↗ arXiv ↗ Hugging Face ↗ Papers with Code
TL;DR#
Many multimodal large language models (MLLMs) struggle with medical image analysis due to insufficient data and a lack of understanding of the relationships between different image features. This paper explores the concept of compositional generalization (CG) where models learn to combine fundamental elements (like image modality, anatomical area, and task) to understand novel combinations of images. This is a significant issue because models trained on existing datasets don’t easily transfer their knowledge to new, unseen medical images.
The researchers created Med-MAT, a large dataset of labeled medical images carefully categorized by modality, anatomy, and task. They evaluated various MLLMs on Med-MAT and found that those leveraging CG performed significantly better at classifying unseen images, especially when the training data was limited. This shows that understanding the relationships between different image features and leveraging them through CG is key to building better-generalizing models. This research also demonstrates that CG is effective across different MLLM backbones, increasing the applicability of the proposed method.
Key Takeaways#
Why does it matter?#
This paper is crucial because it introduces a novel framework, compositional generalization (CG), to enhance the generalization capabilities of multimodal large language models (MLLMs) in medical imaging. It addresses the limitations of existing MLLMs that struggle with limited data and highlights the importance of data selection strategies. This provides valuable insights for researchers and opens up new avenues for building more robust and efficient medical AI systems.
Visual Insights#

🔼 This figure illustrates the concept of compositional generalization (CG) using examples of medical images. The ‘Train’ column shows examples of images the model has been trained on, categorized by Modality (MRI or CT), Anatomical Area (brain or lung), and Task (cancer detection or not). The ‘Test’ column shows unseen combinations of these elements, such as a CT scan of a cat’s brain (combining CT Modality, brain anatomical area, and an unseen task). The idea is that if the model truly understands the elements, it should be able to generalize to unseen combinations of those elements. The success of the model in classifying the test images demonstrates the power of compositional generalization.
read the caption
Figure 1: Examples of Compositional Generalization: The model is required to understand unseen images by recombining the fundamental elements it has learned.
| Model | 02 | 03 | 07 | 08 | 09 | 11 | 13 | 14 | 15 | 16 | 18 | 19 | 21 | 22 | 23 | 25 | 26 | 28 | 30 | 31 | 32 | 33 | 35 | 36 | 37 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 22 | 47 | 40 | 25 | 26 | 27 | 28 | 24 | 22 | 24 | 25 | 23 | 49 | 26 | 25 | 24 | 49 | 30 | 49 | 21 | 49 | 20 | 25 | 23 | 19 | |
| Single-task Training | 24 | 49 | 50 | 68 | 65 | 76 | 83 | 53 | 61 | 32 | 29 | 26 | 57 | 53 | 28 | 24 | 57 | 64 | 89 | 60 | 97 | 54 | 29 | 51 | 49 | |
| Multi-task Training | 96 | 89 | 80 | 80 | 79 | 97 | 92 | 88 | 76 | 57 | 88 | 74 | 87 | 86 | 93 | 52 | 98 | 72 | 94 | 61 | 100 | 72 | 75 | 60 | 50 |
🔼 This table presents the accuracy results of three different models on an in-distribution dataset. The models are compared across 37 different subsets of the dataset. For each subset, the table shows the accuracy of a baseline model, a model trained with a single task, and a model trained with multiple tasks. The best score in each subset is highlighted in bold, and the second-best score is underlined. This demonstrates the impact of single-task versus multi-task training on model performance.
read the caption
Table 1: Accuracy of different models on In-Distribution Dataset. Within each segment, bold highlights the best scores, and underlines indicate the second-best.
In-depth insights#
Med-MAT Dataset#
The Med-MAT dataset represents a significant contribution to the field of medical image analysis. Its structured organization around MAT-Triplets (Modality, Anatomical Area, Task) facilitates the exploration of compositional generalization (CG) in multimodal large language models (MLLMs). This structured approach allows researchers to investigate how MLLMs learn to combine knowledge from different sources and apply it to novel medical image scenarios. The dataset’s size and diversity—106 medical datasets, encompassing 11 modalities, 14 anatomical areas, and 13 medical tasks—make it a powerful tool for evaluating the generalization capabilities of MLLMs in the medical domain. The curated QA pairs, converted to visual question-answering format, streamline the training and evaluation process. Med-MAT’s publicly available nature promotes transparency and collaborative research. However, careful consideration of the datasets’ inherent biases and limitations is crucial for responsible use. Future research could explore additional modalities, incorporate temporal aspects, and enhance the dataset’s diversity further.
Compositional Gen#
The concept of “Compositional Generalization” in the context of multimodal large language models (MLLMs) applied to medical imaging is a significant contribution. It posits that the ability of MLLMs to understand novel combinations of medical images stems from their capacity to recombine learned fundamental elements. These elements, defined as the MAT-Triplet (Modality, Anatomical Area, and Task), provide a structured framework to analyze the model’s generalization capabilities. The research demonstrates that MLLMs leverage compositional generalization to understand unseen medical images, and that this is a key driver of generalization in multi-task training. This framework offers valuable insights into dataset selection for improving MLLM performance, particularly with limited data. Furthermore, the consistency of this compositional generalization across different MLLM backbones highlights its versatility and broad applicability. This research is crucial for advancing the use of MLLMs in medical applications where data scarcity is a major challenge. The proposed MAT-Triplet and the concept of compositional generalization significantly enhance our understanding of how MLLMs learn and generalize in the medical domain.
Multi-task Training#
Multi-task learning, in the context of the provided research paper, is a crucial technique for enhancing the generalization capabilities of multimodal large language models (MLLMs) applied to medical imaging. The paper highlights that training MLLMs on multiple tasks simultaneously, rather than focusing on single tasks, leads to superior performance on unseen datasets. This improvement stems from the models’ ability to leverage knowledge learned from related tasks to improve their understanding of novel combinations of modalities, anatomical areas, and tasks. The effectiveness of multi-task training is directly linked to the presence of compositional generalization (CG). The paper suggests that CG is a key driver of the generalization observed in multi-task settings, allowing the model to effectively recombine learned elements to understand unseen images. However, the study emphasizes that while multi-task learning generally enhances performance, a careful consideration of the relationships between tasks is vital for optimal results. Simply combining many unrelated tasks may not always lead to improvement; a structured approach which focuses on combinations leveraging CG is crucial for successful generalization.
Data Efficiency#
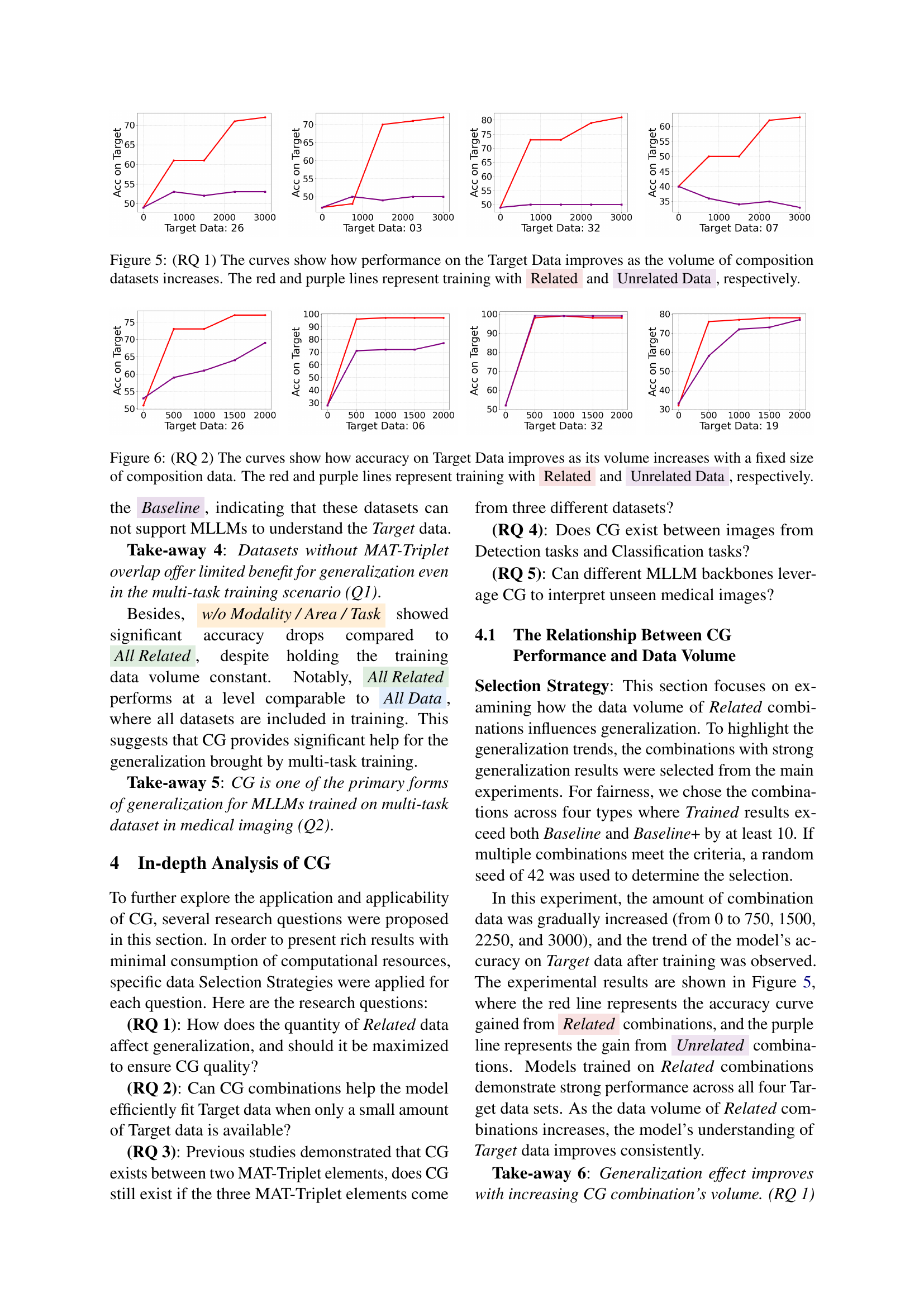
The research paper explores data efficiency in the context of compositional generalization (CG) for multimodal large language models (MLLMs) applied to medical imaging. A key finding is that CG significantly improves data efficiency, enabling MLLMs to generalize well even with limited training data for specific medical tasks. This is demonstrated through experiments showing that models trained with datasets exhibiting CG achieve higher accuracy on unseen data compared to models trained on randomly selected datasets. The study highlights the importance of carefully curating training datasets, emphasizing the selection of data that shares the same MAT-Triplet (Modality, Anatomical Area, Task) to leverage the power of CG. This approach is shown to be particularly effective for low-resource settings, where obtaining substantial amounts of data for each medical condition can be challenging. Furthermore, CG’s benefits are consistent across different MLLM architectures, suggesting that it represents a fundamental mechanism that enhances the models’ generalization capabilities. Therefore, leveraging CG appears to be a crucial strategy to improve data efficiency in training MLLMs for medical imaging tasks.
