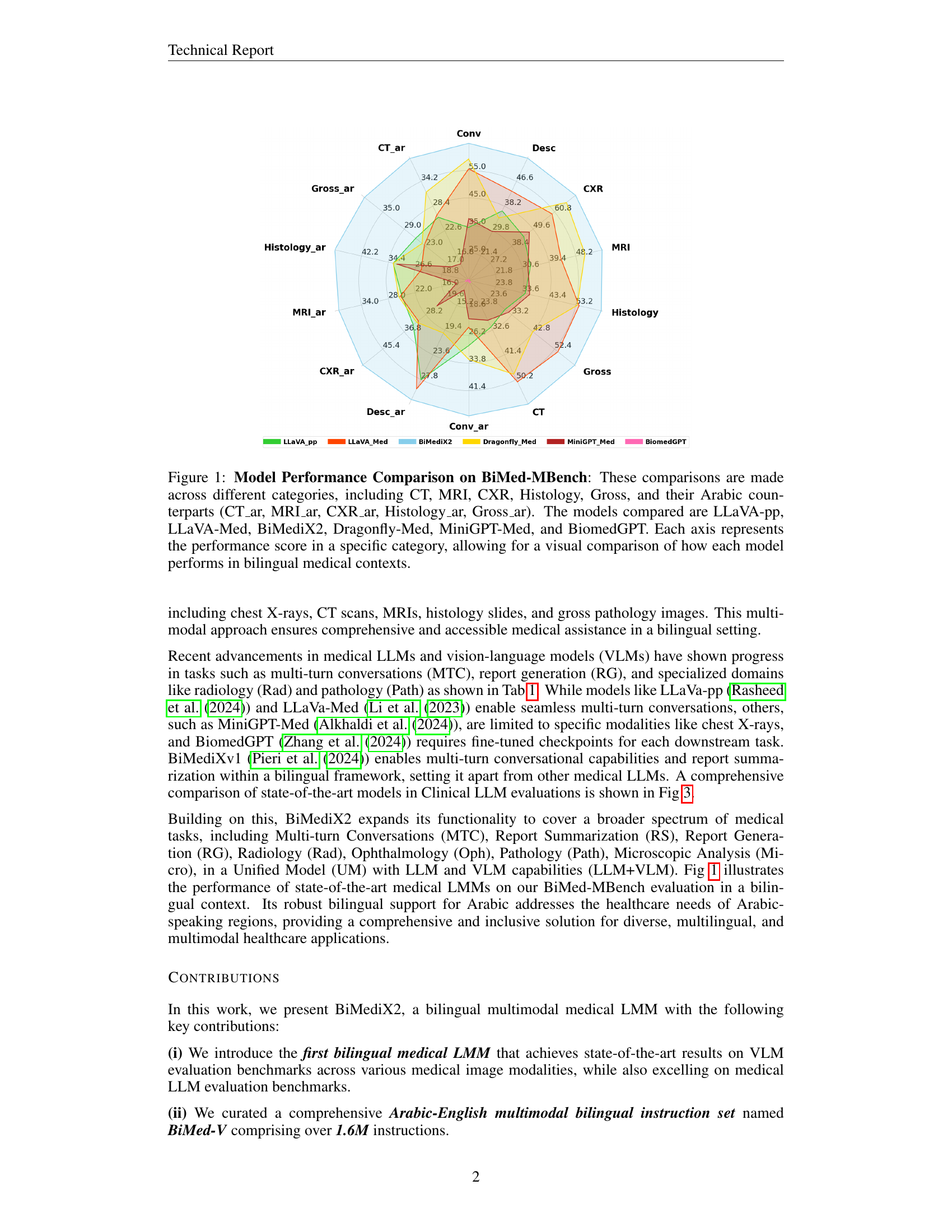
🔼 This radar chart compares the performance of several large multimodal models (LMMs) on the BiMed-MBench, a bilingual (Arabic-English) medical benchmark. The models evaluated include LLaVA-pp, LLaVA-Med, BiMediX2, Dragonfly-Med, MiniGPT-Med, and BiomedGPT. Performance is assessed across different medical image categories: Computed Tomography (CT), Magnetic Resonance Imaging (MRI), Chest X-Ray (CXR), Histology, and Gross pathology, in both English and Arabic. Each axis of the chart represents a specific category, and the model’s score on that category determines its distance from the center. This visualization allows for easy comparison of model performance across different modalities and languages.
read the caption
Figure 1: Model Performance Comparison on BiMed-MBench: These comparisons are made across different categories, including CT, MRI, CXR, Histology, Gross, and their Arabic counterparts (CT_ar, MRI_ar, CXR_ar, Histology_ar, Gross_ar). The models compared are LLaVA-pp, LLaVA-Med, BiMediX2, Dragonfly-Med, MiniGPT-Med, and BiomedGPT. Each axis represents the performance score in a specific category, allowing for a visual comparison of how each model performs in bilingual medical contexts.
