TL;DR#
Retroperitoneal tumor segmentation is challenging due to the tumors’ irregular shapes, complex anatomy, and close proximity to vital organs. Manual segmentation is time-consuming and prone to errors. Current automated methods, like those using Transformers, often suffer from high computational costs and limitations in capturing long-range dependencies in images.
This research introduces ViLU-Net, a new architecture improving upon U-Net. ViLU-Net incorporates Vision-xLSTM modules, which are more efficient than transformers for long-range dependency modeling. The model is evaluated on a new dataset of retroperitoneal CT scans and a publicly available dataset, showing superior performance compared to other state-of-the-art methods. The authors made their code publicly available, facilitating further research and development.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image analysis and deep learning due to its introduction of a novel CT dataset for retroperitoneal tumor segmentation and a novel ViLU-Net architecture. The open-source code and improved accuracy in tumor segmentation make it highly relevant for clinical applications and future research, especially in exploring multi-modal approaches for enhanced segmentation performance.
Visual Insights#

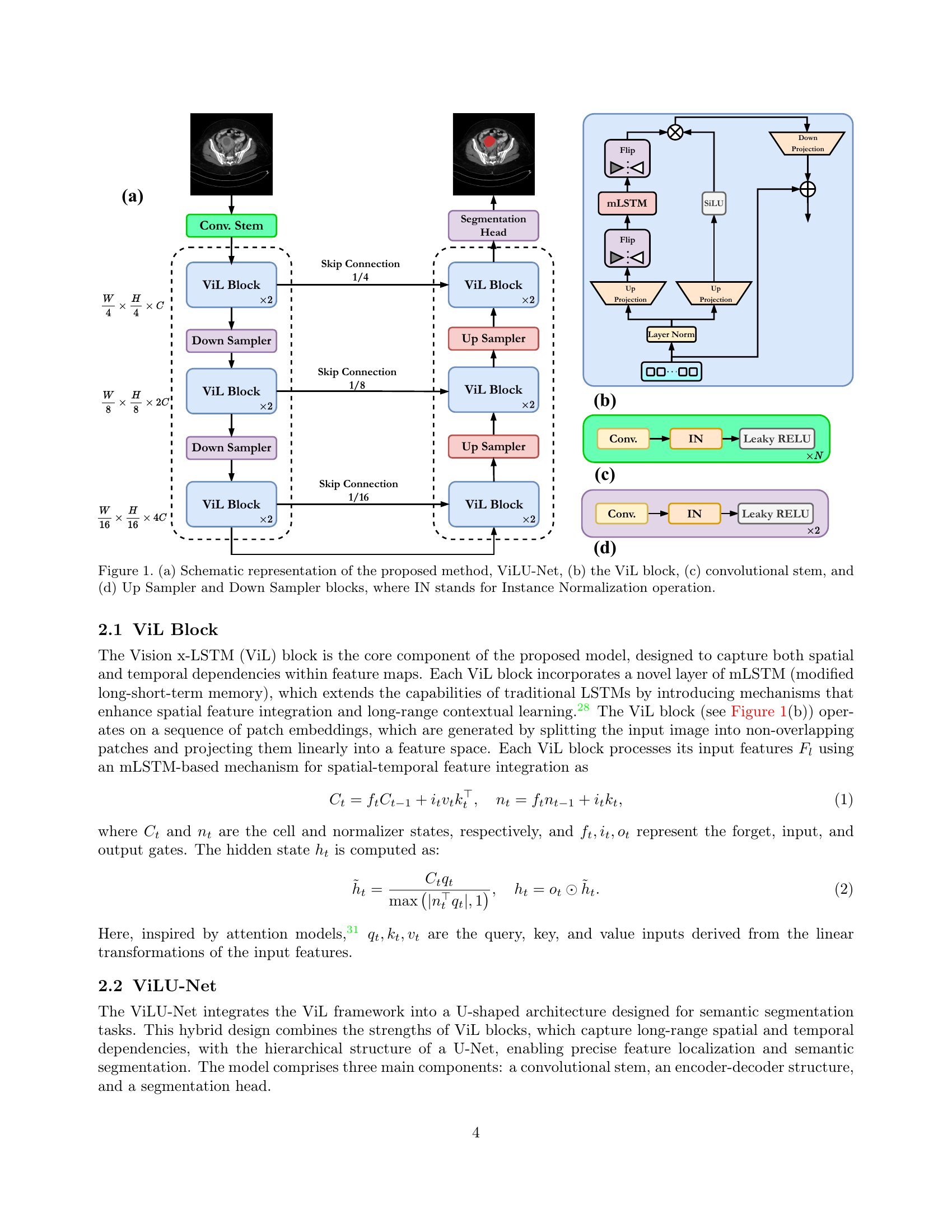
🔼 Figure 1 provides a detailed breakdown of the ViLU-Net architecture. (a) presents a high-level schematic of the entire network, showing the flow of data through the convolutional stem, encoder, decoder, and segmentation head. (b) zooms in on the ViL (Vision x-LSTM) block, the core component of the network, highlighting its internal structure which includes mLSTM (modified long short-term memory) layers for spatial and temporal feature integration. (c) illustrates the convolutional stem, the initial part of the network that processes the input image. Finally, (d) details the Up and Down Sampler blocks, crucial for the encoder and decoder’s functionality, respectively, showing how instance normalization (IN) is applied.