Future Directions#
Future research should prioritize expanding the Med-MAT dataset to encompass a wider range of medical modalities, anatomical areas, and tasks, improving its representativeness of real-world clinical scenarios. This would enhance the generalizability and robustness of the compositional generalization findings. Investigating the interplay between different types of medical data is crucial. For instance, exploring how combining data from imaging modalities with textual clinical notes or genomic data could further boost the performance of MLLMs. Furthermore, a more detailed analysis of the factors influencing the effectiveness of compositional generalization, such as data quality, volume, and task diversity, is warranted. This could involve systematically manipulating these factors in controlled experiments. Finally, exploring the integration of compositional generalization into existing clinical workflows is key to realizing the full potential of MLLMs in healthcare. This includes evaluating MLLM performance on complex real-world medical tasks and examining ways to address potential biases and ethical concerns related to the deployment of AI in medical settings.
More visual insights#
More on figures

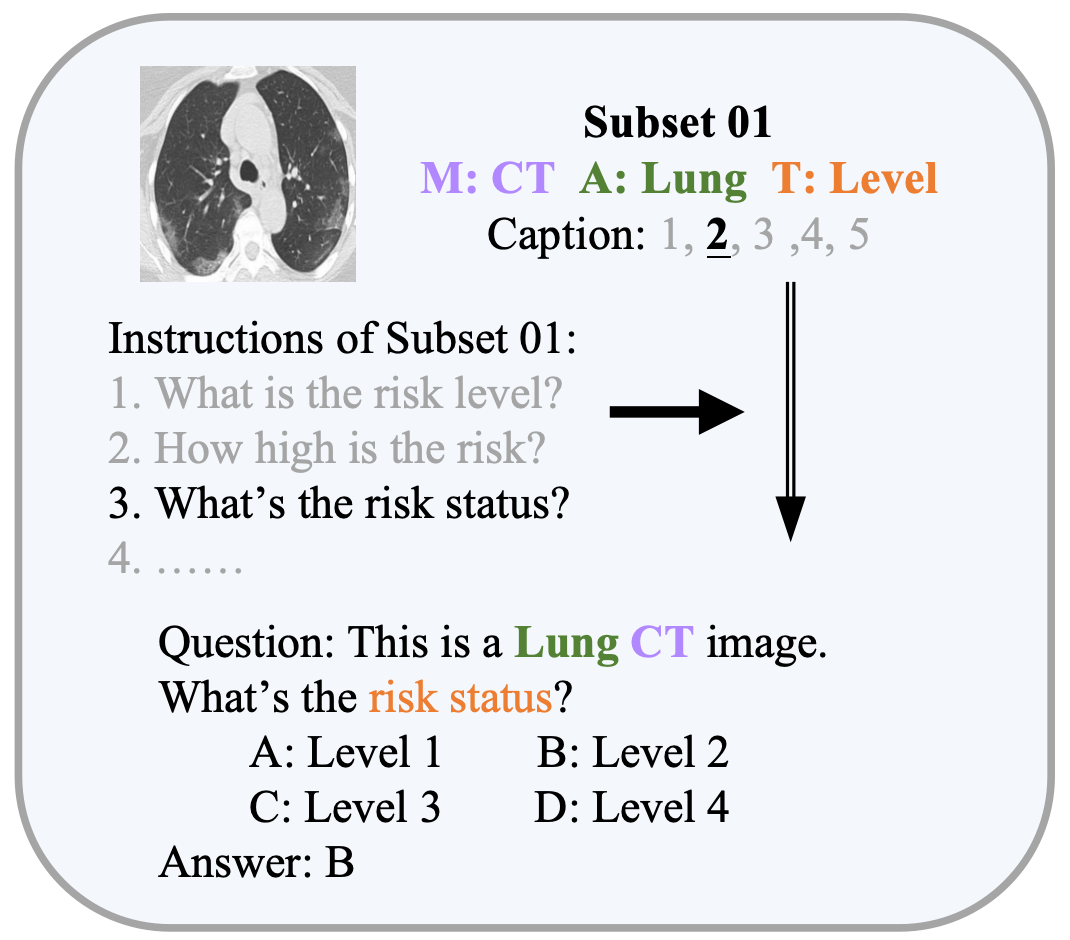
🔼 This figure illustrates the creation of the Med-MAT dataset. It starts with 106 individual medical image datasets. These datasets are categorized and grouped based on their MAT-Triplet (Modality, Anatomical Area, and Task), resulting in 53 subsets. Each subset consists of various modalities, anatomical areas, and medical tasks. The QA pairs construction is shown, where image-label pairs are converted into question-answer pairs suitable for MLLM training. This process ensures each sample in Med-MAT is clearly defined by its MAT-Triplet, enabling research on compositional generalization.
read the caption
Figure 2: The process of integrating a vast amount of labeled medical image data to create Med-MAT.
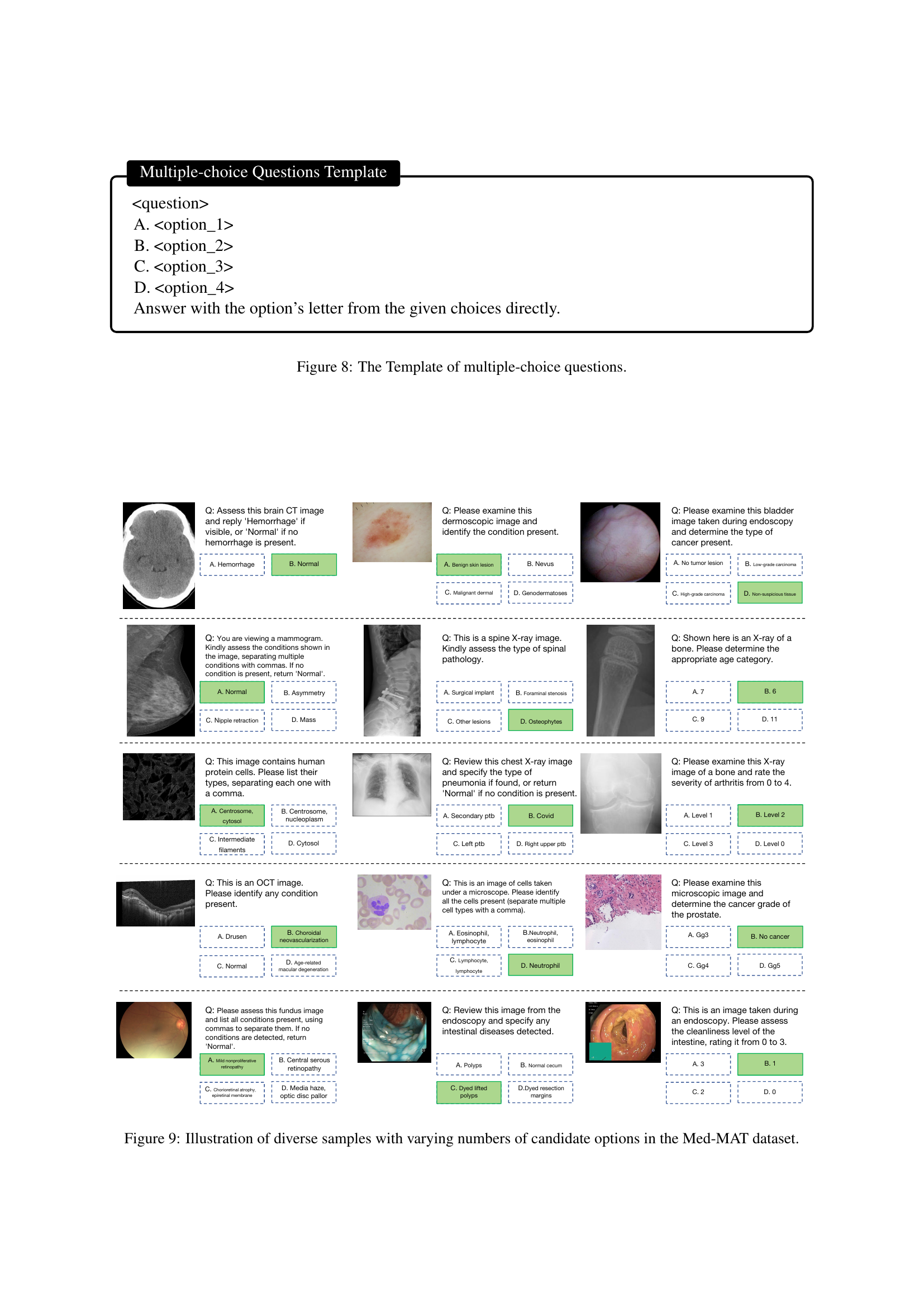
🔼 This figure illustrates the process of converting image-label pairs in Med-MAT into a visual question-answering (VQA) format suitable for training and evaluating multimodal large language models (MLLMs). The process involves assigning multiple instructions to each subset of images, converting each image-label pair into a single-choice question with four options, and randomly selecting distractor options from other labels within the subset. The integration of the ImageWikiQA dataset helps mitigate potential evaluation biases from varying option counts. The figure provides a detailed breakdown of the transformation from raw medical images and captions to structured questions and answers, which are essential for the MLLM training and testing.