| Model | MTC | RS | RG | Rad | Oph | Path | Micro | LLM+VLM | Bil (Ar) |
|---|---|---|---|---|---|---|---|---|---|
| Meditron (Chen et al. (2023)) | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 |
| Med42 (Christophe et al. (2024)) | \usym2713 | \usym2713 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 |
| OpenBioLLM (Ankit Pal (2024)) | \usym2713 | \usym2713 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 |
| Llama3.1 (Meta (2024)) | \usym2713 | \usym2713 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 |
| BiMediXv1 (Pieri et al. (2024)) | \usym2713 | \usym2713 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2717 | \usym2713 |
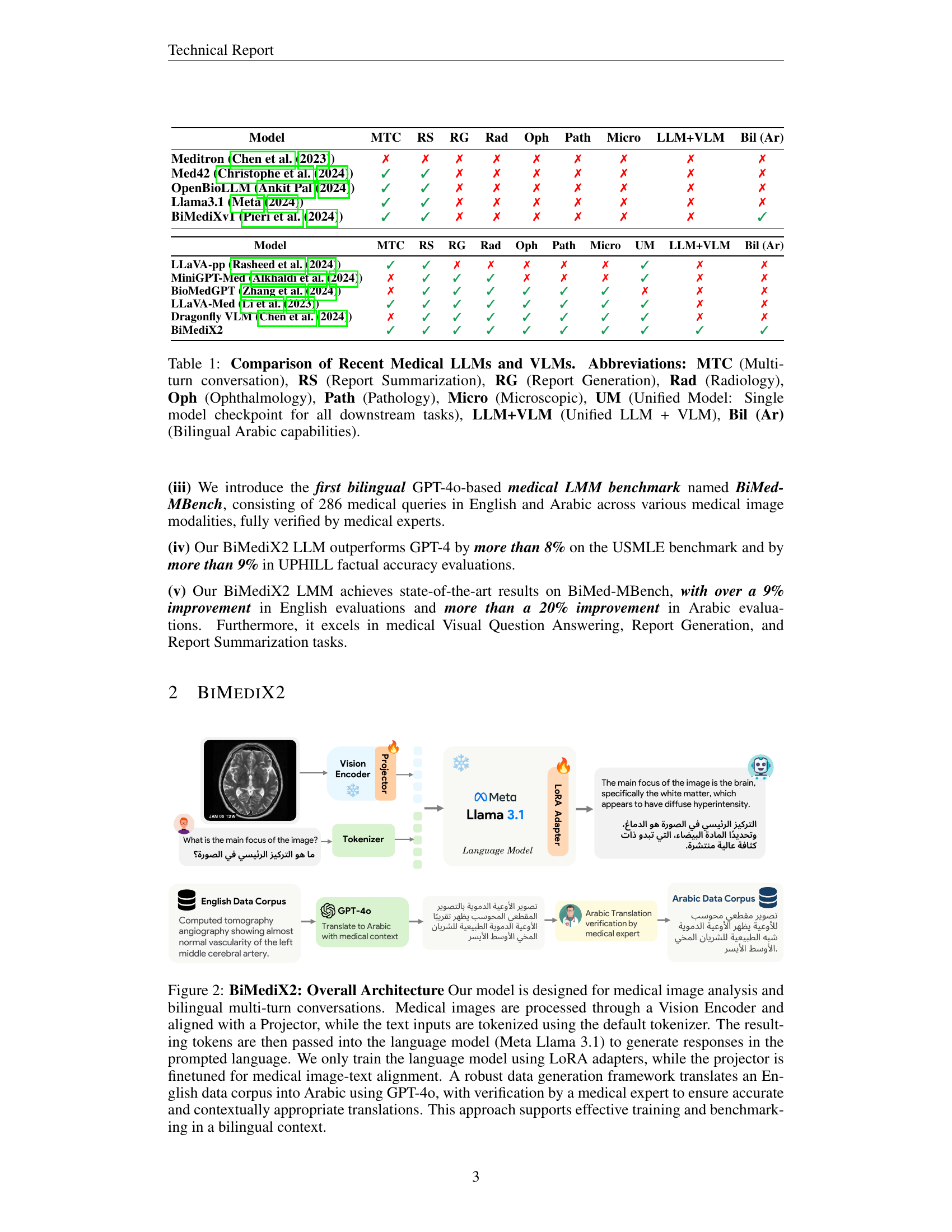
🔼 This table provides a comparison of recent Medical Large Language Models (LLMs) and Vision-Language Models (VLMs), highlighting their capabilities across various medical tasks and modalities. The table uses abbreviations like MTC for Multi-turn conversation, RS for Report Summarization, and others as explained in the caption. It also indicates whether these models offer a Unified Model (UM) approach (single checkpoint for all tasks) or a combined LLM+VLM architecture, and whether they have Bilingual (Arabic) capabilities.
read the caption
Table 1: Comparison of Recent Medical LLMs and VLMs. Abbreviations: MTC (Multi-turn conversation), RS (Report Summarization), RG (Report Generation), Rad (Radiology), Oph (Ophthalmology), Path (Pathology), Micro (Microscopic), UM (Unified Model: Single model checkpoint for all downstream tasks), LLM+VLM (Unified LLM + VLM), Bil (Ar) (Bilingual Arabic capabilities).
In-depth insights#
Bilingual Medical LMM#
BiMediX2, a novel bilingual Medical Large Multimodal Model (LMM), addresses the critical need for inclusive healthcare solutions. By supporting both Arabic and English, it bridges a significant gap in medical AI accessibility. This bilingual capability allows diverse populations to engage with advanced medical information and diagnostics, promoting health equity. BiMediX2 leverages a unified architecture, integrating text and visual modalities for comprehensive medical understanding. Trained on a massive bilingual dataset, BiMed-V, it excels in tasks like medical image analysis and multi-turn conversations. The development of the first bilingual GPT-40 based medical LMM benchmark, BiMed-MBench, facilitates rigorous evaluation and comparison with existing models, demonstrating BiMediX2’s superior performance. While challenges like hallucinations remain, BiMediX2 represents a substantial advancement, paving the way for more inclusive and effective global healthcare.
BiMediX2 Architecture#
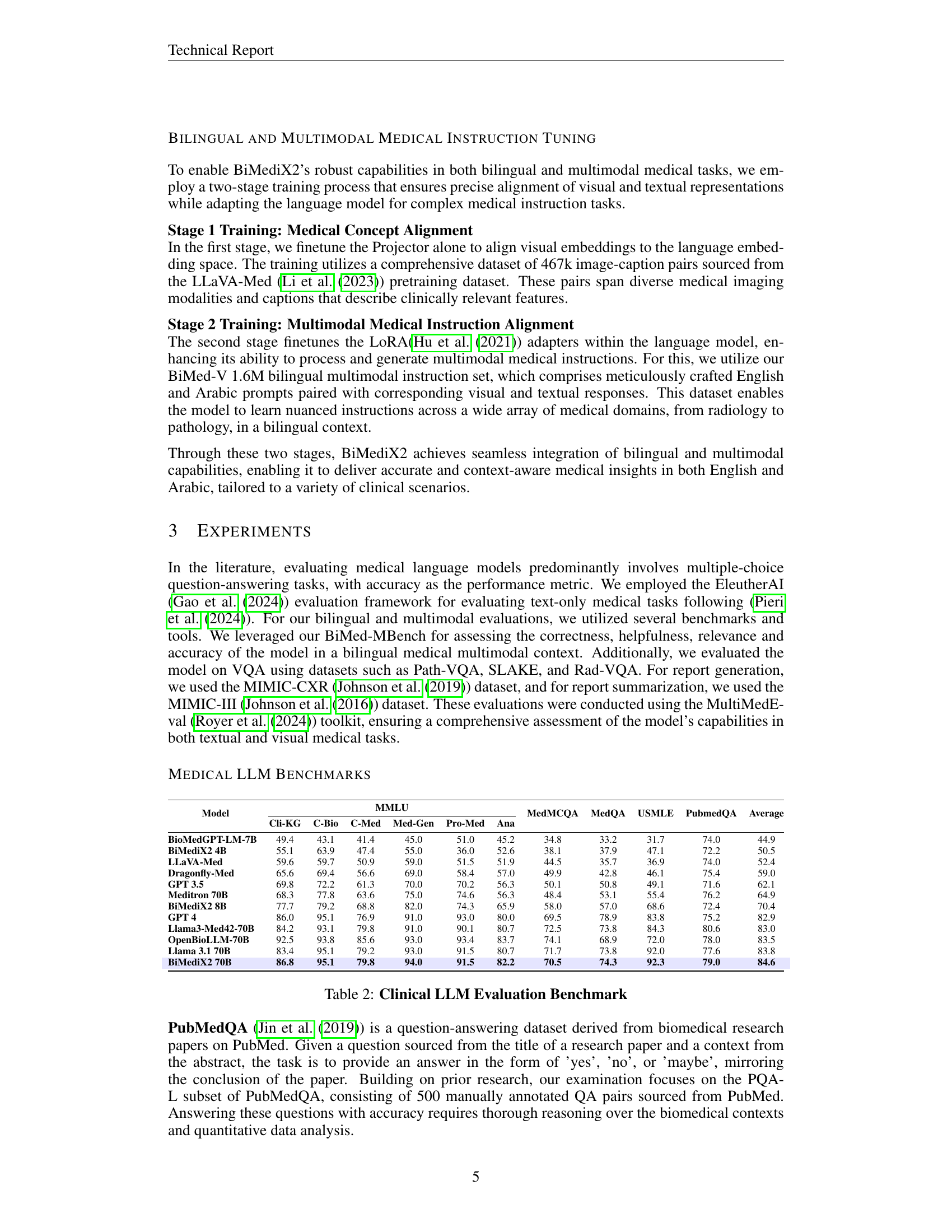
BiMediX2’s architecture effectively integrates textual and visual data for enhanced medical analysis. A Vision Encoder processes medical images, creating visual embeddings. Simultaneously, text inputs are converted into textual embeddings using a tokenizer and the LLaMA 3.1 language model. A Projector aligns these two modalities, mapping visual features to corresponding textual concepts. This unified approach facilitates tasks like image captioning and visual question answering in a bilingual (Arabic-English) context. LoRA adapters enable efficient fine-tuning of the language model while preserving computational resources. This design promotes multi-turn conversations about medical images, fostering a more interactive and informative diagnostic experience. The project’s innovative bilingual dataset and benchmark further enhance its ability to provide inclusive and comprehensive healthcare solutions.
