TL;DR#
Breast cancer survival prediction is challenging due to tumor heterogeneity, making it difficult to extract representative features from whole slide images (WSIs). High-resolution images demand substantial computational resources, leading to a tension between maintaining resolution and learning representative features. To solve the issues, this paper proposes PathoHR, a pipeline enhancing any size of pathological images for effective feature learning. It incorporates a high-resolution ViT to enhance patch-wise WSI representation and evaluates similarity metrics to optimize representation learning.
PathoHR’s pipeline enables smaller image patches to achieve equivalent or superior prediction accuracy compared to raw larger patches, significantly reducing computational overhead. It offers an optimized way of integrating enhanced image resolution with optimized feature learning. It also introduces a streamlined approach for processing enhanced smaller image patches to achieve superior prediction accuracy while reducing computational demands.
Key Takeaways#
Why does it matter?#
This study introduces a novel approach to enhance breast cancer survival prediction, potentially improving diagnosis and treatment strategies. It paves the way for more accurate and efficient computational pathology by integrating high-resolution imaging with optimized feature learning, addressing current limitations in the field.
Visual Insights#

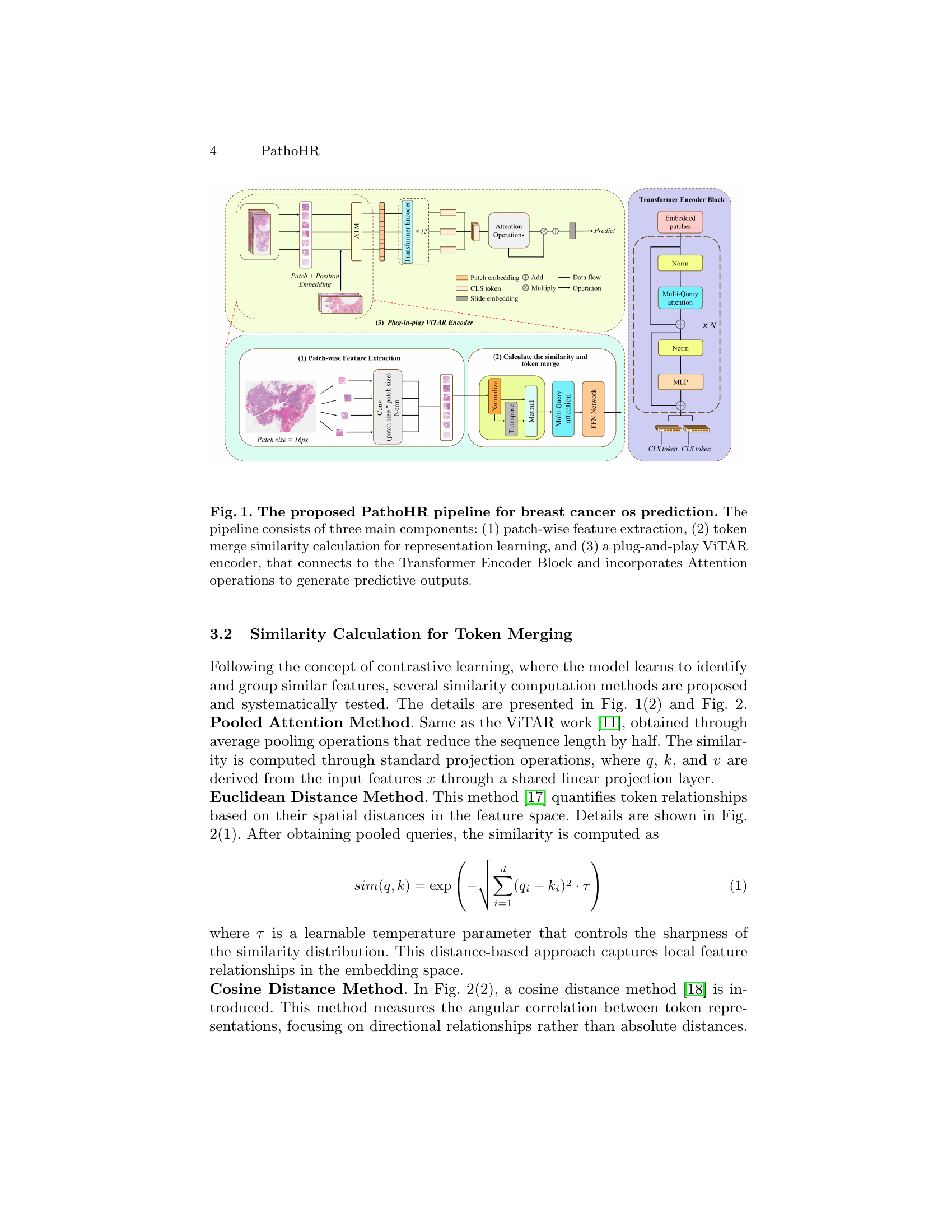
🔼 The figure illustrates the PathoHR pipeline for predicting breast cancer survival. It’s broken into three main stages: 1) Patch-wise Feature Extraction: Whole Slide Images (WSIs) are segmented into smaller patches, and features are extracted from each patch using a pre-trained UNet encoder. 2) Similarity Calculation and Token Merging: Multiple similarity metrics (Euclidean distance, cosine distance, attention score, and semantic similarity) are employed to measure the similarity between patches. Similar patches are then merged using an adaptive token merging mechanism, improving feature representation. 