TL;DR#
Medical image segmentation demands specialized models. Large foundation models offer flexibility but are costly to fine-tune. Parameter-Efficient Fine-Tuning (PEFT) methods like LORA update weights efficiently but can underfit. SVD methods comprehensively update but lack flexibility. The complex nature of medical image segmentation calls for models that are specifically designed to capture detailed, domain-specific features. Large foundation models offer considerable flexibility, yet the cost of fine-tuning these models remains a significant barrier.
This paper introduces SALT, a method that adapts singular values using trainable parameters and low-rank updates. This hybrid approach combines the advantages of LoRA and SVD, enabling effective adaptation without increasing model size or depth. Evaluated on five medical datasets, SALT outperforms state-of-the-art PEFT methods by 2-5% in Dice with only 3.9% trainable parameters. The proposed approach enables effective adaptation without relying on increasing model size or depth.
Key Takeaways#
Why does it matter?#
This paper introduces SALT, a parameter-efficient fine-tuning method, that can enhance medical image segmentation. The proposed solution addresses the limitations of existing methods and offers a promising direction for future research in medical imaging and foundation model adaptation.
Visual Insights#

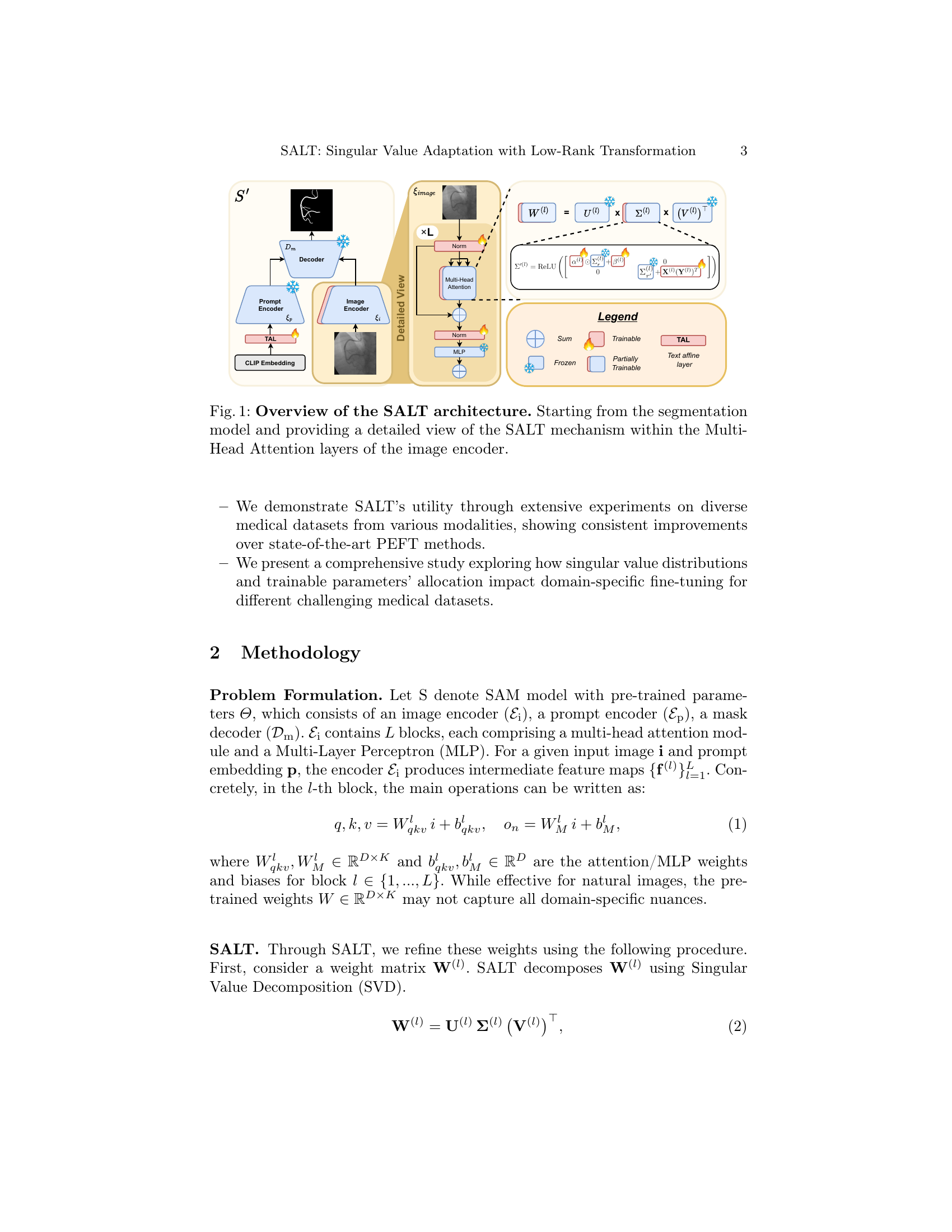
🔼 Figure 1 illustrates the SALT (Singular Value Adaptation with Low-Rank Transformation) architecture, which enhances the Segment Anything Model (SAM) for medical image segmentation. The diagram shows the overall model workflow, starting with the input image and prompt, progressing through the image encoder, Multi-Head Attention layers, and decoder to produce the final segmentation mask. The detailed view focuses on the SALT mechanism integrated into the Multi-Head Attention layers of the image encoder. This mechanism is key to the algorithm’s parameter efficiency and ability to adapt the SAM to the unique characteristics of medical images. SALT achieves this by selectively scaling and shifting the most influential singular values while simultaneously adding a low-rank update for the remaining values, thus improving performance without increasing the overall model size.
read the caption
Figure 1: Overview of the SALT architecture. Starting from the segmentation model and providing a detailed view of the SALT mechanism within the Multi-Head Attention layers of the image encoder.