BiMed-V Dataset#
The BiMed-V dataset is a multilingual, multimodal medical instruction set. Its 1.6M samples enhance medical image-text alignment and multimodal understanding. It leverages existing datasets like PMC-OA, Rad-VQA, Path-VQA, and SLAKE, supplemented with custom data and repurposed LLaVA-Med examples. Bilingual support is a key feature, with 163k Arabic samples generated via GPT-40 translation and expert validation. This hybrid approach minimizes reliance on human experts while ensuring quality. The inclusion of BiMediXv1’s text-based clinical data strengthens language understanding. BiMed-V enables advanced medical image-text alignment and conversational applications, addressing the need for inclusive healthcare solutions.
Multimodal Eval#
BiMediX2, a bilingual (Arabic-English) medical Large Multimodal Model (LMM), undergoes a multimodal evaluation to assess its proficiency in processing and understanding medical images along with textual queries. The evaluation leverages various benchmarks and datasets including BiMed-MBench, a novel bilingual medical benchmark, and established VQA datasets like Rad-VQA, SLAKE, and Path-VQA. Performance metrics include accuracy, recall, F1-score, and BLEU scores for tasks such as Visual Question Answering, report generation (using MIMIC-CXR), and report summarization (using MIMIC-III). The model’s ability to interpret diverse imaging modalities like X-rays, CT scans, MRIs, and histology slides, coupled with its bilingual capabilities, is rigorously tested, providing a comprehensive assessment of its potential in real-world medical applications. The robust evaluation framework underscores the emphasis on accuracy, clinical relevance, and language proficiency. This helps in creating more inclusive and effective medical AI solutions.
Arabic & MedImg Focus#
BiMediX2’s Arabic focus addresses a critical gap in medical AI, serving Arabic-speaking populations. This inclusivity broadens access to advanced medical insights and fosters more equitable healthcare. The model’s training on a large, bilingual dataset, including translated and expert-verified medical texts and images, enhances its understanding of Arabic medical terminology and nuances. BiMediX2’s strength in medical image analysis, combined with its bilingual capabilities, empowers healthcare professionals in Arabic-speaking regions to leverage cutting-edge technology for improved diagnosis, treatment planning, and patient care. This inclusivity in medical AI represents a significant step toward reducing health disparities and promoting global health equity. Further research will explore regional dialects and cultural contexts to enhance BiMediX2’s sensitivity and relevance within diverse Arabic-speaking communities.
More visual insights#
More on figures

🔼 BiMediX2 processes medical images through a Vision Encoder and aligns them with text using a Projector. Text is tokenized and input to Llama 3.1, which generates responses in the prompted language. The language model is trained using LoRA adapters, and the projector is fine-tuned for medical image-text alignment. An English data corpus is translated to Arabic using GPT-40 and verified by medical experts, facilitating bilingual training and benchmarking.
read the caption
Figure 2: BiMediX2: Overall Architecture Our model is designed for medical image analysis and bilingual multi-turn conversations. Medical images are processed through a Vision Encoder and aligned with a Projector, while the text inputs are tokenized using the default tokenizer. The resulting tokens are then passed into the language model (Meta Llama 3.1) to generate responses in the prompted language. We only train the language model using LoRA adapters, while the projector is finetuned for medical image-text alignment. A robust data generation framework translates an English data corpus into Arabic using GPT-4o, with verification by a medical expert to ensure accurate and contextually appropriate translations. This approach supports effective training and benchmarking in a bilingual context.

🔼 This figure presents a comparison of the state-of-the-art models in Clinical Large Language Model (LLM) benchmarks. Each bar represents a different model and its height represents the average performance across various clinical datasets (Cli-KG, C-Bio, C-Med, Med-Gen, Pro-Med, Ana, MedMCQA, MedQA, USMLE, PubmedQA). BiMediX2 models (4B, 8B, and 70B) are compared against existing LLM models (BioMedGPT, LLaVA-Med, Dragonfly-Med, GPT 3.5 & 4, Meditron, Llama3-Med42, OpenBioLLM, Llama 3.1). Overall, the results show that larger models generally exhibit higher average performance and the proposed models demonstrate competitive performance against existing models on this benchmark.
read the caption
Figure 3: State of the art comparison of models in Clinical LLM Benchmarks

🔼 This figure compares the performance of several large language models (LLMs), including BiMediX2, on the UPHILL OpenQA benchmark. The UPHILL benchmark tests the factual accuracy of LLMs in handling health-related queries, specifically focusing on the models’ ability to correctly refute false medical claims. The x-axis shows the model names, and the y-axis represents the overall factual accuracy percentage achieved by each model on the benchmark.
read the caption
Figure 4: Performance comparison on UPHILL OpenQA (Kaur et al. (2023)), assessing the model’s ability to address false medical claims at different presupposition levels.