read the caption
Figure 3: The QA formatting process of Med-MAT.

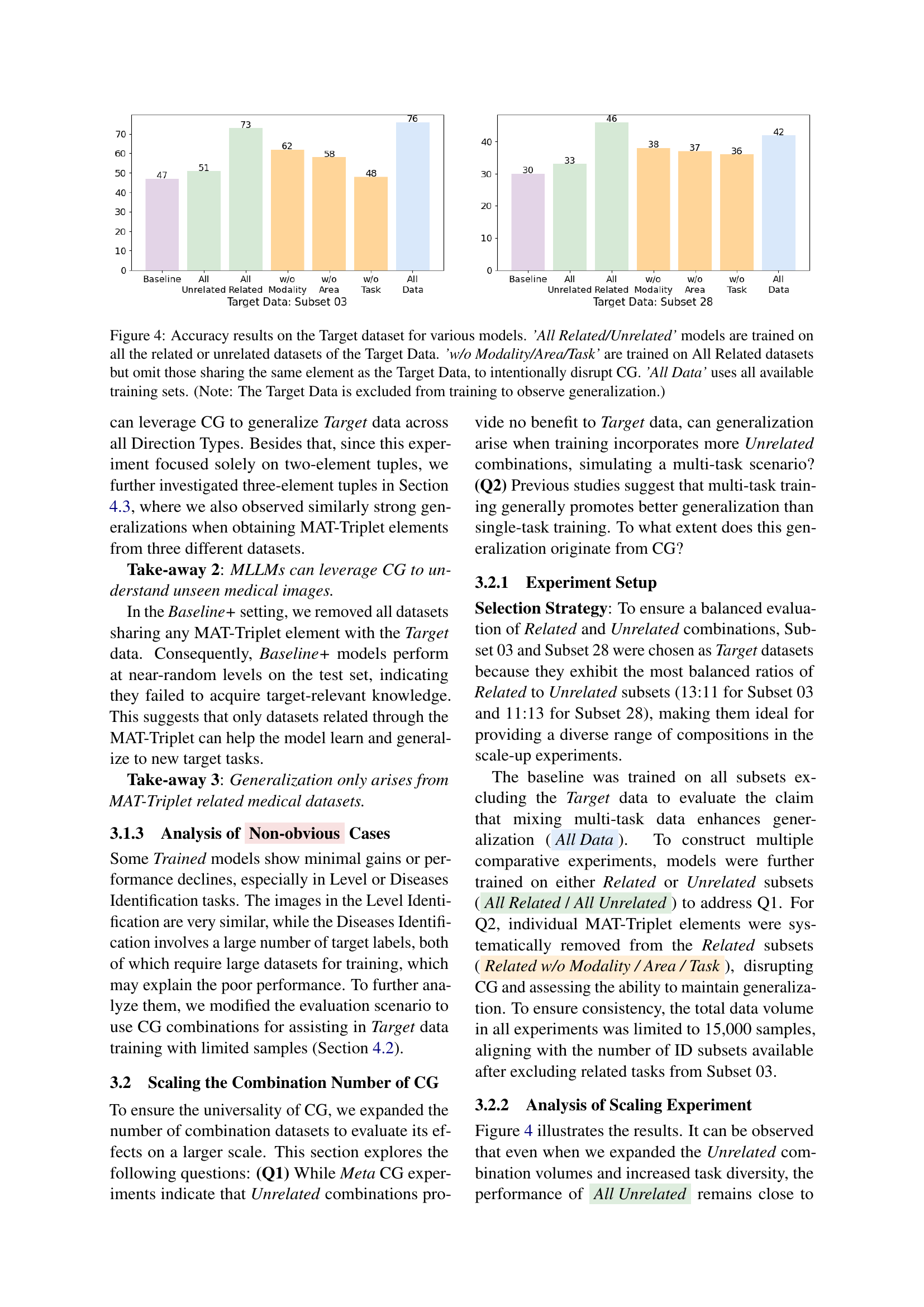
🔼 Figure 4 presents a comparative analysis of the generalization performance of several multimodal large language models (MLLMs) on unseen medical image data. The models were trained using different strategies, and their accuracy on a target dataset is shown. Specifically, the figure displays results for models trained on all related datasets, all unrelated datasets, all related datasets excluding those that share a Modality, Anatomical Area, or Task with the target dataset (to disrupt compositional generalization, or CG), and models trained on all available datasets. The target data itself was excluded from training to assess genuine generalization ability. This visual comparison helps illustrate the impact of compositional generalization on the model’s capacity to generalize.
read the caption
Figure 4: Accuracy results on the Target dataset for various models. ’All Related/Unrelated’ models are trained on all the related or unrelated datasets of the Target Data. ’w/o Modality/Area/Task’ are trained on All Related datasets but omit those sharing the same element as the Target Data, to intentionally disrupt CG. ’All Data’ uses all available training sets. (Note: The Target Data is excluded from training to observe generalization.)
More on tables
| Model | 01 | 04 | 05 | 06 | 10 | 12 | 17 | 20 | 24 | 27 | 29 | 34 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 32 | 25 | 33 | 33 | 48 | 27 | 33 | 13 | 34 | 37 | 31 | 20 |
| Multi-task Training | 39 | 26 | 70 | 31 | 58 | 38 | 61 | 40 | 35 | 41 | 55 | 50 |
🔼 This table presents the accuracy of different models on an Out-of-Distribution (OOD) dataset. The OOD dataset consists of medical images not seen during the model’s training phase. The table compares the performance of a baseline model, a single-task training model, and a multi-task training model. Bold text highlights the best accuracy scores achieved for each task by any of the models, indicating which model performed best on each OOD dataset subset.
read the caption
Table 2: Accuracy of different models on Out-Of-Distribution Dataset. Bold highlights the best scores.
