TL;DR#
Generating 3D molecules is vital for drug discovery, yet it’s challenging due to the need to manage atom types, bonds, and 3D coordinates while maintaining SE(3) equivariance. Existing methods use separate latent spaces, reducing efficiency. They often rely on intricate networks to ensure the geometric and rotational invariance that is required. Moreover, prior works overlooked the reconstruction error, leading to stability issues in generated molecules. This leads to models being complex while at the same time not very accurate.
To solve these challenges, the authors propose Unified Variational Auto-Encoder for 3D Molecular Latent Diffusion Modeling (UAE-3D). UAE-3D compresses 3D molecules into a unified latent space, ensuring near-zero reconstruction error and employing SE(3) augmentations to ensure equivariance without complex baked-in designs. The model integrates modalities through a Relational Transformer and uses a Diffusion Transformer (DiT) for latent generation. This unified approach allows the model to outperform previous methods. The experiments done in GEOM-Drugs and QM9 datasets demonstrates that UAE-3D sets new benchmarks in efficiency and quality for both de novo and conditional generation of 3D molecules.
Key Takeaways#
Why does it matter?#
This research introduces UAE-3D for unified molecular representation, significantly improving 3D molecule generation. It offers enhanced efficiency and geometric accuracy, setting new benchmarks and opening avenues for advanced molecular design and drug discovery research.
Visual Insights#

🔼 Figure 1 illustrates the different approaches to 3D molecular representation. (a) shows the three distinct modalities of a 3D molecule: atom types, chemical bonds, and 3D coordinates. (b) depicts conventional methods that use separate latent spaces for each modality, resulting in fragmented representations. This separation is often done because the 3D coordinates require SE(3) equivariance while the others do not. (c) introduces the proposed UAE-3D model, which uses a unified latent space to jointly encode all three modalities, maintaining SE(3) equivariance for the 3D coordinates.
read the caption
Figure 1: (a) A 3D molecule is characterized by features of distinct modalities. (b) Conventional approaches use separate latent spaces for equivariant (3D coordinates) and invariant (2D features) components, leading to fragmented molecular representations. (c) Our UAE-3D establishes a unified SE(3)-equivariant latent space that jointly encodes both modalities
| Metric | GeoLDM Xu et al. (2023) | UAE-3D (Ours) |
| Atom Accuracy (%) | 98.6 | 100.0 |
| Bond Accuracy (%) | 96.2 | 100.0 |
| Coordinate RMSD (Å) | 0.1830 | 0.0002 |
🔼 This table compares the performance of UAE-3D and GeoLDM’s VAEs in reconstructing 3D molecular structures. It shows the reconstruction accuracy for atom types (Atom Accuracy), bond types (Bond Accuracy), and the geometric deviation between the reconstructed and ground truth 3D coordinates (Coordinate RMSD). Lower RMSD values indicate better geometric accuracy. The metrics provide a detailed assessment of the models’ ability to accurately capture the structural information of 3D molecules.
read the caption
Table 1: Comparing the reconstruction error between UAE-3D and GeoLDM’s VAE. Atom/Bond accuracy measures correct atom/bond type predictions. Coordinate RMSD quantifies geometric deviation from the ground truth 3D coordinates.
In-depth insights#
Unified Latents#
The concept of unified latents in molecular representation learning signifies a shift towards more integrated and efficient models. Instead of maintaining separate latent spaces for different modalities like atom types, bond information, and 3D coordinates, a unified latent space aims to encode all these aspects into a single, compressed representation. This approach has several potential benefits. First, it can lead to more compact models with fewer parameters, reducing computational costs during training and inference. Second, it may facilitate better capture of the complex relationships between different molecular properties. For example, the 3D geometry of a molecule is often closely related to its chemical properties and reactivity. By encoding all of these aspects into a single latent space, the model can more easily learn these relationships and generate molecules with desired properties. However, creating an effective unified latent space is challenging. It requires carefully designing the model architecture and loss function to ensure that all the important information is encoded and that the resulting latent space is smooth and well-behaved. Furthermore, it is important to maintain SE(3) equivariance, which means that the model’s predictions should be invariant to rotations and translations of the input molecule. Despite these challenges, the potential benefits of unified latents make it an important area of research in molecular representation learning.