| Model | Rank | %Trainable | ARCADE | DIAS | ROSE | XRay-Angio | Drive | Average | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dice | HD95 | Dice | HD95 | Dice | HD95 | Dice | HD95 | Dice | HD95 | Dice | HD95 | |||
| Traditional DL Methods | ||||||||||||||
| U-Net [20] | – | 100% / 6.5M | 0.36 | 157.14 | 0.13 | 262.66 | 0.63 | 8.07 | 0.71 | 30.40 | 0.18 | 84.43 | 0.40 | 108.54 |
| UNETR [5] | – | 100% / 91M | 0.48 | 162.44 | 0.31 | 183.97 | 0.63 | 8.96 | 0.74 | 20.62 | 0.46 | 33.58 | 0.52 | 81.91 |
| RegUnet [20] | – | 100% / 7.8M | 0.11 | 247.79 | 0.35 | 172.09 | 0.22 | 58.08 | 0.42 | 79.51 | 0.43 | 72.97 | 0.31 | 126.08 |
| SegNet [1] | – | 100% / 29M | 0.14 | 231.11 | 0.13 | 295.06 | 0.38 | 9.09 | 0.09 | 110.94 | 0.14 | 79.03 | 0.18 | 145.05 |
| DeepLabV3+ [3] | – | 100% / 45M | 0.55 | 103.22 | 0.55 | 77.66 | 0.52 | 12.90 | 0.74 | 14.03 | 0.46 | 27.79 | 0.56 | 47.12 |
| Segformer [24] | – | 100% / 43M | 0.53 | 113.98 | 0.57 | 73.64 | 0.56 | 11.21 | 0.75 | 16.55 | 0.48 | 36.98 | 0.58 | 50.47 |
| SAM-Based PEFT Methods (241M) | ||||||||||||||
| SAM w/ text prompts | – | – | 0.00 | - | 0.00 | - | 0.00 | - | 0.04 | - | 0.00 | - | 0.01 | - |
| S-SAM [16] | – | 0.40% / 1M | 0.76 | 52.44 | 0.67 | 42.36 | 0.65 | 12.41 | 0.75 | 27.56 | 0.70 | 15.81 | 0.71 | 30.12 |
| LoRA [7] | 4 | 0.64% / 1.5M | 0.78 | 39.58 | 0.61 | 27.81 | 0.59 | 16.26 | 0.73 | 26.78 | 0.62 | 21.04 | 0.66 | 26.29 |
| LoRA [7] | 256 | 14.08% / 33.9M | 0.81 | 36.89 | 0.63 | 34.36 | 0.62 | 14.62 | 0.76 | 24.84 | 0.68 | 19.00 | 0.70 | 25.94 |
| SALT (ours) | 4 | 0.46% / 1.1M | 0.78 | 51.74 | 0.69 | 39.58 | 0.66 | 12.19 | 0.75 | 29.55 | 0.72 | 16.04 | 0.72 | 29.82 |
| SALT (ours) | 256 | 3.9% / 9.4M | 0.81 | 42.50 | 0.71 | 27.35 | 0.67 | 12.64 | 0.77 | 23.67 | 0.75 | 13.20 | 0.74 | 23.87 |
🔼 This table presents a quantitative comparison of different deep learning (DL) models and parameter-efficient fine-tuning (PEFT) methods for medical image segmentation. It evaluates the performance of traditional DL models (U-Net, UNETR, RegUnet, SegNet, DeepLabV3+, Segformer) against SAM-based PEFT approaches (S-SAM and LoRA), and finally the proposed SALT method. The models are assessed across five diverse medical image segmentation datasets, using Dice coefficient (Dice) and 95th percentile Hausdorff Distance (HD95) as evaluation metrics. The table displays the Dice scores (higher is better) and HD95 values (lower is better) achieved by each method on each dataset, alongside the percentage of trainable parameters for each PEFT method. This allows for comparison of performance relative to the number of parameters modified during fine-tuning.