3) Plug-and-play ViTAR Encoder: The merged patches are fed into a high-resolution Vision Transformer (ViT) architecture that processes these features to generate predictions for patient survival outcomes. The ViTAR encoder uses adaptive token merging and fuzzy positional encoding to handle various input patch sizes and maintain high accuracy with less computational cost. The entire pipeline facilitates the efficient use of high-resolution images for more accurate survival predictions, addressing the challenge of tumor heterogeneity.
read the caption
Figure 1: The proposed PathoHR pipeline for breast cancer os prediction. The pipeline consists of three main components: (1) patch-wise feature extraction, (2) token merge similarity calculation for representation learning, and (3) a plug-and-play ViTAR encoder, that connects to the Transformer Encoder Block and incorporates Attention operations to generate predictive outputs.
| Models | AUC | ACC | F1 | Recall | Precision |
| Intra(24) | 0.8 | 0.95313 | 0.83025 | 0.95313 | 0.90845 |
| Intra(16) | 0.76786 | 0.97436 | 0.91077 | 0.87179 | 0.89451 |
| Intra with pool | 0.71622 | 0.94872 | 0.92375 | 0.94872 | 0.90007 |
| Intra with cos | 0.90741 | 0.97436 | 0.96170 | 0.97436 | 0.94938 |
| Intra with euclidean | 0.82413 | 0.97436 | 0.92375 | 0.94872 | 0.92915 |
| Intra with semantic | 0.90541 | 0.89286 | 0.92375 | 0.89286 | 0.94938 |
| Intra with attention score | 0.62353 | 0.94872 | 0.92375 | 0.94872 | 0.90007 |
| Intra with ToMe | 0.78947 | 0.97436 | 0.96170 | 0.97436 | 0.94938 |
🔼 This table presents the validation performance results for predicting overall survival (OS) in breast cancer pathology. The performance is evaluated using several metrics (AUC, ACC, F1 Recall, and Precision) and compares two different input patch sizes: 24x24 and 16x16 pixels. The results show how different model variations (e.g., using different similarity metrics) and patch sizes affect the accuracy of OS prediction. The table helps in understanding the impact of feature extraction methods and patch sizes on the model’s predictive capability.
read the caption
Table 1: Validation performance on os prediction for breast cancer pathology.(24) and (16) denote input patch sizes of 24×24 and 16×16, respectively.