SE(3) Equivariance#
SE(3) equivariance is crucial for molecular modeling because it ensures that the model’s predictions are invariant to rotations and translations of the input molecule. In essence, if you rotate or translate a molecule, the model should still predict the same properties or generate a structurally equivalent molecule. Failing to account for this can lead to inaccurate results and physically unrealistic structures. Many approaches bake in 3D equivariance into the neural network, while UAE-3D trains a neural network to “learn” 3D equivariance through tailor-made SE(3) augmentations, encouraging the transformation on the input coordinates are reflected equivariantly on the output coordinates.
Diffusion Models#
Diffusion models progressively add noise to data until it becomes pure noise, then learn to reverse this process to generate new samples. This offers a flexible generative framework, operating by iteratively refining an initial random sample. In molecular generation, diffusion models have shown promise by offering a way to produce complex 3D structures, but challenges remain in ensuring chemical validity. By starting with noise and gradually building structure, they avoid some limitations of other generative methods. They often work in conjunction with variational autoencoders (VAEs), which compress data into a latent space where the diffusion process takes place, improving efficiency and stability.
Geometric Accuracy#
Geometric accuracy in molecular generation is crucial for producing realistic and stable 3D structures. Achieving high geometric accuracy means the generated molecules closely adhere to known chemical principles and spatial constraints. This involves precisely predicting bond lengths, bond angles, and dihedral angles, ensuring that the molecule’s conformation is energetically favorable and chemically valid. Models with poor geometric accuracy may produce molecules with strained bonds or unfavorable conformations, leading to instability or non-existence. Advancements in geometric accuracy directly impact the usefulness of generated molecules in downstream applications like drug discovery and material design, where precise 3D structure is essential for accurate binding predictions and property estimations. The breakthrough lies in using unified latent spaces which are highly effective for 3D geometric modeling for accurate 3D molecule generation.
Efficient Models#
The pursuit of efficient models is central to modern machine learning, especially when dealing with resource-intensive tasks like 3D molecular generation. Efficiency can refer to several aspects: training time, sampling speed, and computational cost. For 3D molecules, managing the complexity of multi-modal data (atom types, bonds, coordinates) while maintaining equivariance is a significant challenge. Efficient models might involve decoupling the training process, as seen in the paper, where a VAE is trained separately before the diffusion model. This allows for targeted optimization and can significantly reduce overall training time. Simplified architectures and unified latent spaces contribute to efficient sampling by reducing the computational overhead during inference. Techniques like adaptive layer normalization can further enhance diffusion model efficiency by dynamically adjusting to varying noise scales.