read the caption
Figure 1: (a) Schematic representation of the proposed method, ViLU-Net, (b) the ViL block, (c) convolutional stem, and (d) Up Sampler and Down Sampler blocks, where IN stands for Instance Normalization operation.
 |  |  |  |  |  |
 |  |  |  |  |  |
| (a) Input Image | (b) Ground Truth | (c) nnU-Net [33] | (d) SwinUNETR [32] | (e) U-Mamba [27] | (f) ViLU-Net (Ours) |
🔼 This table presents a quantitative comparison of different deep learning models’ performance on a 3D abdominal organ segmentation task using a CT dataset. Metrics include Dice Similarity Coefficient (DSC), Normalized Surface Distance (NSD), Hausdorff Distance (HD), and Intersection over Union (IoU). The results are shown for several state-of-the-art U-Net variants (nnU-Net, SwinUNETR, and U-Mamba) and the authors’ proposed ViLU-Net method. This allows for a direct comparison of the effectiveness and accuracy of each model in segmenting various abdominal organs from 3D CT scan data.
read the caption
Table 1: Results summary of 3D organ segmentation on the abdomen CT dataset.
In-depth insights#
Retroperitoneal AI#
Retroperitoneal AI represents a significant advancement in medical image analysis, particularly focusing on the challenges of identifying and segmenting retroperitoneal tumors. The complexity of the retroperitoneal space, with its close proximity to critical structures, makes manual segmentation extremely time-consuming and error-prone. AI-powered solutions offer the potential to automate this process, leading to faster and more accurate diagnoses and treatment planning. However, developing robust and reliable AI models requires addressing the unique challenges of retroperitoneal imaging, including the variability in tumor size, shape, and location, and the presence of artifacts and noise. The success of such AI models heavily relies on access to high-quality, annotated datasets, which are often scarce and difficult to obtain. Future research should focus on developing more efficient and accurate AI models, potentially through the use of hybrid deep learning architectures that combine the strengths of convolutional neural networks and transformers, and the exploration of multi-modal imaging data to improve diagnostic accuracy and patient outcomes.
U-Net Variants#
The exploration of U-Net variants reveals a rich landscape of architectural modifications designed to improve performance and efficiency in medical image segmentation. Core innovations involve integrating advanced components like Vision Transformers (ViTs) for enhanced global context modeling, or State Space Models (SSMs) such as Mamba, to address the computational limitations of traditional Transformers, thereby enabling scalability and efficient processing, particularly crucial for handling high-resolution medical images. The use of Extended Long Short-Term Memory (xLSTM) networks offers another compelling alternative by providing a competitive edge in sequence modeling with linear computational and memory efficiency, complementing the strengths of CNNs. These modifications aim to tackle challenges posed by the complex anatomy, irregular shapes, and critical structure associations often present in medical images, ultimately improving the accuracy and speed of tumor segmentation, while mitigating the issues associated with manual segmentation. ViLU-Net, a novel architecture proposed in the study, provides a prime example of combining these advancements, highlighting the synergy of ViTs and xLSTMs within a U-Net framework. This ongoing research and development of U-Net variants underscore a persistent pursuit of robust and efficient segmentation tools for addressing diverse challenges in medical imaging.