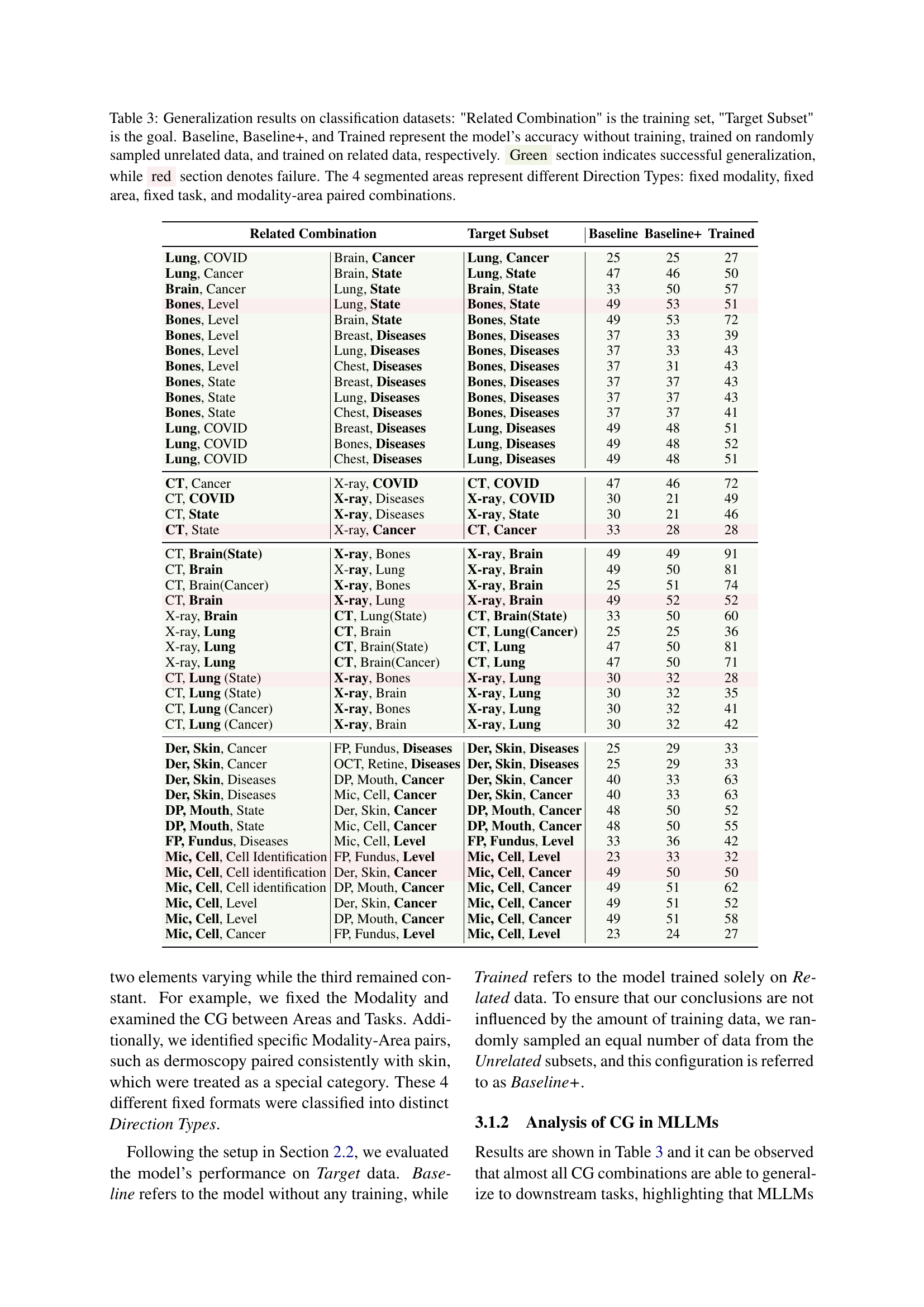
| Related Combination | Target Subset | Baseline | Baseline+ | Trained |
|---|---|---|---|---|
| Lung, COVID | Brain, Cancer | Lung, Cancer | 25 | 25 |
| Lung, Cancer | Brain, State | Lung, State | 47 | 46 |
| Brain, Cancer | Lung, State | Brain, State | 33 | 50 |
| Bones, Level | Lung, State | Bones, State | 49 | 53 |
| Bones, Level | Brain, State | Bones, State | 49 | 53 |
| Bones, Level | Breast, Diseases | Bones, Diseases | 37 | 33 |
| Bones, Level | Lung, Diseases | Bones, Diseases | 37 | 33 |
| Bones, Level | Chest, Diseases | Bones, Diseases | 37 | 31 |
| Bones, State | Breast, Diseases | Bones, Diseases | 37 | 37 |
| Bones, State | Lung, Diseases | Bones, Diseases | 37 | 37 |
| Bones, State | Chest, Diseases | Bones, Diseases | 37 | 37 |
| Lung, COVID | Breast, Diseases | Lung, Diseases | 49 | 48 |
| Lung, COVID | Bones, Diseases | Lung, Diseases | 49 | 48 |
| Lung, COVID | Chest, Diseases | Lung, Diseases | 49 | 48 |
| CT, Cancer | X-ray, COVID | CT, COVID | 47 | 46 |
| CT, COVID | X-ray, Diseases | X-ray, COVID | 30 | 21 |
| CT, State | X-ray, Diseases | X-ray, State | 30 | 21 |
| CT, State | X-ray, Cancer | CT, Cancer | 33 | 28 |
| CT, Brain(State) | X-ray, Bones | X-ray, Brain | 49 | 49 |
| CT, Brain | X-ray, Lung | X-ray, Brain | 49 | 50 |
| CT, Brain(Cancer) | X-ray, Bones | X-ray, Brain | 25 | 51 |
| CT, Brain | X-ray, Lung | X-ray, Brain | 49 | 52 |
| X-ray, Brain | CT, Lung(State) | CT, Brain(State) | 33 | 50 |
| X-ray, Lung | CT, Brain | CT, Lung(Cancer) | 25 | 25 |
| X-ray, Lung | CT, Brain(State) | CT, Lung | 47 | 50 |
| X-ray, Lung | CT, Brain(Cancer) | CT, Lung | 47 | 50 |
| CT, Lung (State) | X-ray, Bones | X-ray, Lung | 30 | 32 |
| CT, Lung (State) | X-ray, Brain | X-ray, Lung | 30 | 32 |
| CT, Lung (Cancer) | X-ray, Bones | X-ray, Lung | 30 | 32 |
| CT, Lung (Cancer) | X-ray, Brain | X-ray, Lung | 30 | 32 |
| Der, Skin, Cancer | FP, Fundus, Diseases | Der, Skin, Diseases | 25 | 29 |
| Der, Skin, Cancer | OCT, Retine, Diseases | Der, Skin, Diseases | 25 | 29 |
| Der, Skin, Diseases | DP, Mouth, Cancer | Der, Skin, Cancer | 40 | 33 |
| Der, Skin, Diseases | Mic, Cell, Cancer | Der, Skin, Cancer | 40 | 33 |
| DP, Mouth, State | Der, Skin, Cancer | DP, Mouth, Cancer | 48 | 50 |
| DP, Mouth, State | Mic, Cell, Cancer | DP, Mouth, Cancer | 48 | 50 |
| FP, Fundus, Diseases | Mic, Cell, Level | FP, Fundus, Level | 33 | 36 |
| Mic, Cell, Cell Identification | FP, Fundus, Level | Mic, Cell, Level | 23 | 33 |
| Mic, Cell, Cell identification | Der, Skin, Cancer | Mic, Cell, Cancer | 49 | 50 |
| Mic, Cell, Cell identification | DP, Mouth, Cancer | Mic, Cell, Cancer | 49 | 51 |
| Mic, Cell, Level | Der, Skin, Cancer | Mic, Cell, Cancer | 49 | 51 |
| Mic, Cell, Level | DP, Mouth, Cancer | Mic, Cell, Cancer | 49 | 51 |
| Mic, Cell, Cancer | FP, Fundus, Level | Mic, Cell, Level | 23 | 24 |
🔼 Table 3 presents the results of a controlled experiment evaluating compositional generalization (CG) in a medical image classification task. The table shows the accuracy of a multimodal large language model (MLLM) on various target datasets (unseen during training). Each row represents a different target dataset and its associated training set. The ‘Related Combination’ column specifies the training set of datasets containing images with similar characteristics (same Modality, Anatomical Area, and Task). The ‘Target Subset’ column identifies the dataset tested. Three accuracy scores are provided: Baseline (no training), Baseline+ (trained on randomly selected unrelated datasets), and Trained (trained on the ‘Related Combination’ dataset). The green sections highlight successful generalization (accuracy improved by using related data), while red sections indicate unsuccessful generalization (no improvement or decrease in accuracy by using related data). The table is divided into four sections that represent different combinations of fixed elements (modality, area, or task) within the MAT-Triplet, allowing for analysis of how compositional generalization varies based on the combination of similar data.
read the caption
Table 3: Generalization results on classification datasets: 'Related Combination' is the training set, 'Target Subset' is the goal. Baseline, Baseline+, and Trained represent the model’s accuracy without training, trained on randomly sampled unrelated data, and trained on related data, respectively. Green section indicates successful generalization, while red section denotes failure. The 4 segmented areas represent different Direction Types: fixed modality, fixed area, fixed task, and modality-area paired combinations.
| Related Combination | Target Subset | Baseline | Trained |
|---|---|---|---|
| CT - Subset02 | Brain - Subset22 | Cancer - Subset07 | CT, Brain, Cancer |
| CT - Subset03 | Brain - Subset22 | Cancer - Subset21 | CT, Brain, Cancer |
| CT - Subset02 | Brain - Subset22 | State - Subset09 | CT, Brain, State |
| CT - Subset03 | Brain - Subset22 | State - Subset26 | CT, Brain, State |
| X-ray - Subset25 | Lung - Subset03 | Diseases - Subset02 | X-ray, Lung, Diseases |
| X-ray - Subset26 | Lung - Subset03 | Diseases - Subset02 | X-ray, Lung, Diseases |
| X-ray - Subset26 | Lung - Subset03 | Diseases - Subset08 | X-ray, Lung, Diseases |
| X-ray - Subset26 | Breast - Subset24 | Diseases - Subset02 | X-ray, Breast, Diseases |
| X-ray - Subset28 | Breast - Subset24 | Diseases - Subset08 | X-ray, Breast, Diseases |
🔼 This table presents the results of a controlled experiment designed to investigate compositional generalization (CG) in multi-modal large language models (MLLMs). Three datasets were selected, each providing different combinations of MAT-Triplet elements (Modality, Anatomical area, and Task). The experiment tested whether the model could generalize to a target subset (a new combination of MAT-Triplet elements) by training only on related combinations of MAT-Triplet elements. The table shows the baseline performance (no training), and the accuracy of the model trained on related combinations. Green indicates successful generalization (the model correctly predicted the target task using only related training data), while red indicates failure.