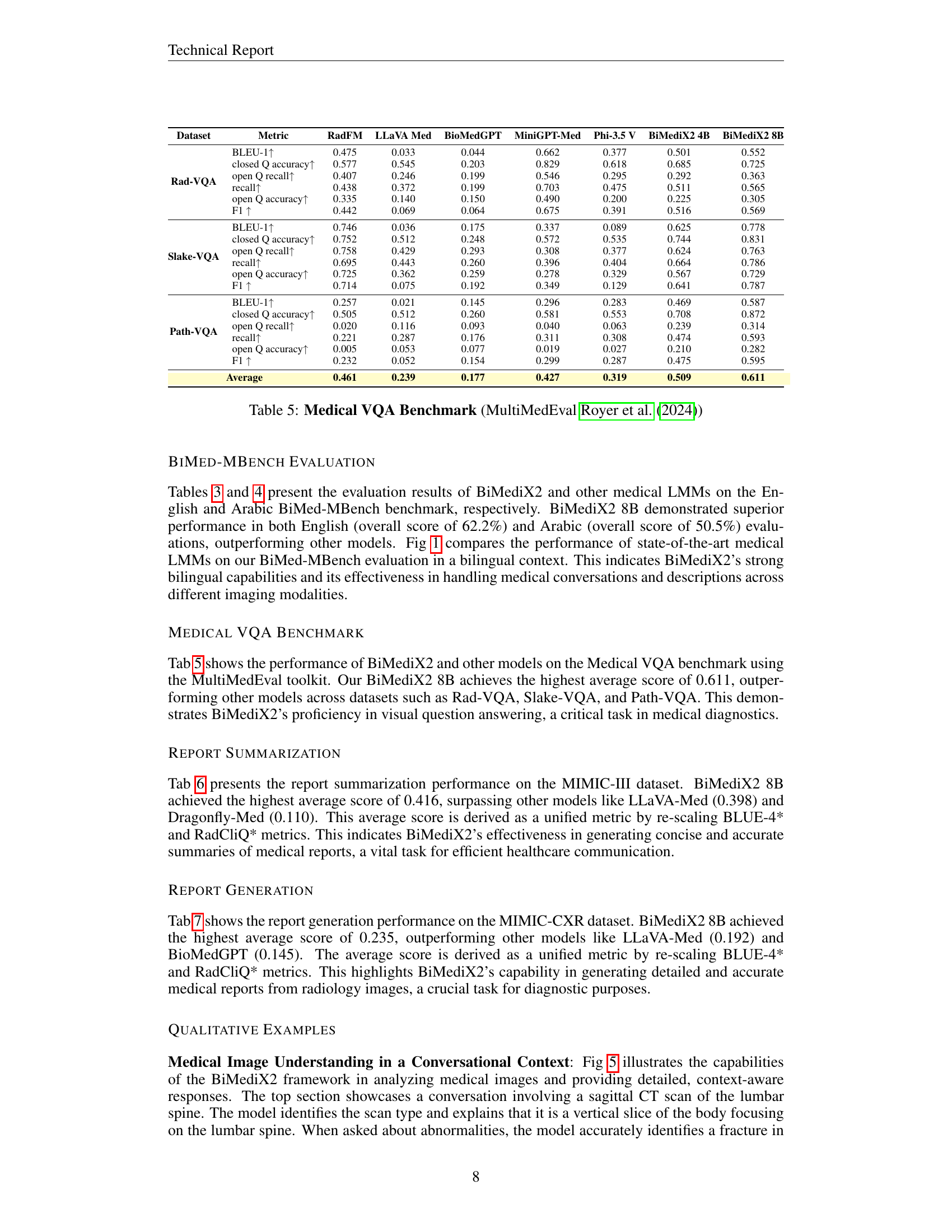
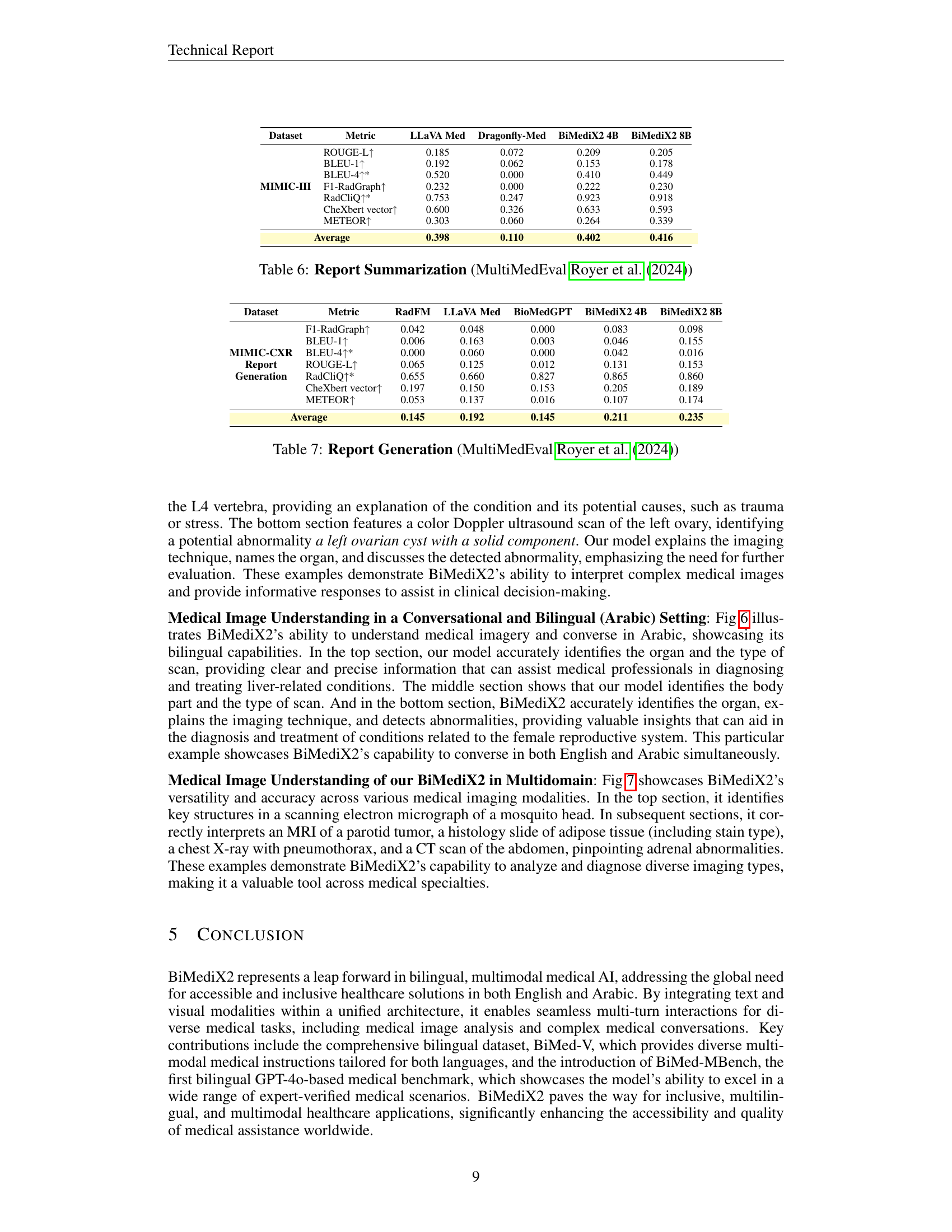
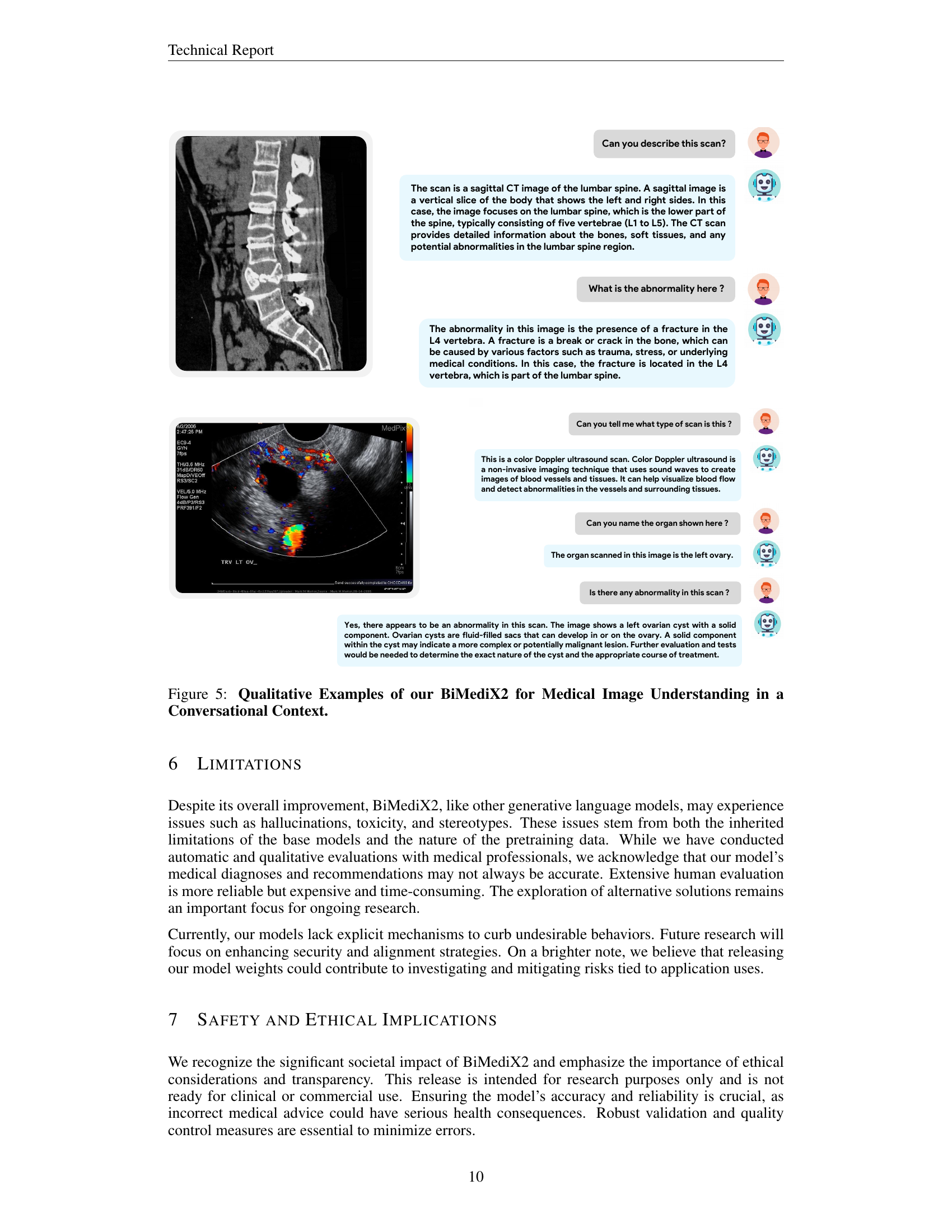
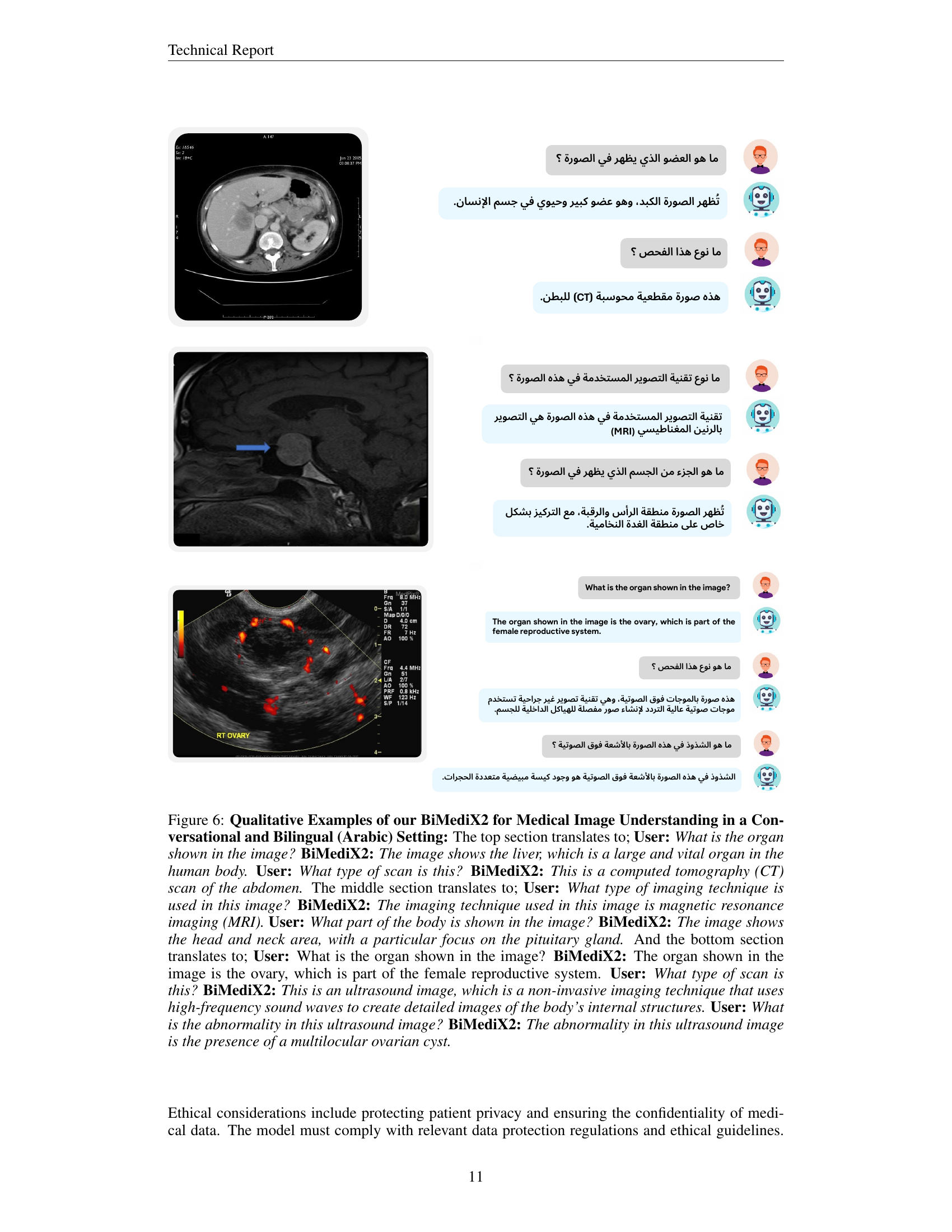
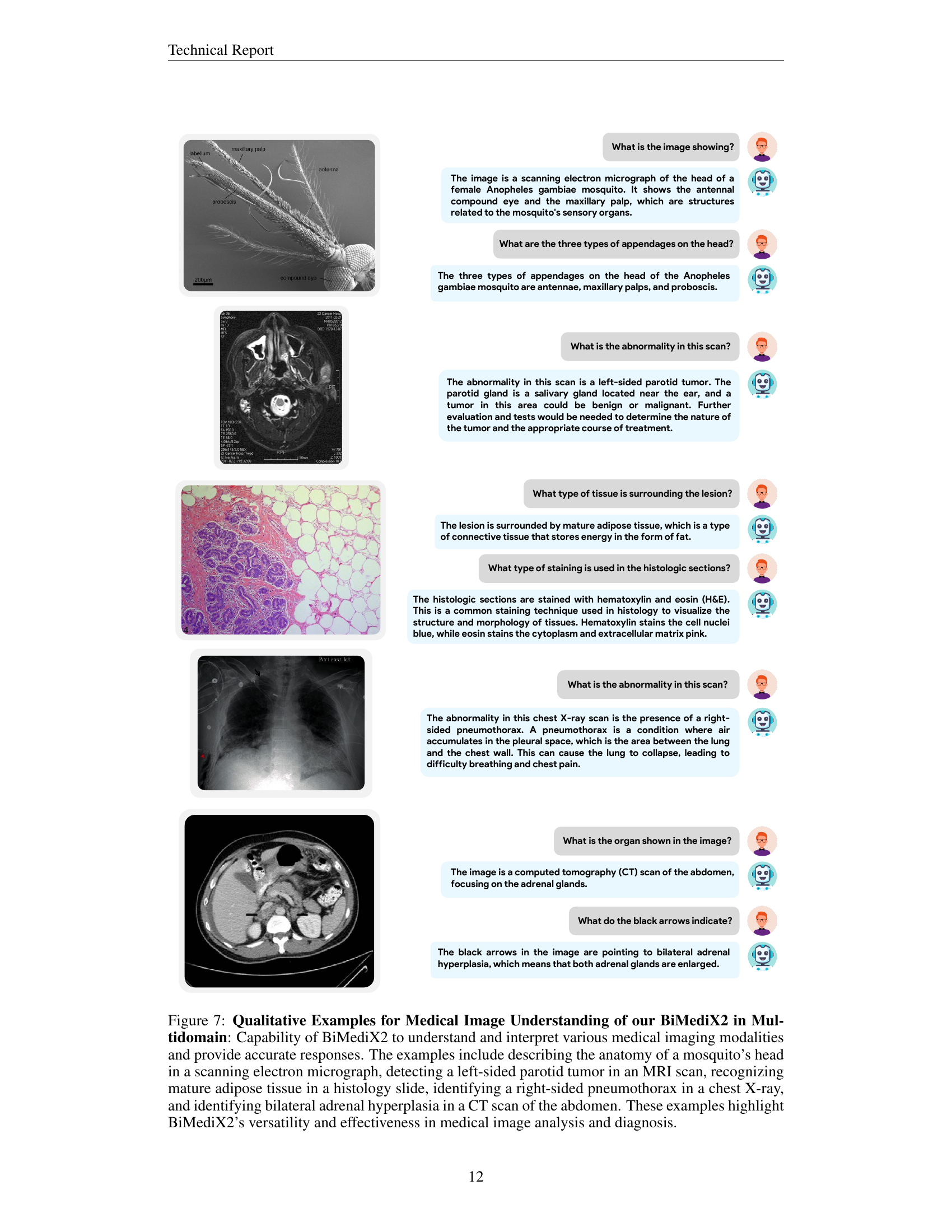
🔼 Figure 5 showcases BiMediX2’s capabilities in analyzing medical images within a conversational context. The top part presents a conversation about a sagittal CT scan of the lumbar spine, where BiMediX2 identifies the scan type, describes the anatomy, and pinpoints a fracture in the L4 vertebra, explaining potential causes. The bottom section features a color Doppler ultrasound of the left ovary. Here, the model explains the technique, identifies the organ, and notes a potential left ovarian cyst with a solid component, highlighting the need for further evaluation. These examples illustrate BiMediX2’s ability to interpret complex medical images and engage in informative medical conversations.
read the caption
Figure 5: Qualitative Examples of our BiMediX2 for Medical Image Understanding in a Conversational Context.
More on tables
| Model | MTC | RS | RG | Rad | Oph | Path | Micro | UM | LLM+VLM | Bil (Ar) |
|---|---|---|---|---|---|---|---|---|---|---|
| LLaVA-pp (Rasheed et al. (2024)) | 2713 | 2713 | 2717 | 2717 | 2717 | 2717 | 2717 | 2713 | 2717 | 2717 |