More visual insights#
More on figures

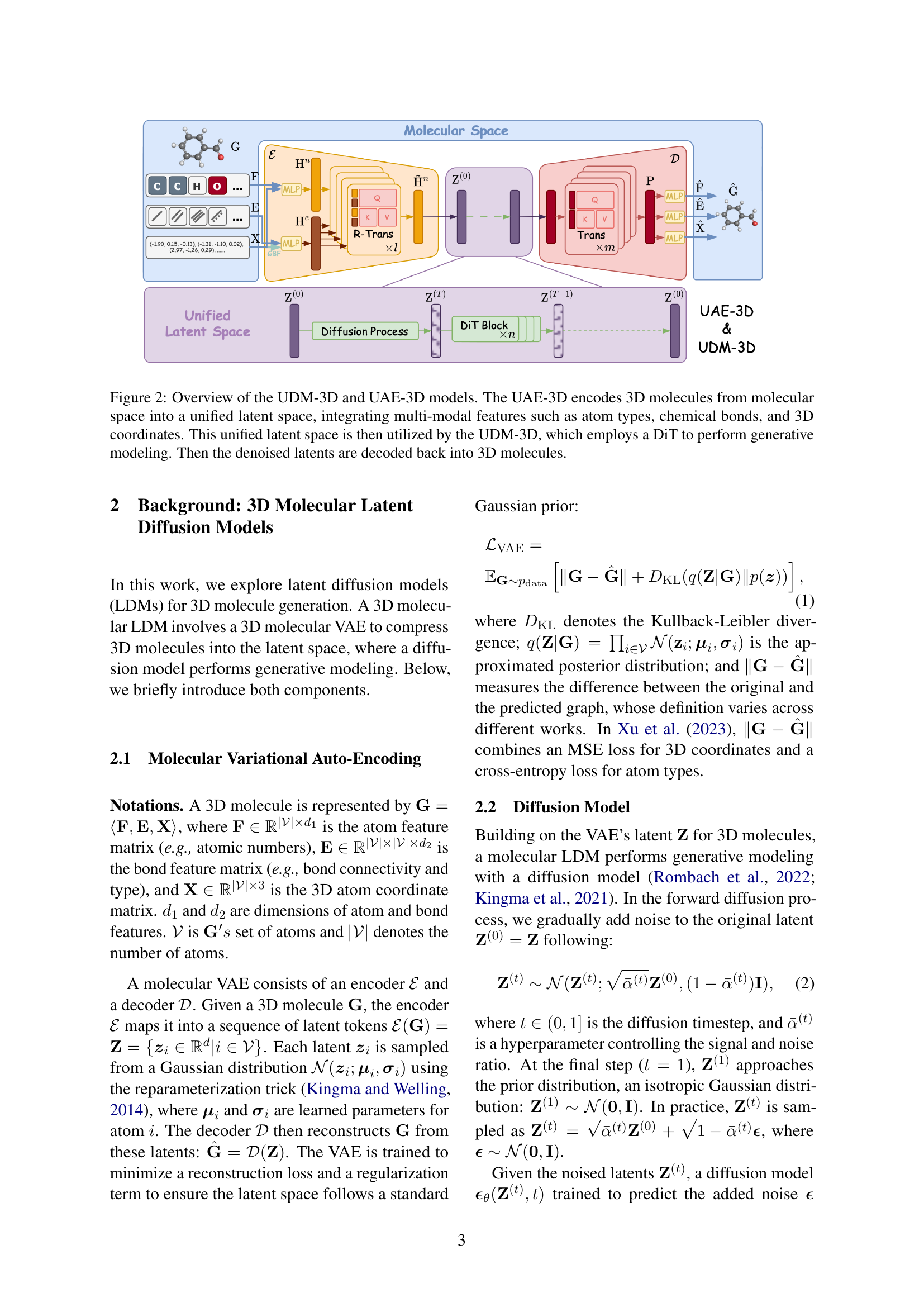
🔼 The figure illustrates the architecture of the UDM-3D model, which consists of two main components: UAE-3D and DiT. UAE-3D acts as a Variational Autoencoder (VAE), taking as input a 3D molecule represented by its multi-modal features (atom types, chemical bonds, and 3D coordinates). It encodes these features into a unified latent space. This latent representation is then fed into the DiT (Diffusion Transformer), a diffusion model that performs generative modeling in the latent space. The DiT outputs denoised latent representations, which are subsequently decoded by the UAE-3D’s decoder to generate a new 3D molecule.
read the caption
Figure 2: Overview of the UDM-3D and UAE-3D models. The UAE-3D encodes 3D molecules from molecular space into a unified latent space, integrating multi-modal features such as atom types, chemical bonds, and 3D coordinates. This unified latent space is then utilized by the UDM-3D, which employs a DiT to perform generative modeling. Then the denoised latents are decoded back into 3D molecules.