read the caption
Table 4: Generalization results from 3 datasets providing different elements of MAT-Triplet (RQ 3). 'Related Combination' is the training set, 'Target Subset' is the goal. Baseline, and Trained represent the model’s accuracy without training and trained on Related data, respectively. Green section indicates successful generalization, while red section denotes failure.
| Related Combination | Target Subset | Target Subset | Baseline | Trained |
|---|---|---|---|---|
| Lung, Lung Det | Bones, Diseases | Lung, Diseases | 49 | 52 |
| Lung, Lung Det | Breast, Diseases | Lung, Diseases | 49 | 54 |
| Bones, Spinal Error Det | Breast, Diseases | Bones, Diseases | 20 | 30 |
| Bones, Spinal Error Det | Lung, Diseases | Bones, Diseases | 20 | 33 |
| MRI, Diseases Det | End, Level | End, Diseases | 24 | 27 |
| X-ray, Lung Det | CT, COVID | X-ray, COVID | 23 | 26 |
| Der, Skin, Cancer Det | FP, Fundus, Diseases | Der, Skin, Diseases | 24 | 29 |
| Mic, Cell, Cancer Det | CT, Kidney, Diseases | Mic, Cell, Diseases | 24 | 26 |
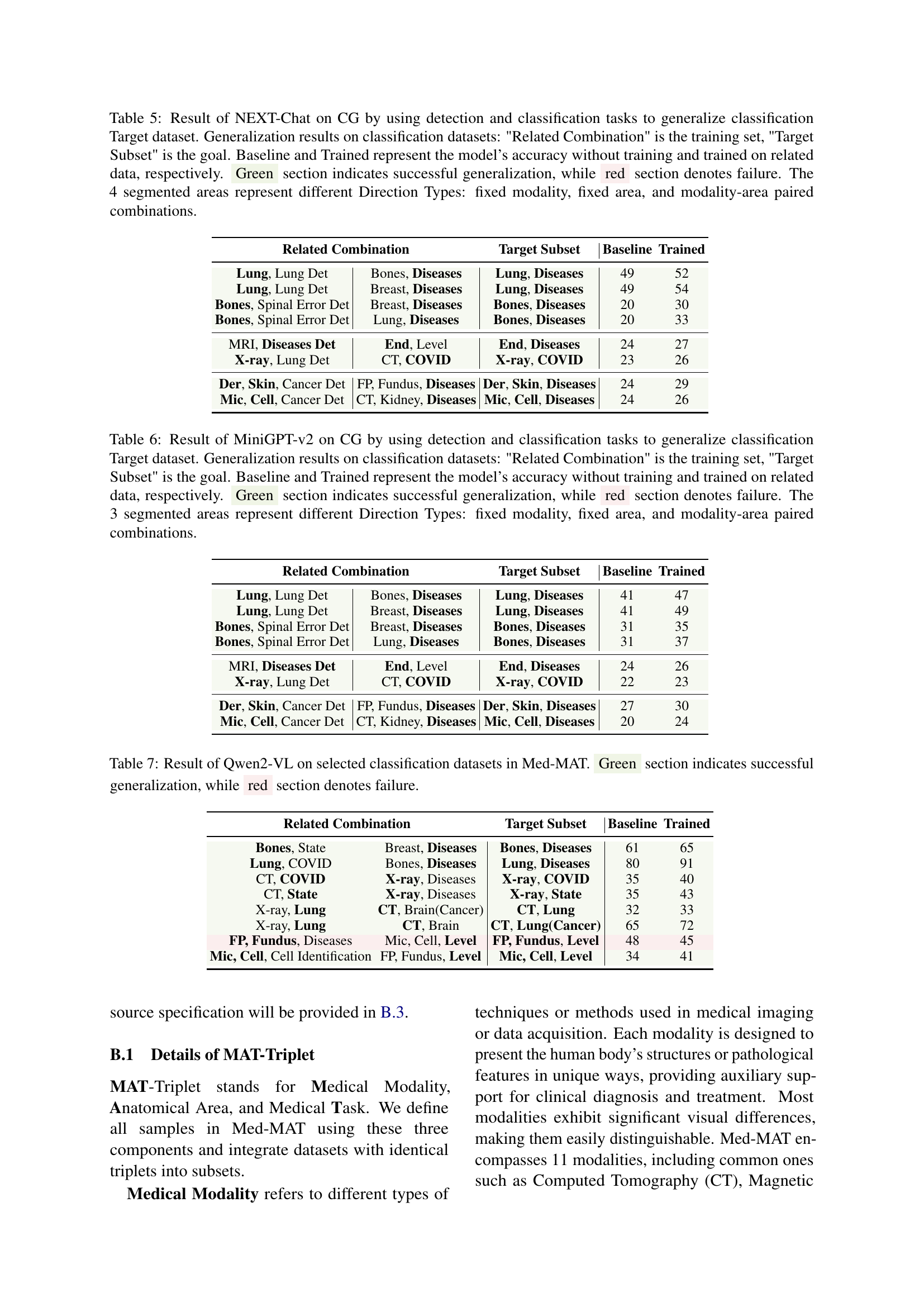
🔼 Table 5 presents the results of using the NEXT-Chat model to assess compositional generalization (CG). The experiment focuses on whether training with a combination of detection and classification datasets improves the model’s ability to generalize to unseen classification tasks. The table shows different combinations of related datasets used for training (‘Related Combination’), the target dataset tested (‘Target Subset’), and the model’s accuracy with no training (‘Baseline’), and after training on related data (‘Trained’). The results are categorized into four ‘Direction Types’ based on how the related and target datasets were related: fixed modality, fixed area, fixed task, and paired modality-area combinations. Green highlights successful generalization, while red indicates failure. This helps understand how different relationships between training and target datasets impact generalization performance.
read the caption
Table 5: Result of NEXT-Chat on CG by using detection and classification tasks to generalize classification Target dataset. Generalization results on classification datasets: 'Related Combination' is the training set, 'Target Subset' is the goal. Baseline and Trained represent the model’s accuracy without training and trained on related data, respectively. Green section indicates successful generalization, while red section denotes failure. The 4 segmented areas represent different Direction Types: fixed modality, fixed area, and modality-area paired combinations.
| Related Combination | Target Subset | Target Subset | Baseline | Trained |
|---|---|---|---|---|
| Lung, Lung Det | Bones, Diseases | Lung, Diseases | 41 | 47 |
| Lung, Lung Det | Breast, Diseases | Lung, Diseases | 41 | 49 |
| Bones, Spinal Error Det | Breast, Diseases | Bones, Diseases | 31 | 35 |
| Bones, Spinal Error Det | Lung, Diseases | Bones, Diseases | 31 | 37 |
| MRI, Diseases Det | End, Level | End, Diseases | 24 | 26 |
| X-ray, Lung Det | CT, COVID | X-ray, COVID | 22 | 23 |
| Der, Skin, Cancer Det | FP, Fundus, Diseases | Der, Skin, Diseases | 27 | 30 |
| Mic, Cell, Cancer Det | CT, Kidney, Diseases | Mic, Cell, Diseases | 20 | 24 |
🔼 This table presents the results of using MiniGPT-v2, a multimodal large language model, to perform a compositional generalization task. The model was trained on various combinations of datasets (‘Related Combination’) with a shared MAT-Triplet (Modality, Anatomical Area, Task) to predict the accuracy on a target dataset (‘Target Subset’) with unseen combinations of these elements. ‘Baseline’ represents the model’s accuracy without any training on related datasets, while ‘Trained’ reflects the accuracy after training with related datasets. The table highlights successful (‘Green’) and unsuccessful (‘Red’) generalization, categorized by three ‘Direction Types’ that reflect variations in dataset selection: fixed modality, fixed area, and modality-area paired combinations. This helps understand how the model’s ability to generalize depends on the relationships between the training and test data based on shared MAT-Triplets.
read the caption
Table 6: Result of MiniGPT-v2 on CG by using detection and classification tasks to generalize classification Target dataset. Generalization results on classification datasets: 'Related Combination' is the training set, 'Target Subset' is the goal. Baseline and Trained represent the model’s accuracy without training and trained on related data, respectively. Green section indicates successful generalization, while red section denotes failure. The 3 segmented areas represent different Direction Types: fixed modality, fixed area, and modality-area paired combinations.