xLSTM in U-Net#
The integration of xLSTM (Extended Long Short-Term Memory) within the U-Net architecture presents a compelling approach to medical image segmentation. xLSTM’s ability to efficiently handle long-range dependencies offers a significant advantage over traditional LSTMs, particularly in processing high-resolution medical images where capturing contextual information across large spatial distances is crucial. By incorporating xLSTM blocks, the modified U-Net can better model complex spatial relationships within the image, leading to more accurate and detailed segmentations. This is particularly beneficial for tasks like tumor segmentation where precise boundary delineation is essential and the shapes of tumors can be irregular. Furthermore, xLSTM’s linear computational complexity compared to the quadratic scaling of self-attention mechanisms in Transformers makes it a more practical and scalable choice for deployment on resource-constrained clinical settings. The combination of U-Net’s powerful encoder-decoder structure with xLSTM’s efficient long-range dependency modeling thus creates a robust and efficient framework capable of delivering high-performing segmentations while being suitable for practical applications. The results demonstrated in the study highlight xLSTM’s potential to improve both the accuracy and computational efficiency of U-Net for various medical image segmentation tasks.
Abdomen CT Results#
The abdomen CT results section likely presents a quantitative analysis of the proposed ViLU-Net model’s performance on a publicly available abdominal CT dataset, comparing it against other state-of-the-art (SOTA) methods such as nnU-Net, SwinUNETR, and U-Mamba. Key metrics like Dice Similarity Coefficient (DSC), Normalized Surface Distance (NSD), Hausdorff Distance (HD), and Intersection over Union (IoU) would be reported to provide a comprehensive evaluation. A high DSC score, indicating strong overlap between automated and manual segmentations, would suggest successful performance. Low NSD and HD scores, representing the average and maximum distances between the segmented surfaces, respectively, would signify accurate boundary delineation. A high IoU score would further confirm the model’s precision. The results likely demonstrate ViLU-Net’s competitive performance compared to SOTA models, possibly achieving superior accuracy in specific aspects or for particular organs, thereby highlighting the model’s efficacy in abdominal organ segmentation and its potential clinical relevance. The section might also include visual examples showcasing qualitative differences between the segmentation maps generated by various models, further strengthening the analysis and enhancing the readers’ understanding.
Future Directions#
The “Future Directions” section of this research paper would naturally focus on extending the capabilities of the ViLU-Net model and exploring its clinical applications. Multi-modal fusion, integrating data from MRI, PET, and CT scans, is a crucial direction. This would provide a more complete picture of a patient’s condition and potentially improve segmentation accuracy. Another key area would be improving computational efficiency to enable real-time processing, crucial for clinical workflows. Addressing challenges of data heterogeneity, like variations in image resolution and scanner types, is essential. Furthermore, research should concentrate on developing robust methods to handle noisy or incomplete datasets. This will enhance the generalizability and robustness of ViLU-Net for diverse clinical scenarios. Finally, exploring interpretability methods is important to build trust and provide clinicians with insights into the model’s decision-making process. This could involve techniques like visualizing attention maps or generating explainable AI (XAI) outputs.
More visual insights#
More on figures

🔼 Figure 2 presents a visual comparison of abdominal organ segmentation results obtained using different methods. The images show the ground truth segmentations alongside the results produced by various methods, including ViLU-Net, a novel U-Net modification using Vision-xLSTM blocks. The comparison highlights the ability of ViLU-Net to accurately segment intricate soft tissues and structures within the abdominal region, exhibiting superior performance compared to other methods.
read the caption
Figure 2: Visualized segmentation examples of abdominal organ segmentation in CT. The ViLU-Net excels at differentiating intricate soft tissues within the abdominal region.