| MiniGPT-Med (Alkhaldi et al. (2024)) | 2717 | 2713 | 2713 | 2713 | 2717 | 2717 | 2717 | 2713 | 2717 | 2717 |
| BioMedGPT (Zhang et al. (2024)) | 2717 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2717 | 2717 | 2717 |
| LLaVA-Med (Li et al. (2023)) | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2717 | 2717 |
| Dragonfly VLM (Chen et al. (2024)) | 2717 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2717 | 2717 |
| BiMediX2 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 | 2713 |
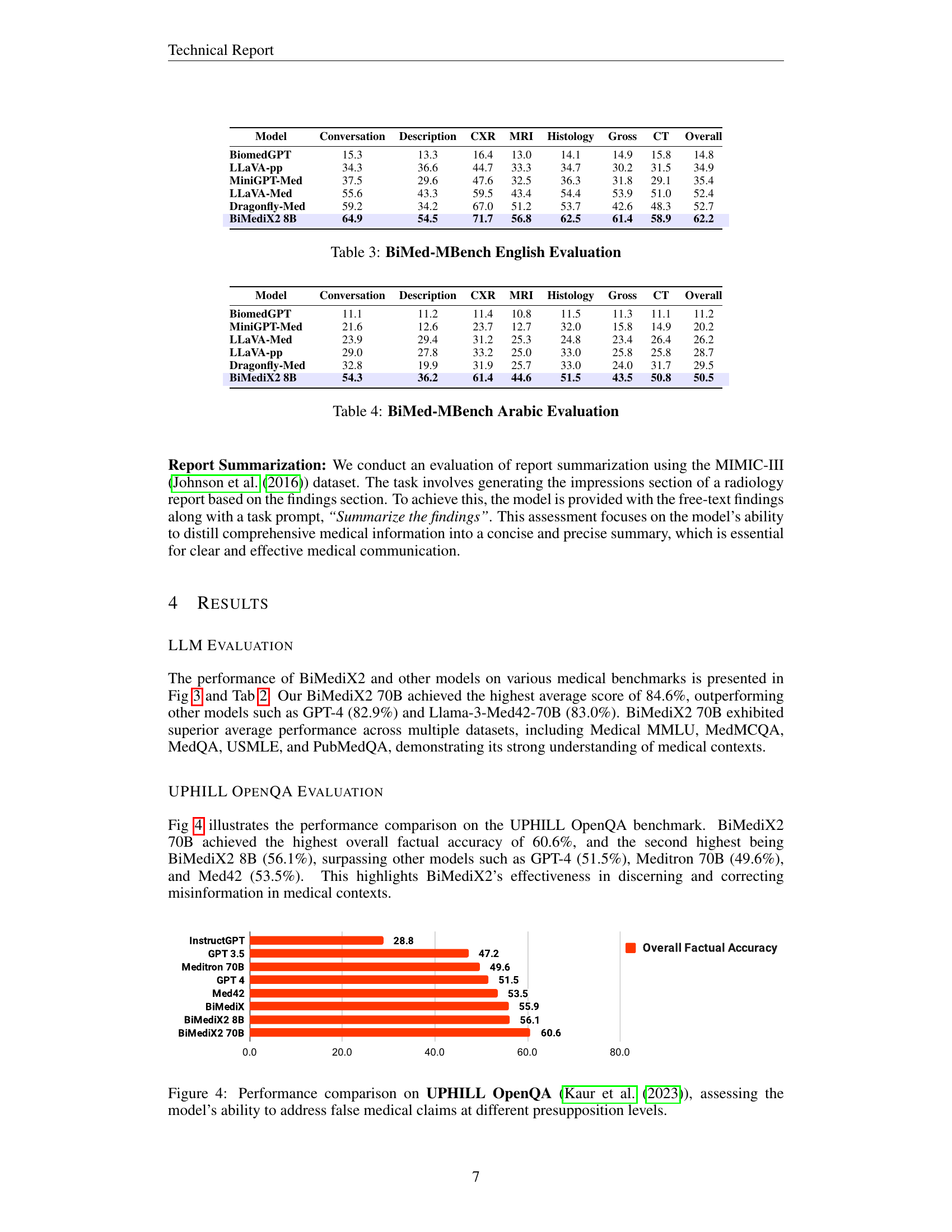
🔼 This table presents a comparison of various large language models (LLMs) on a set of clinical benchmarks. These benchmarks cover various medical question answering tasks like MedMCQA, MedQA, USMLE, and PubmedQA, as well as general medical knowledge and reasoning across multiple domains like Clinical Knowledge, College Biology, and others. The models are evaluated on these benchmarks, and their performance is represented by scores (e.g., accuracy, F1, etc.). This comparison allows researchers to assess the strengths and weaknesses of different LLMs in handling medical information, and BiMediX2 models in different sizes are compared against other existing models.
read the caption
Table 2: Clinical LLM Evaluation Benchmark