In-depth insights#
PathoHR Pipeline#
The PathoHR pipeline is a multi-faceted framework designed to enhance breast cancer survival prediction using high-resolution pathology images. It addresses the challenges of tumor heterogeneity and computational demands through innovative feature extraction and representation learning. The pipeline likely incorporates a plug-and-play high-resolution Vision Transformer (ViT) to improve patch-wise WSI representation. It will also involve a systematic evaluation of similarity metrics to better capture tumor characteristics. Key to its efficiency is the ability to work effectively with smaller image patches, reducing computational overhead without sacrificing prediction accuracy. The pipeline probably includes processing Whole Slide Images (WSIs) to extract low-dimensional feature representations and leveraging a weakly supervised attention-based model with a pre-trained UNI encoder for feature extraction. Adaptive token merging will likely be a key component.
Similarity Metrics#
The research paper delves into the crucial aspect of similarity metrics within the context of breast cancer survival prediction using high-resolution pathological images. Several methods are proposed and tested to enhance feature representation learning. These include the Pooled Attention Method, Euclidean Distance Method, and Cosine Distance Method. The paper highlights the integration of these similarity metrics, such as the transformer architecture benefiting from the attention mechanism. It also shows how different similarity metrics influence the model’s ability to capture essential features from WSIs, impacting survival prediction accuracy. The study suggests that future research should explore the interplay between similarity metrics and residual connections to optimize pathology-based survival prediction.
ViTAR Integration#
ViTAR integration, leveraging Vision Transformers with dynamic resolution adaptation, presents a compelling approach for enhancing breast cancer survival prediction. ViTAR’s ability to process images at any resolution addresses a critical challenge in computational pathology, where image resolutions vary significantly. By incorporating adaptive token merging and fuzzy position encoding, ViTAR significantly reduces computational costs while maintaining high accuracy, crucial for large-scale histology image analysis. The plug-and-play nature of ViTAR allows seamless integration with existing architectures, enhancing patch-based WSIs to extract fine-grained morphological details essential for accurate survival prediction. The focus on resolution robustness through controlled positional uncertainty further strengthens the model’s ability to generalize across diverse datasets and imaging conditions, potentially improving the reliability and clinical applicability of breast cancer survival predictions. ViTAR’s dual-mechanism architecture efficiently processes multi-resolution inputs, adaptively managing token density while preserving spatial coherence, ensuring consistent performance across varying image scales, making it a valuable asset in computational pathology.
Multi-Resolution#
Multi-resolution techniques in medical image analysis, particularly within computational pathology, address challenges arising from tumor heterogeneity and computational demands. High-resolution images provide detailed morphological information crucial for accurate predictions, but processing such data requires substantial resources. Approaches involve cropping whole slide images (WSIs) into smaller patches, introducing the challenge of effectively learning similarity features across patches to capture overall tumor characteristics. Methods like ViTAR, which is able to process images at any resolution through adaptive token merging and fuzzy position encoding, help reduce computational costs while maintaining high accuracy. The integration of multi-resolution processing with advanced feature representation learning aims to enhance pathological image analysis by combining detailed information with efficient processing, crucial for improved cancer survival prediction.
Model Enhancements#
The study showcases that enhancements brought by the proposed PathoHR strategies indicate that independently integrating residual connections or similarity comparisons can improve os prediction accuracy. Optimal results come by either using cosine similarity without residual connections or using only pooling with residual links. This suggests a potential counteractive relationship between these components. Future work could explore this interaction effect, combining residual connections with selective similarity metrics for improved survival prediction.