| Related Combination | Target Subset | Baseline | Trained |
|---|---|---|---|
| Bones, State | Breast, Diseases | Bones, Diseases | 61 |
| Lung, COVID | Bones, Diseases | Lung, Diseases | 80 |
| CT, COVID | X-ray, Diseases | X-ray, COVID | 35 |
| CT, State | X-ray, Diseases | X-ray, State | 35 |
| X-ray, Lung | CT, Brain(Cancer) | CT, Lung | 32 |
| X-ray, Lung | CT, Brain | CT, Lung(Cancer) | 65 |
| FP, Fundus, Diseases | Mic, Cell, Level | FP, Fundus, Level | 48 |
| Mic, Cell, Cell Identification | FP, Fundus, Level | Mic, Cell, Level | 34 |
🔼 Table 7 presents the results of using the Qwen2-VL model on a subset of classification datasets from the Med-MAT dataset. The table showcases the model’s performance in generalizing to unseen data based on training with related and unrelated data. Green highlights successful generalization, while red denotes failure. Each row represents a pair of related and target datasets, indicating which combinations successfully promote generalization to the target dataset.
read the caption
Table 7: Result of Qwen2-VL on selected classification datasets in Med-MAT. Green section indicates successful generalization, while red section denotes failure.
| Related Combination | Target Subset | Baseline | Trained |
|---|---|---|---|
| Bones, State, Breast, Diseases | Bones, Diseases | 52 | 59 |
| Lung, COVID, Bones, Diseases | Lung, Diseases | 64 | 75 |
| CT, COVID, X-ray, Diseases | X-ray, COVID | 33 | 38 |
| CT, State, X-ray, Diseases | X-ray, State | 33 | 41 |
| X-ray, Lung, CT, Brain(Cancer) | CT, Lung | 31 | 29 |
| X-ray, Lung, CT, Brain | CT, Lung(Cancer) | 49 | 57 |
| FP, Fundus, Diseases, Mic, Cell, Level | FP, Fundus, Level | 55 | 61 |
| Mic, Cell, Cell Identification, FP, Fundus, Level | Mic, Cell, Level | 10 | 32 |
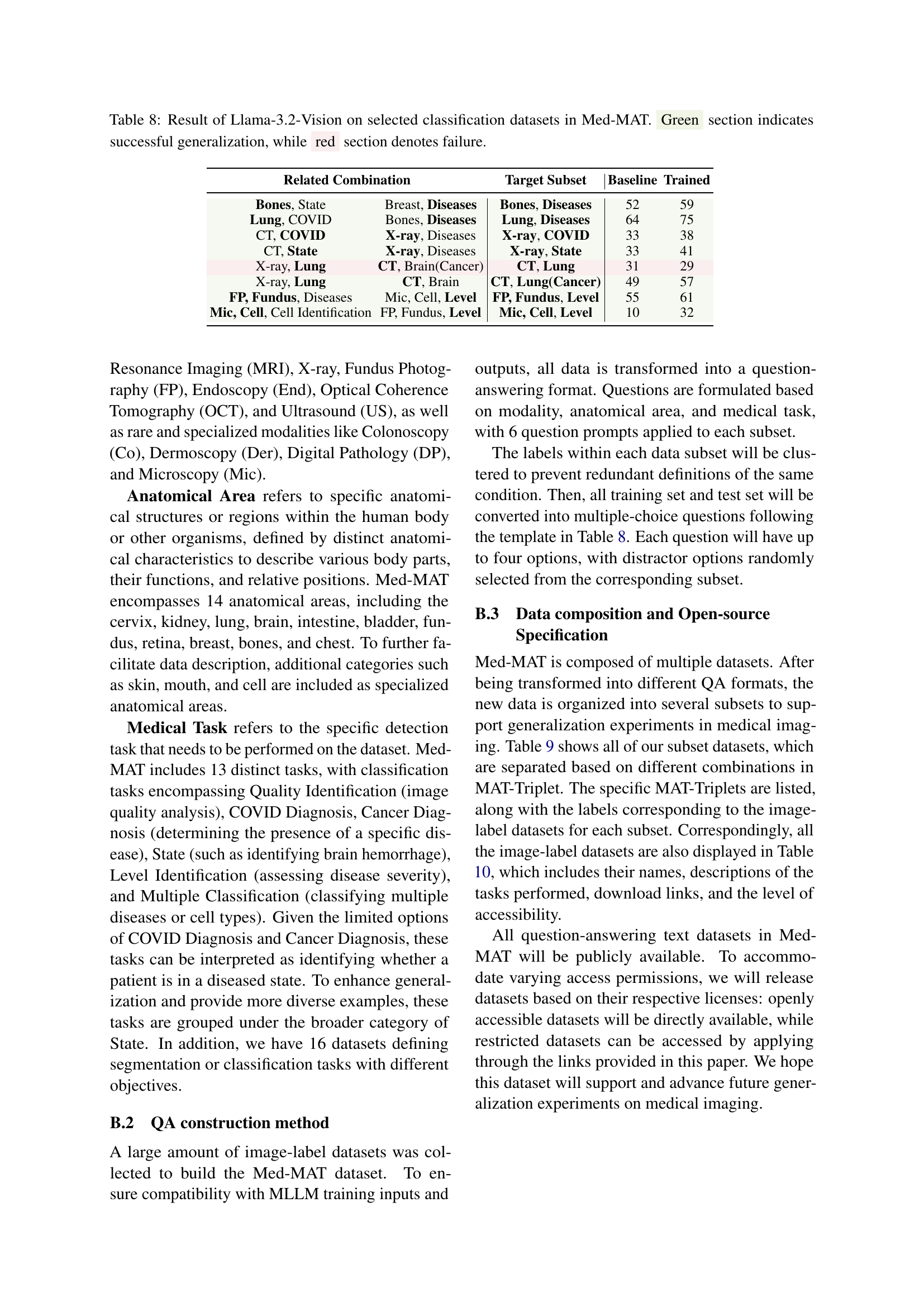
🔼 Table 8 presents the results of using the Llama-3.2-Vision model on a subset of classification datasets from the Med-MAT dataset. The table shows the model’s performance on unseen data (‘Target Subset’) after being trained on related datasets (‘Related Combination’). The results are categorized to show successful generalization (green) or failure (red). The categories represent different types of relationships between the training and target datasets, based on fixed modalities, anatomical areas, tasks, or combinations of these factors.
read the caption
Table 8: Result of Llama-3.2-Vision on selected classification datasets in Med-MAT. Green section indicates successful generalization, while red section denotes failure.
| Subset No. | Modality | Anatomical Area | Task | Datasets No. |
|---|---|---|---|---|
| 01 | Co | Cervix | Cervical Picture Quality Evaluation | 1 |
| 02 | CT | Kidney | Kidney Diseases Classification | 2 |
| 03 | CT | Lung | COVID-19 Classification | 3,4,6 |
| 04 | CT | Lung | Lung Cancer Classification | 5 |
| 05 | CT | Brain | Brain Hemorrhage Classification | 7 |
| 06 | CT | Brain | Brain Cancer Classification | 8 |
| 07 | Der | Skin | Melanoma Type Classification | 10 |
| 08 | Der | Skin | Skin Diseases Classification | 9, 11-15, 71, 72, 74 |
| 09 | DP | Mouth | Teeth Condition Classification | 16 |
| 10 | DP | Mouth | Oral Cancer Classification | 17 |
| 11 | End | Intestine | Intestine Cleanliness Level | 18 |
| 12 | End | Bladder | Cancer Degree Classification | 19 |
| 13 | End | Intestine | Intestine Diseases Classification | 20 |
| 14 | FP | Fundus | Eye Diseases Classification | 21-23, 26-28, 31, 32, 75 |
| 15 | FP | Fundus | Multiple-labels Eye Diseases Classification | 24, 25, 68 |
| 16 | FP | Fundus | Blindness Level | 29 |
| 17 | FP | Fundus | Retinal Images Quality Evaluation | 30 |
| 18 | Mic | Cell | Cell Type Classification | 33, 36-38, 39-41, 44, 65, 70 |
| 19 | Mic | Cell | Prostate Cancer Degree Classification | 34 |
| 20 | Mic | Cell | Multiple-labels Blood Cell Classification | 35 |
| 21 | Mic | Cell | Cancer Classification | 42, 67 |
| 22 | MRI | Brain | Head Diseases Classification | 44, 45 |
| 23 | OCT | Retina | Retina Diseases Classification | 46, 47 |
| 24 | US | Breast | Breast Cancer Classification | 48 |
| 25 | X-ray | Bones | Degree Classification of Knee | 49, 53 |
| 26 | X-ray | Bones | Fractured Classification | 50, 51 |
| 27 | X-ray | Bones | Vertebrae Diseases Classification | 52 |
| 28 | X-ray | Lung | COVID-19 and Pneumonia Classification | 54-57, 60, 62, 81 |
| 29 | X-ray | Breast | Breast Diseases Classification | 58, 78 |
| 30 | X-ray | Lung | Tuberculosis Classification | 59, 79 |
| 31 | X-ray | Chest | Multiple-labels Chest Classification | 61, 73, 76, 77, 80, 85, 87 |
| 32 | X-ray | Brain | Tumor Classification | 63 |
| 33 | Mic | Cell | Multi-labels Diseases | 84 |
| 34 | FP | Fundus | Level Identification | 66 |
| 35 | X-ray | Bones | Level Identification | 69 |
| 36 | X-ray | Bones | Spinal lesion Classification | 86 |
| 37 | X-ray | Breast | Multi-labels Diseases | 82 |
| 38 | Der | Skin | Lesion Det/Seg | 88-91 |
| 39 | End | Intestine | PolyP Det/Seg | 92-93 |
| 40 | End | Intestine | Surgical Procedures Det/Seg | 94 |
| 41 | End | Intestine | Multi-labels Det/Seg | 95 |
| 42 | Mic | Cell | Cancer Cell Det/Seg | 96 |
| 43 | US | Chest | Cancer Det/Seg | 97 |
| 44 | US | Thyroid | Thyroid Nodule Region Det/Seg | 98 |
| 45 | MRI | Intestine | Multi-labels Det/Seg | 103 |
| 46 | MRI | Liver | Liver Det/Seg | 104, 105 |
| 47 | X-ray | Lung | Lung Det/Seg | 99 |
| 48 | X-ray | Lung | Pneumothorax Det/Seg | 106 |
| 49 | X-ray | Bones | Spinal Anomaly Det | 100 |
| 50 | X-ray | Chest | Multi-labels Det | 101, 102 |