| Model | Cli-KG | C-Bio | C-Med | Med-Gen | Pro-Med | Ana | MedMCQA | MedQA | USMLE | PubmedQA | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BioMedGPT-LM-7B | 49.4 | 43.1 | 41.4 | 45.0 | 51.0 | 45.2 | 34.8 | 33.2 | 31.7 | 74.0 | 44.9 |
| BiMediX2 4B | 55.1 | 63.9 | 47.4 | 55.0 | 36.0 | 52.6 | 38.1 | 37.9 | 47.1 | 72.2 | 50.5 |
| LLaVA-Med | 59.6 | 59.7 | 50.9 | 59.0 | 51.5 | 51.9 | 44.5 | 35.7 | 36.9 | 74.0 | 52.4 |
| Dragonfly-Med | 65.6 | 69.4 | 56.6 | 69.0 | 58.4 | 57.0 | 49.9 | 42.8 | 46.1 | 75.4 | 59.0 |
| GPT 3.5 | 69.8 | 72.2 | 61.3 | 70.0 | 70.2 | 56.3 | 50.1 | 50.8 | 49.1 | 71.6 | 62.1 |
| Meditron 70B | 68.3 | 77.8 | 63.6 | 75.0 | 74.6 | 56.3 | 48.4 | 53.1 | 55.4 | 76.2 | 64.9 |
| BiMediX2 8B | 77.7 | 79.2 | 68.8 | 82.0 | 74.3 | 65.9 | 58.0 | 57.0 | 68.6 | 72.4 | 70.4 |
| GPT 4 | 86.0 | 95.1 | 76.9 | 91.0 | 93.0 | 80.0 | 69.5 | 78.9 | 83.8 | 75.2 | 82.9 |
| Llama3-Med42-70B | 84.2 | 93.1 | 79.8 | 91.0 | 90.1 | 80.7 | 72.5 | 73.8 | 84.3 | 80.6 | 83.0 |
| OpenBioLLM-70B | 92.5 | 93.8 | 85.6 | 93.0 | 93.4 | 83.7 | 74.1 | 68.9 | 72.0 | 78.0 | 83.5 |
| Llama 3.1 70B | 83.4 | 95.1 | 79.2 | 93.0 | 91.5 | 80.7 | 71.7 | 73.8 | 92.0 | 77.6 | 83.8 |
| BiMediX2 70B | 86.8 | 95.1 | 79.8 | 94.0 | 91.5 | 82.2 | 70.5 | 74.3 | 92.3 | 79.0 | 84.6 |