🔼 The bar chart compares the training time of four different models: EDM, GeoLDM, JODO, and the proposed UDM-3D model. UDM-3D demonstrates a significantly shorter training time compared to the other three models, highlighting its efficiency. The total training time for UDM-3D is broken down into the time spent training the Variational Autoencoder (VAE, UAE-3D) and the Diffusion Transformer (DiT, UDM-3D) separately.
read the caption
(a) Training time comparison.

🔼 The bar chart compares the sampling speed of different models for generating 3D molecules. UDM-3D is significantly faster than other models such as EDM, GeoLDM, and JODO, demonstrating its enhanced efficiency in generating 3D molecules.
read the caption
(b) Sampling speed comparison.
More on tables
| 2D-Metric | AtomStable | V&C | V&U | V&U&N | SNN | Frag | Scaf | FCD |
| Train | 1.000 | 1.000 | 1.000 | 0.000 | 0.585 | 0.999 | 0.584 | 0.251 |
| CDGS | 0.991 | 0.285 | 0.285 | 0.285 | 0.262 | 0.789 | 0.022 | 22.051 |
| JODO | 1.000 | 0.874 | 0.905 | 0.902 | 0.417 | 0.993 | 0.483 | 2.523 |
| MiDi* | 0.968 | 0.633 | 0.654 | 0.652 | 0.392 | 0.951 | 0.196 | 7.054 |
| UDM-3D, ours | 1.000 | 0.879 | 0.913 | 0.958 | 0.525 | 0.989 | 0.540 | 0.692 |
| 3D-Metric | AtomStable | FCD | Bond length | Bond angle | Dihedral angle | |||