| 51 | FP | Fundus | Vessel Seg | 107 |
| 52 | FP | Fundus | Optic Disc and Cup Seg | 108 |
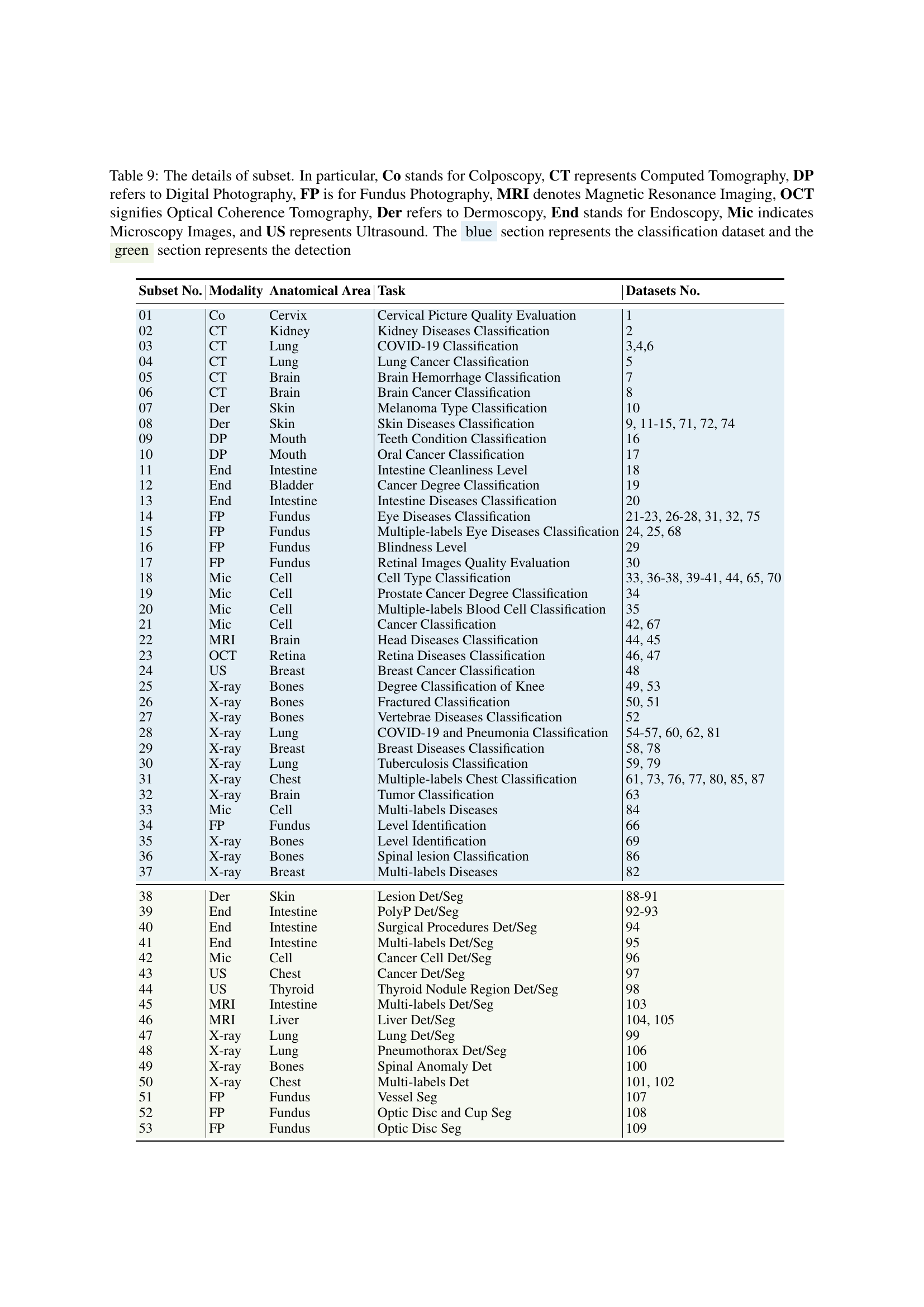
🔼 Table 9 details the composition of the Med-MAT dataset, showing how it’s divided into subsets. Each subset contains medical images categorized by three elements: Modality (e.g., CT scan, MRI), Anatomical Area (e.g., lung, brain), and Task (e.g., cancer detection, disease classification). The table lists each subset, specifying its modality, anatomical area, task, and the number of datasets included. It also uses color-coding to distinguish between classification and detection tasks within the subsets. Abbreviations for modalities (e.g., Co for Colposcopy, CT for Computed Tomography, etc.) are provided in the caption to aid understanding.
read the caption
Table 9: The details of subset. In particular, Co stands for Colposcopy, CT represents Computed Tomography, DP refers to Digital Photography, FP is for Fundus Photography, MRI denotes Magnetic Resonance Imaging, OCT signifies Optical Coherence Tomography, Der refers to Dermoscopy, End stands for Endoscopy, Mic indicates Microscopy Images, and US represents Ultrasound. The blue section represents the classification dataset and the green section represents the detection
| No. | Name | Description | Citation |
|---|---|---|---|
| 1 | Intel & MobileODT Cervical Screening | Cervix Type in Screening | BenO et al. (2017) |
| 2 | CT Kindney Dataset | Normal or Cyst or Tumor | Islam et al. (2022a) |
| 3 | SARS-COV-2 Ct-Scan | COVID19, Classification Dataset | Soares et al. (2020) |
| 4 | COVID CT COVID-CT | COVID19, Classification Dataset. | Zhao et al. (2020) |
| 5 | Chest CT-Scan | Cancer Classification | SunneYi (2021) |
| 6 | COVID-19-CT SCAN IMAGES | COVID19, Classification | wjXiaochuangw (2019) |
| 7 | Head CT | Head Hemorrhage | Kitamura (2018) |
| 8 | CT of Brain | Head Cancer | Data (2023) |
| 9 | MED-NODE | Melanoma or Naevus | Giotis et al. (2015) |
| 10 | ISIC 2020 | Melanoma, Benign or Malignant | Rotemberg et al. (2021) |
| 11 | PED-UFES-20 | Skin Multi Classification | Pacheco et al. (2020) |
| 12 | Web-scraped Skin Image | Skin Desease Multi Classification | Islam et al. (2022b) |
| 13 | ISBI 2016 | Skin Lesion Classification | Gutman et al. (2016) |
| 14 | ISIC 2019 | Skin Desease Multi Classification | Combalia et al. (2019) |
| 15 | Skin Cancer ISIC | Skin Cancer Multi Classification | Katanskiy (2019) |
| 16 | Dental Condition Dataset | Teeth condition classification | Sajid (2024) |
| 17 | Oral Cancer Dataset | Oral cancer Classification | RASHID (2024) |
| 18 | The Nerthus Dataset | Cleanliness level | Pogorelov et al. (2017a) |
| 19 | Endoscopic Bladder Tissue | Canser Degree Classification | Lazo et al. (2023) |
| 20 | Kvasir | Multi Disease Classification | Pogorelov et al. (2017b) |
| 21 | ACRIMA | Glaucoma | Ovreiu et al. (2021) |
| 22 | Augemnted ocular diseases AOD | Multi Classification of eye diseases | Бақтыбекұлы (2021) |
| 23 | JSIEC | Multi Classification of eye diseases | Cen et al. (2021) |
| 24 | Multi-Label Retinal Diseases | Multi Classification of eye diseases | Rodríguez et al. (2022) |
| 25 | RFMiD 2.0 | Multi Classification of eye diseases | Panchal et al. (2023) |
| 26 | ToxoFundus(Data Processed Paper) | Ocular toxoplasmosis | Cardozo et al. (2023) |
| 27 | ToxoFundus(Data Raw 6class All) | Ocular toxoplasmosis | Cardozo et al. (2023) |
| 28 | Adam dataset | Age-related Macular Degeneration | Liang (2021) |
| 29 | APTOS 2019 Blindness | Blindness Level Identification 0 4 | Karthik et al. (2019) |
| 30 | DRIMBD | Quality Testing of Retinal Images | Prentasic et al. (2013) |
| 31 | Glaucoma Detection | Glaucoma Classification | Zhang and Das (2022) |
| 32 | AIROGS | Glaucoma Classification | de Vente et al. (2023) |
| 33 | ICPR-HEp-2 | Multi Classification | Qi et al. (2016) |
| 34 | SICAPv2 | Cancer Degree Classification | Silva-Rodríguez et al. (2020) |
| 35 | Blood Cell Images | Blood Cell Classificaion (Multi) | Mooney (2017) |
| 36 | BreakHis | Cell type and beginormag | Bukun (2019) |
| 37 | Chaoyang | Multi Classification of pathologists | Zhu et al. |
| 38 | HuSHeM | Sperm Head Morphology Classificaion | Shaker (2018) |
| 39 | Bone Marrow Cell Classification | Bone Marrow Cell Classification | Matek et al. (2021) |
| 40 | NCT-CRC-HE-100K | Multi Classification | Kather et al. (2018) |
| 41 | Malignant Lymphoma Classification | Multi Classification | Orlov et al. (2010a) |
| 42 | Histopathologic Cancer Detection | Cancer Classification | Cukierski (2018) |
| 43 | LC25000 | Multi Classification of Lung and Colon | Zhu (2022) |
| 44 | Brain Tumor 17 Classes | Multi Classification | Feltrin (2022) |
| 45 | Tumor Classification | Pituitary or Glioma or Meningioma or Notumor | Nickparvar (2021a) |
| 46 | Malignant Lymphoma Classification | Multi Classification of eye diseases | Orlov et al. (2010b) |
| 47 | Retinal OCT-C8 | Multi Classification of eye diseases | Subramanian et al. (2022) |
| 48 | BUSI | Breast Cancer | Al-Dhabyani et al. (2020) |
| 49 | Digital Knee X-Ray Images | Degree Classification of Knee | Gornale and Patravali (2020) |
| 50 | Bone Fracture Multi-Region X-ray Data | Fractured Classification | Nickparvar (2021b) |
| 51 | Fracture detection | Fractured Classification | Batra (2024) |
| 52 | The vertebrae X-ray image | Vertebrae | Fraiwan et al. (2022) |
| 53 | Knee Osteoarthritis Dataset | Knee Osteoarthritis with severity grading | Chen (2018) |