read the caption
Table 1: Performance comparison of traditional DL and SAM-based PEFT methods using Dice and HD95 metrics on a variety of medical segmentation datasets.
In-depth insights#
Efficient Tuning#
Efficient tuning is critical in adapting large models to specific tasks, especially when computational resources are limited. Parameter-efficient fine-tuning (PEFT) methods, like LoRA, address this by updating only a small subset of parameters, reducing computational overhead. However, PEFT methods may struggle to capture domain-specific nuances with very low ranks, leading to underfitting. The challenge lies in balancing parameter efficiency with the ability to effectively learn complex patterns. Methods like SVD offer comprehensive updates but can lack flexibility and show variable performance. A hybrid approach could offer benefits, selectively updating the most crucial parameters while using low-rank adaptations for the remaining subspace, potentially achieving a better trade-off between efficiency and accuracy. The goal is robust adaptation without significantly increasing model size.
SALT Architecture#
SALT’s architecture, as depicted in the research paper, presents a novel approach to parameter-efficient fine-tuning, particularly in the context of medical image segmentation. The core idea revolves around selectively adapting the most influential singular values obtained through Singular Value Decomposition (SVD) of weight matrices within a pre-trained model, such as the Segment Anything Model (SAM). This adaptation is achieved using trainable scale and shift parameters, allowing for fine-grained control over the contribution of these dominant singular values. Complementing this selective adaptation is a low-rank update applied to the remaining subspace, leveraging techniques like Low-Rank Adaptation (LoRA). This hybrid approach intelligently combines the strengths of both SVD and LoRA, enabling effective domain adaptation without drastically increasing the number of trainable parameters or relying on deeper or wider models. By strategically allocating trainable parameters to the most critical components of the weight matrices, SALT achieves a balance between preserving pre-trained knowledge and capturing domain-specific nuances, ultimately leading to improved segmentation performance on medical images.
Medical PEFT SOTA#
The paper addresses the challenge of adapting large foundation models, like SAM, to medical image segmentation through parameter-efficient fine-tuning (PEFT). It acknowledges that while models like U-Net and nnU-Net achieve SOTA performance, they often rely on large parameter counts, limiting adaptability. The limitations of directly applying SAM to medical data are also highlighted, showing inferiority to specialized models. Existing PEFT methods, such as LoRA, may struggle to capture both dominant and nuanced features specific to medical images. To overcome these, the paper introduces a novel PEFT framework that synergizes SVD and low-rank adaptation. The SALT approach selectively scales critical singular values, while applying trainable low-rank transformations to residual components. The research emphasizes balancing minimal parameter overhead, computational feasibility, and preserving pre-trained knowledge while addressing unique medical imaging challenges. This enables efficient adaptation to medical domains with minimal parameter overhead, leading to improved segmentation accuracy and robustness in low-resource settings.