| Train | 0.861 | 13.73 | 1.56E-04 | 1.81E-04 | 1.56E-04 | |||
| EDM | 0.831 | 31.29 | 4.29E-01 | 4.96E-01 | 1.46E-02 | |||
| JODO | 0.845 | 19.99 | 8.49E-02 | 1.15E-02 | 6.68E-04 | |||
| MiDi* | 0.750 | 23.14 | 1.17E-01 | 9.57E-02 | 4.46E-03 | |||
| GeoLDM | 0.843 | 30.68 | 3.91E-01 | 4.22E-01 | 1.69E-02 | |||
| UDM-3D, ours | 0.852 | 17.36 | 9.89E-03 | 5.11E-03 | 1.74E-04 | |||
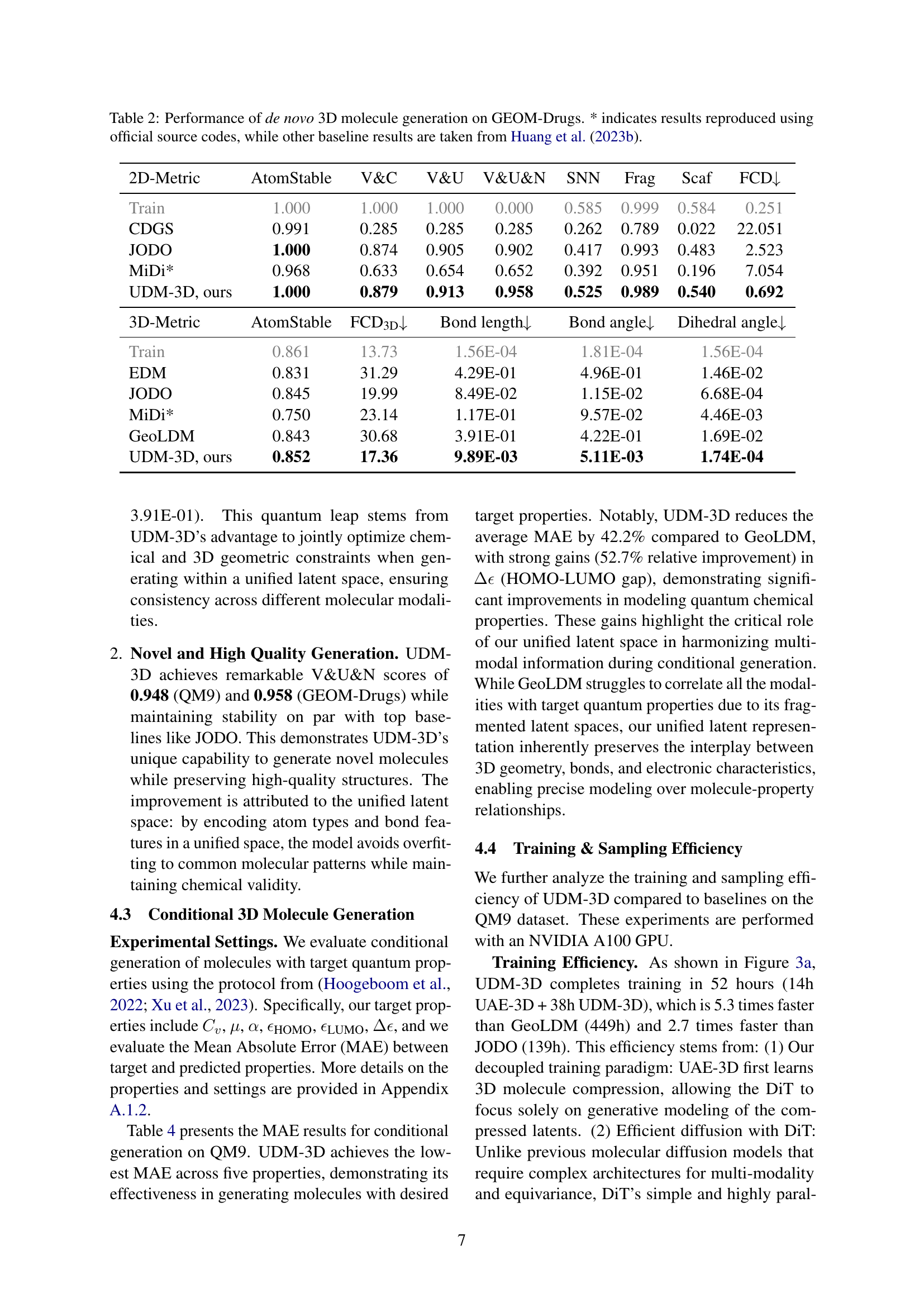
🔼 Table 2 presents a comprehensive comparison of various de novo 3D molecular generation models on the GEOM-Drugs dataset. The table evaluates the performance of different models using a range of 2D and 3D metrics. 2D metrics assess aspects such as the chemical validity, uniqueness, and novelty of the generated molecules. 3D metrics focus on the accuracy of the generated 3D structures. The results highlight the performance of the proposed UDM-3D model against existing state-of-the-art methods. The asterisk (*) indicates that the results for certain baselines were reproduced using the original source codes, ensuring a fair comparison. Other baseline results were obtained from Huang et al. (2023b).
read the caption
Table 2: Performance of de novo 3D molecule generation on GEOM-Drugs. * indicates results reproduced using official source codes, while other baseline results are taken from Huang et al. (2023b).