🔼 This table presents the evaluation results of BiMediX2 and other large multimodal models on the English portion of the BiMed-MBench dataset. The table includes performance metrics for different categories (Conversation, Description, CXR, MRI, Histology, Gross, CT, and Overall) and allows for comparison across models like BiomedGPT, LLaVA, MiniGPT-Med, Dragonfly, and versions of BiMediX2.
read the caption
Table 3: BiMed-MBench English Evaluation
| Model | Conversation | Description | CXR | MRI | Histology | Gross | CT | Overall |
|---|---|---|---|---|---|---|---|---|
| BiomedGPT | 15.3 | 13.3 | 16.4 | 13.0 | 14.1 | 14.9 | 15.8 | 14.8 |
| LLaVA-pp | 34.3 | 36.6 | 44.7 | 33.3 | 34.7 | 30.2 | 31.5 | 34.9 |
| MiniGPT-Med | 37.5 | 29.6 | 47.6 | 32.5 | 36.3 | 31.8 | 29.1 | 35.4 |
| LLaVA-Med | 55.6 | 43.3 | 59.5 | 43.4 | 54.4 | 53.9 | 51.0 | 52.4 |
| Dragonfly-Med | 59.2 | 34.2 | 67.0 | 51.2 | 53.7 | 42.6 | 48.3 | 52.7 |
| BiMediX2 8B | 64.9 | 54.5 | 71.7 | 56.8 | 62.5 | 61.4 | 58.9 | 62.2 |
🔼 BiMediX2’s performance on the Arabic portion of the BiMed-MBench benchmark, broken down by categories (Conversation, Description, CXR, MRI, Histology, Gross, CT) and overall score. The table shows BiMediX2 outperforms other models (BiomedGPT, MiniGPT-Med, LLaVA-Med, LLaVA-pp, Dragonfly-Med) in most categories and overall.