Adaptation Study#
While the provided text lacks a specific section titled ‘Adaptation Study,’ the core concept of adaptation permeates the entire paper, focusing on adapting the Segment Anything Model (SAM) for medical image segmentation. The authors address the limitations of directly applying SAM to medical images due to the domain gap, prompting the development of SALT (Singular Value Adaptation with Low-Rank Transformation). SALT is explicitly designed to facilitate adaptation, improving performance compared to traditional fine-tuning methods and other parameter-efficient techniques like LoRA and S-SAM. The research emphasizes the importance of adapting pre-trained models to specialized domains. The study explores how manipulating singular values and integrating low-rank updates can effectively transfer knowledge and enhance segmentation accuracy in medical imaging. This is a comprehensive, insightful approach into understanding adaptation, and the details provided will lead to greater domain adaptation.
Future Extentions#
While the paper doesn’t explicitly discuss ‘Future Extensions,’ we can infer potential areas for future work. One avenue is extending SALT to other foundation models beyond SAM, exploring its applicability to diverse tasks. Dynamic rank selection could optimize parameter allocation by adjusting ranks based on dataset complexity. Investigating the impact of varying scales for the top singular values to enhance adaptability to different feature representations could also improve accuracy. More extensive ablation studies could evaluate the impact of different architectural components to enhance the design of SALT. Another extension of SALT involves refining the weight update using different scaling and low rank adaptation factors.
More visual insights#
More on figures

🔼 Figure 2 illustrates four different parameter-efficient fine-tuning (PEFT) methods for adapting pre-trained models to new datasets. (A) Full fine-tuning, the most computationally expensive method, updates all the model’s parameters. (B) LoRA (Low-Rank Adaptation) uses low-rank matrices for efficient updates, only modifying a small subset of parameters. (C) SVD (Singular Value Decomposition)-based methods update the model’s singular values, representing another efficient update strategy. (D) SALT (Singular Value Adaptation with Low-Rank Transformation), the proposed method, combines the strengths of both LoRA and SVD by selectively fine-tuning the most important singular values with scale and shift parameters, and also applying low-rank updates to the remaining subspace. This hybrid approach aims for better adaptation compared to using only LoRA or SVD.
read the caption
Figure 2: Comparison of PEFT methods: (A) Full fine-tuning updates all W𝑊Witalic_W parameters. (B) LoRA updates low-rank matrices. (C) SVD fine-tunes scale and shift components of W𝑊Witalic_W. (D) SALT (Ours) integrates scale, shift, and low-rank updates for singular values.

🔼 Figure 3 showcases a qualitative comparison of vessel segmentation results across five different medical datasets: ARCADE [17], ROSE [14], DIAS [12], X-ray Angio [2], and DRIVE [22]. Each row represents a single dataset and shows the original image, the ground truth segmentation, and segmentation results from several different methods. These methods include U-Net, UNETR, LoRA, S-SAM (other SAM-based PEFT methods), and the proposed SALT method. The predicted masks are shown in white, while any false negatives (areas missed by the segmentation) are highlighted in red. This visual comparison demonstrates SALT’s performance relative to other state-of-the-art segmentation techniques in diverse medical imaging scenarios.
read the caption
Figure 3: Qualitative Vessel Segmentation. The figure displays segmentation results for five datasets (Arcade [17], Rose [14], Dias [12], X-ray Angio [2], Drive [22]). Columns show the ground truth, SALT, SAM-based PEFT methods, and traditional DL models. Predicted masks are white, and false negatives are red.

🔼 The figure shows an ablation study on SALT configurations and adaptability to SAM2. Subfigure (a) presents a comparison of Dice scores across different combinations of LoRA and SALT ranks for Multi-Head Attention (MHA), convolutional layers, and MLP modules. Circle size corresponds to the percentage of trainable parameters. This illustrates the impact of varying the rank (the number of top singular values modified using scale and shift) on model performance and parameter efficiency.
read the caption
(a)

🔼 The figure shows an ablation study on SALT configurations and their adaptability to SAM2. It compares the performance of different methods (SALT, LoRA, SVD) on two datasets of varying sizes by assessing Dice scores and the number of trainable parameters used. The x-axis indicates various rank combinations for convolutional, MLP, and multi-head attention layers, illustrating the impact of rank selection on performance. The y-axis represents the Dice score. Circle size is proportional to the percentage of trainable parameters.
read the caption
(b)
Full paper#