| 2D-Metric | AtomStable | V&C | V&U | V&U&N | SNN | Frag | Scaf | FCD |
| Train | 0.999 | 0.989 | 0.989 | 0.000 | 0.490 | 0.992 | 0.946 | 0.063 |
| CDGS | 0.997 | 0.951 | 0.936 | 0.860* | 0.493 | 0.973 | 0.784 | 0.798 |
| JODO | 0.999 | 0.990 | 0.960 | 0.780* | 0.522 | 0.986 | 0.934 | 0.138 |
| MiDi* | 0.998 | 0.980 | 0.954 | 0.769 | 0.501 | 0.979 | 0.882 | 0.187 |
| UDM-3D, ours | 0.999 | 0.983 | 0.972 | 0.948 | 0.508 | 0.987 | 0.897 | 0.161 |
| 3D-Metric | AtomStable | FCD | Bond length | Bond angle | Dihedral angle | |||
| Train | 0.994 | 0.877 | 5.44E-04 | 4.65E-04 | 1.78E-04 | |||
| EDM | 0.986 | 1.285 | 1.30E-01 | 1.82E-02 | 6.64E-04 | |||
| MDM | 0.992 | 4.861 | 2.74E-01 | 6.60E-02 | 2.39E-02 | |||
| JODO | 0.992 | 0.885 | 1.48E-01 | 1.21E-02 | 6.29E-04 | |||
| MiDi* | 0.983 | 1.100 | 8.96E-01 | 2.08E-02 | 8.14E-04 | |||
| GeoLDM | 0.989 | 1.030 | 2.40E-01 | 1.00E-02 | 6.59E-04 | |||
| UDM-3D, ours | 0.993 | 0.881 | 7.04E-02 | 9.84E-03 | 3.47E-04 | |||
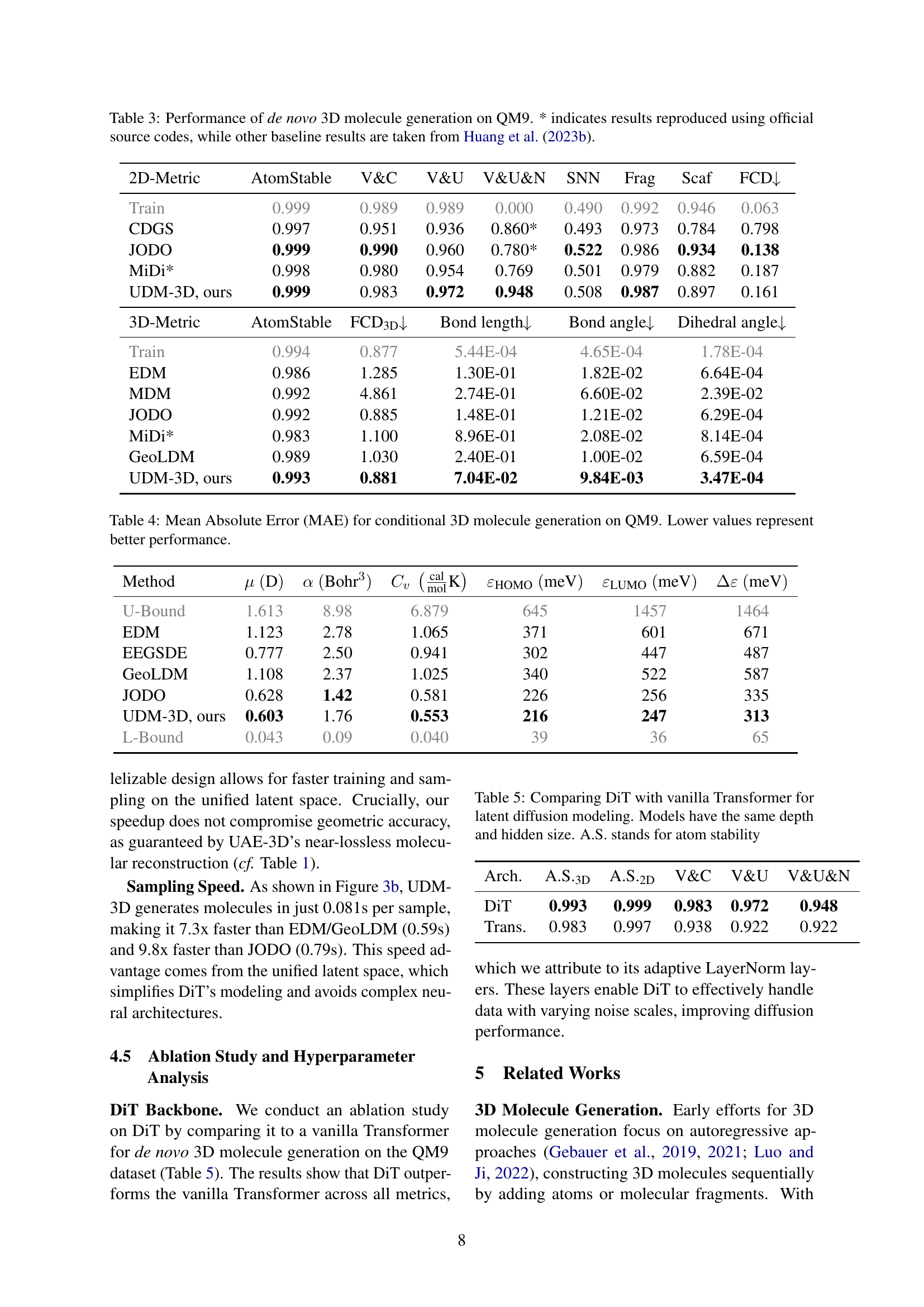
🔼 This table presents the results of de novo 3D molecule generation on the QM9 dataset. It compares the performance of the proposed UDM-3D model against several state-of-the-art baselines. The metrics assess both 2D structural validity (atom stability, validity and completeness, etc.) and 3D geometric accuracy (bond length, bond angle, dihedral angle distributions). The asterisk (*) indicates that results for certain baselines were reproduced using the official source code, whereas other results were obtained from a publication by Huang et al. (2023b). This allows for a more robust and accurate comparison of model performance.
read the caption
Table 3: Performance of de novo 3D molecule generation on QM9. * indicates results reproduced using official source codes, while other baseline results are taken from Huang et al. (2023b).