read the caption
Table 4: BiMed-MBench Arabic Evaluation
| Model | Conversation | Description | CXR | MRI | Histology | Gross | CT | Overall |
|---|---|---|---|---|---|---|---|---|
| BiomedGPT | 11.1 | 11.2 | 11.4 | 10.8 | 11.5 | 11.3 | 11.1 | 11.2 |
| MiniGPT-Med | 21.6 | 12.6 | 23.7 | 12.7 | 32.0 | 15.8 | 14.9 | 20.2 |
| LLaVA-Med | 23.9 | 29.4 | 31.2 | 25.3 | 24.8 | 23.4 | 26.4 | 26.2 |
| LLaVA-pp | 29.0 | 27.8 | 33.2 | 25.0 | 33.0 | 25.8 | 25.8 | 28.7 |
| Dragonfly-Med | 32.8 | 19.9 | 31.9 | 25.7 | 33.0 | 24.0 | 31.7 | 29.5 |
| BiMediX2 8B | 54.3 | 36.2 | 61.4 | 44.6 | 51.5 | 43.5 | 50.8 | 50.5 |
🔼 This table presents a benchmark comparison of different medical Vision-Language Answering (VQA) models using the MultiMedEval toolkit. Performance metrics including BLEU-1, closed question accuracy, open question recall, overall recall, open question accuracy, and F1 score are reported for models including RadFM, LLaVA-Med, BioMedGPT, MiniGPT-Med, Phi-3.5V, and two versions of BiMediX2 (4B and 8B) across three VQA datasets: Rad-VQA, Slake-VQA, and Path-VQA. The average performance across all datasets is also provided for each model.
read the caption
Table 5: Medical VQA Benchmark (MultiMedEval Royer et al. (2024))
| Dataset | Metric | RadFM | LLaVA Med | BioMedGPT | MiniGPT-Med | Phi-3.5 V | BiMediX2 4B | BiMediX2 8B |
|---|---|---|---|---|---|---|---|---|
| Rad-VQA | BLEU-1↑ | 0.475 | 0.033 | 0.044 | 0.662 | 0.377 | 0.501 | 0.552 |
| closed Q accuracy↑ | 0.577 | 0.545 | 0.203 | 0.829 | 0.618 | 0.685 | 0.725 | |
| open Q recall↑ | 0.407 | 0.246 | 0.199 | 0.546 | 0.295 | 0.292 | 0.363 | |
| recall↑ | 0.438 | 0.372 | 0.199 | 0.703 | 0.475 | 0.511 | 0.565 | |
| open Q accuracy↑ | 0.335 | 0.140 | 0.150 | 0.490 | 0.200 | 0.225 | 0.305 | |
| F1 ↑ | 0.442 | 0.069 | 0.064 | 0.675 | 0.391 | 0.516 | 0.569 | |
| Slake-VQA | BLEU-1↑ | 0.746 | 0.036 | 0.175 | 0.337 | 0.089 | 0.625 | 0.778 |
| closed Q accuracy↑ | 0.752 | 0.512 | 0.248 | 0.572 | 0.535 | 0.744 | 0.831 | |
| open Q recall↑ | 0.758 | 0.429 | 0.293 | 0.308 | 0.377 | 0.624 | 0.763 | |
| recall↑ | 0.695 | 0.443 | 0.260 | 0.396 | 0.404 | 0.664 | 0.786 | |
| open Q accuracy↑ | 0.725 | 0.362 | 0.259 | 0.278 | 0.329 | 0.567 | 0.729 | |
| F1 ↑ | 0.714 | 0.075 | 0.192 | 0.349 | 0.129 | 0.641 | 0.787 | |
| Path-VQA | BLEU-1↑ | 0.257 | 0.021 | 0.145 | 0.296 | 0.283 | 0.469 | 0.587 |
| closed Q accuracy↑ | 0.505 | 0.512 | 0.260 | 0.581 | 0.553 | 0.708 | 0.872 | |
| open Q recall↑ | 0.020 | 0.116 | 0.093 | 0.040 | 0.063 | 0.239 | 0.314 | |
| recall↑ | 0.221 | 0.287 | 0.176 | 0.311 | 0.308 | 0.474 | 0.593 | |
| open Q accuracy↑ | 0.005 | 0.053 | 0.077 | 0.019 | 0.027 | 0.210 | 0.282 | |
| F1 ↑ | 0.232 | 0.052 | 0.154 | 0.299 | 0.287 | 0.475 | 0.595 | |
| Average | 0.461 | 0.239 | 0.177 | 0.427 | 0.319 | 0.509 | 0.611 |
🔼 This table presents a benchmark evaluation of different medical Large Language Models (LLMs) on a report summarization task using the MIMIC-III dataset. The models are evaluated on their ability to generate concise and accurate summaries of medical reports based on their ‘findings’ sections, using metrics such as ROUGE-L, BLEU-1, BLEU-4*, F1-RadGraph, RadCliQ+*, CheXbert vector, and METEOR. Higher scores indicate better performance. The table compares the performance of several LLMs, including LLaVA-Med, Dragonfly-Med, and two versions of BiMediX2 (4B and 8B). The results show that BiMediX2 8B achieves the highest average score across all the metrics.
read the caption
Table 6: Report Summarization (MultiMedEval Royer et al. (2024))
| Dataset | Metric | LLaVA Med | Dragonfly-Med | BiMediX2 4B | BiMediX2 8B |
|---|---|---|---|---|---|
| MIMIC-III | ROUGE-L↑ | 0.185 | 0.072 | 0.209 | 0.205 |
| BLEU-1↑ | 0.192 | 0.062 | 0.153 | 0.178 | |
| BLEU-4↑* | 0.520 | 0.000 | 0.410 | 0.449 | |
| F1-RadGraph↑ | 0.232 | 0.000 | 0.222 | 0.230 | |
| RadCliQ↑* | 0.753 | 0.247 | 0.923 | 0.918 | |
| CheXbert vector↑ | 0.600 | 0.326 | 0.633 | 0.593 | |
| METEOR↑ | 0.303 | 0.060 | 0.264 | 0.339 | |
| Average | 0.398 | 0.110 | 0.402 | 0.416 |
🔼 This table presents the results of report generation on the MIMIC-CXR dataset, using metrics like F1-RadGraph, BLEU-1, BLEU-4*, ROUGE-L, RadCliQ*, CheXbert vector, and METEOR. The average score, a unified metric derived by rescaling BLEU-4* and RadCliQ*, is also provided for each model.
read the caption
Table 7: Report Generation (MultiMedEval Royer et al. (2024))
Full paper#