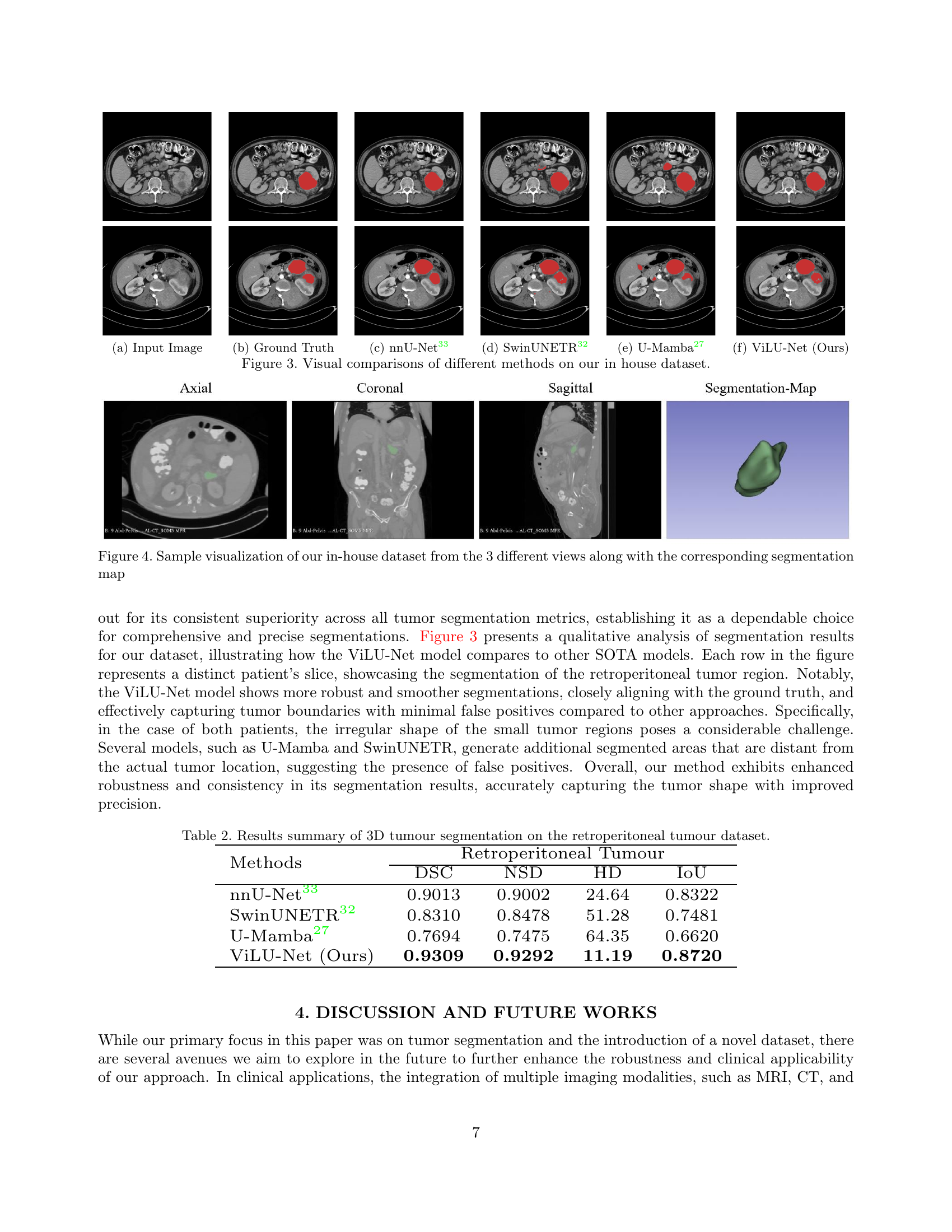
🔼 Figure 3 presents a qualitative comparison of different deep learning models for retroperitoneal tumor segmentation on a custom dataset. The figure displays example slices (axial, coronal, and sagittal views) from the dataset, showing the input CT image, the ground truth segmentation, and the segmentation masks generated by several methods (nnU-Net, SwinUNETR, U-Mamba, and the proposed ViLU-Net). This allows for a direct visual assessment of the relative performance of each method in terms of accuracy, boundary precision, and ability to handle irregular tumor shapes. The results highlight ViLU-Net’s superior performance in accurately segmenting tumor regions with irregular shapes compared to other methods.
read the caption
Figure 3: Visual comparisons of different methods on our in house dataset.
Full paper#