| Method | ||||||
| U-Bound | 1.613 | 8.98 | 6.879 | 645 | 1457 | 1464 |
| EDM | 1.123 | 2.78 | 1.065 | 371 | 601 | 671 |
| EEGSDE | 0.777 | 2.50 | 0.941 | 302 | 447 | 487 |
| GeoLDM | 1.108 | 2.37 | 1.025 | 340 | 522 | 587 |
| JODO | 0.628 | 1.42 | 0.581 | 226 | 256 | 335 |
| UDM-3D, ours | 0.603 | 1.76 | 0.553 | 216 | 247 | 313 |
| L-Bound | 0.043 | 0.09 | 0.040 | 39 | 36 | 65 |
🔼 This table presents the Mean Absolute Error (MAE) achieved by different methods on the QM9 dataset for conditional 3D molecule generation. The goal is to generate 3D molecules with specific target quantum chemical properties. Lower MAE values indicate better performance, meaning the generated molecules’ properties are closer to the target values. The properties considered include dipole moment (μ), polarizability (α), heat capacity (Cv), HOMO energy (εHOMO), LUMO energy (εLUMO), and HOMO-LUMO gap (Δε).
read the caption
Table 4: Mean Absolute Error (MAE) for conditional 3D molecule generation on QM9. Lower values represent better performance.
| Arch. | A.S. | A.S. | V&C | V&U | V&U&N |
| DiT | 0.993 | 0.999 | 0.983 | 0.972 | 0.948 |
| Trans. | 0.983 | 0.997 | 0.938 | 0.922 | 0.922 |
🔼 This table compares the performance of two different model architectures, Diffusion Transformer (DiT) and a vanilla Transformer, when used for latent diffusion modeling in 3D molecular generation. Both models have the same depth and hidden size to ensure a fair comparison. The results are evaluated based on atom stability (A.S.) in 2D and 3D spaces, and other key metrics for the validity and uniqueness of generated molecules: V&C, V&U, and V&U&N.
read the caption
Table 5: Comparing DiT with vanilla Transformer for latent diffusion modeling. Models have the same depth and hidden size. A.S. stands for atom stability
Full paper#