| 54 | Shenzhen Chest X-Ray Set | COVID19, Classification Dataset. | Jaeger et al. (2014) |
| 55 | Chest X-ray PD | COVID and Pneumonia | Asraf and Islam (2021) |
| 56 | COVID-19 CHEST X-RAY DATABASE | COVID and Pneumonia | Chowdhury et al. (2020) |
| 57 | COVIDGR | COVID19, Classification | Tabik et al. (2020) |
| 58 | MIAS | Multi Classification of Breast | Mader (2017) |
| 59 | Tuberculosis Chest X-Ray Database | Tuberculosis | Rahman et al. (2020) |
| 60 | Pediatric Pneumonia Chest X-Ray | Pneumonia Classification | Kermany (2018) |
🔼 Table 10 provides detailed information on the 109 medical datasets used in the study. For each dataset, it lists the dataset name, a brief description of its contents (e.g., type of medical images, specific diseases or conditions), and the corresponding citation from the literature where the dataset is originally described. This table is crucial for understanding the breadth and diversity of the data used to train and evaluate the multimodal large language models (MLLMs) in the paper, particularly concerning compositional generalization.
read the caption
Table 10: The details of the medical datasets are provided
| No. | Name | Description | Citation |
|---|---|---|---|
| 61 | Random Sample of NIH Chest X-Ray Dataset | Multi Classificaiton of Chest | Wang et al. (2017) |
| 62 | CoronaHack-Chest X-Ray | Pnemonia Classifcition with Virus type | Praveen (2019) |
| 63 | Brain Tumor Dataset | Tumor Classification | Viradiya (2020) |
| 64 | Fitzpatrick 17k (Nine Labels) | Multi Classification | Groh et al. (2021) |
| 65 | BioMediTech | Multi Classification | Nanni et al. (2016) |
| 66 | Diabetic retinopathy | Diabetic Retinopathy Level | Benítez et al. (2021) |
| 67 | Leukemia | Cancer Classification | Codella et al. (2019) |
| 68 | ODIR-5K | Multiple Labels Classification | University (2019) |
| 69 | Arthrosis | Bone Age Classification | Zha (2021) |
| 70 | HSA-NRL | Multi Classification of pathologists | Zhu et al. (2021) |
| 71 | ISIC 2018 (Task 3) | Multi Classification | Codella et al. (2019) |
| 72 | ISIC 2017 (Task 3) | Multi Classification | Codella et al. (2018) |
| 73 | ChestX-Det | Multi Classification | Lian et al. (2021) |
| 74 | Monkeypox Skin Lesion Dataset | Only Monkeypox | Ali et al. (2022) |
| 75 | Cataract Dataset | Multi Classification | JR2NGB (2019) |
| 76 | ChestX-rays IndianaUniversity | Multi-label Classification | Raddar (2019) |
| 77 | CheXpert v1.0 small | Multi-label Classification | Arevalo (2020) |
| 78 | CBIS-DDSM | Multi Classification | Lee et al. (2017) |
| 79 | NLM-TB | Tuberculosis | Karaca (2022) |
| 80 | ChestXray-NIHCC | Multi-label Classification | Summers and Ronald (2020) |
| 81 | COVIDx CXR-4 | COVID19, Classification | Wang et al. (2020) |
| 82 | VinDr-Mammo | Multi-label Classification | Nguyen et al. (2023) |
| 83 | PBC dataset normal DIB | Multi Classification | Acevedo et al. (2020) |
| 84 | Human Protein Atlas | Multi-label Classification (Only green) | Le et al. (2022) |
| 85 | RSNA Pneumonia Detection Challenge 2018 | Multi-label Classification | Anouk Stein et al. (2018) |
| 86 | VinDr-SpineXR | Multi Classification of Bones Diseases | Pham et al. (2021) |
| 87 | VinDr-PCXR | Multi-label Classification | Pham et al. (2022) |
| 88 | PH2 | Melanoma Segmentation | Mendonca et al. (2015) |
| 89 | ISBI 2016 (Task3B) | Melanoma Segmentation | Gutman et al. (2016) |
| 90 | ISIC 2016 (Task 1) | Melanoma Segmentation | Gutman et al. (2016) |
| 91 | ISIC 2017 | Melanoma Segmentation | Codella et al. (2018) |
| 92 | CVC-ClinicDB | Polyp Segmentation | Bernal et al. (2015) |
| 93 | [Kvasir-SEG](https://datasets.simula.no/kvasir-seg/, https://github.com/DebeshJha/2020-MediaEval-Medico-polyp-segmentation/tree/master) | Polyp segmentation | Jha et al. (2020) |
| 94 | m2caiseg | Surgical Instrument Segmentation | Maqbool et al. (2020) |
| 95 | EDD 2020 | Multiple Diseases Segmentation in Intestine | Ali et al. (2020) |
| 96 | SICAPv2 | Cancer Cells Segmentation | Silva-Rodríguez et al. (2020) |
| 97 | BUSI | Cancer Segmentation | Hesaraki (2022) |
| 98 | TN3K | Thyroid Nodule Segmentation | Gong et al. (2022) |
| 99 | NLM-TB | Lung Segmentation (With left or right) | Gong et al. (2021) |
| 100 | VinDr-SpineXR | Spinal X-ray Anaomaly Detection | Pham et al. (2021) |
| 101 | VinDr-PCXR | Multiple Diseases Segmentation in Chest | Pham et al. (2022) |
| 102 | ChestX-Det | Multiple Diseases Segmentation in Chest | Lian et al. (2021) |
| 103 | UW-Madison Gl Tract Image Segmentation | Surgical Instrument Segmentation | Lee et al. (2024) |
| 104 | Duke Liver Dataset MRI v1 | Liver Segmentation | Macdonald et al. (2020) |
| 105 | Duke Liver Dataset MRI v2 | Liver Segmentation | Macdonald et al. (2020) |
| 106 | SIIM-ACR Pneumothorax Segmentation | Pneumothorax Segmentation | Zawacki et al. (2019) |
| 107 | FIVES | Fundus Vascular Segmentation | Jin et al. (2022) |
| 108 | RIM-ONE DL | Optic Disc and Cup Segmentation | Batista et al. (2020) |
| 109 | PALM19 | Optic Disc Segmentation | Fu et al. (2019) |
🔼 Table 11 provides a continuation of the dataset descriptions started in Table 10. It lists additional medical image datasets used in the study, detailing their names, descriptions of the medical tasks involved (e.g., classification, segmentation, detection), and the citation for each dataset’s source. The table is crucial for understanding the breadth and diversity of data used to evaluate the model’s capabilities and generalization performance.
read the caption
Table 11: Continued from Table 10.
Full paper#