More visual insights#
More on figures

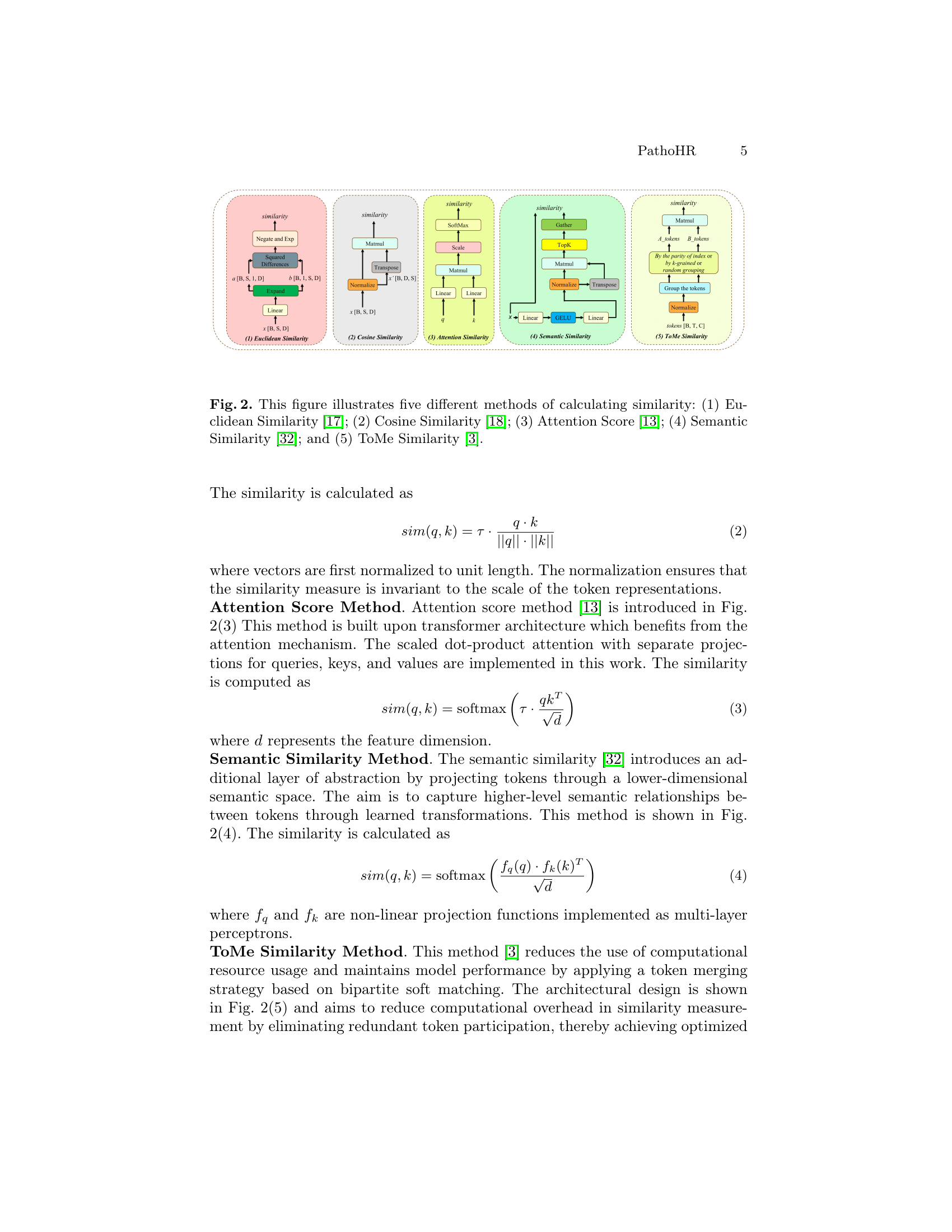
🔼 Figure 2 compares five different methods for calculating the similarity between image patches: Euclidean distance, cosine similarity, attention score, semantic similarity, and ToMe similarity. Each method is visually represented, showing its unique approach to computing similarity and its integration within the overall architecture. The figure highlights the different ways these methods process and combine features to identify similar patches within the whole-slide images, which is crucial for effective representation learning in the downstream classification task.
read the caption
Figure 2: This figure illustrates five different methods of calculating similarity: (1) Euclidean Similarity [17]; (2) Cosine Similarity [18]; (3) Attention Score [13]; (4) Semantic Similarity [32]; and (5) ToMe Similarity [3].

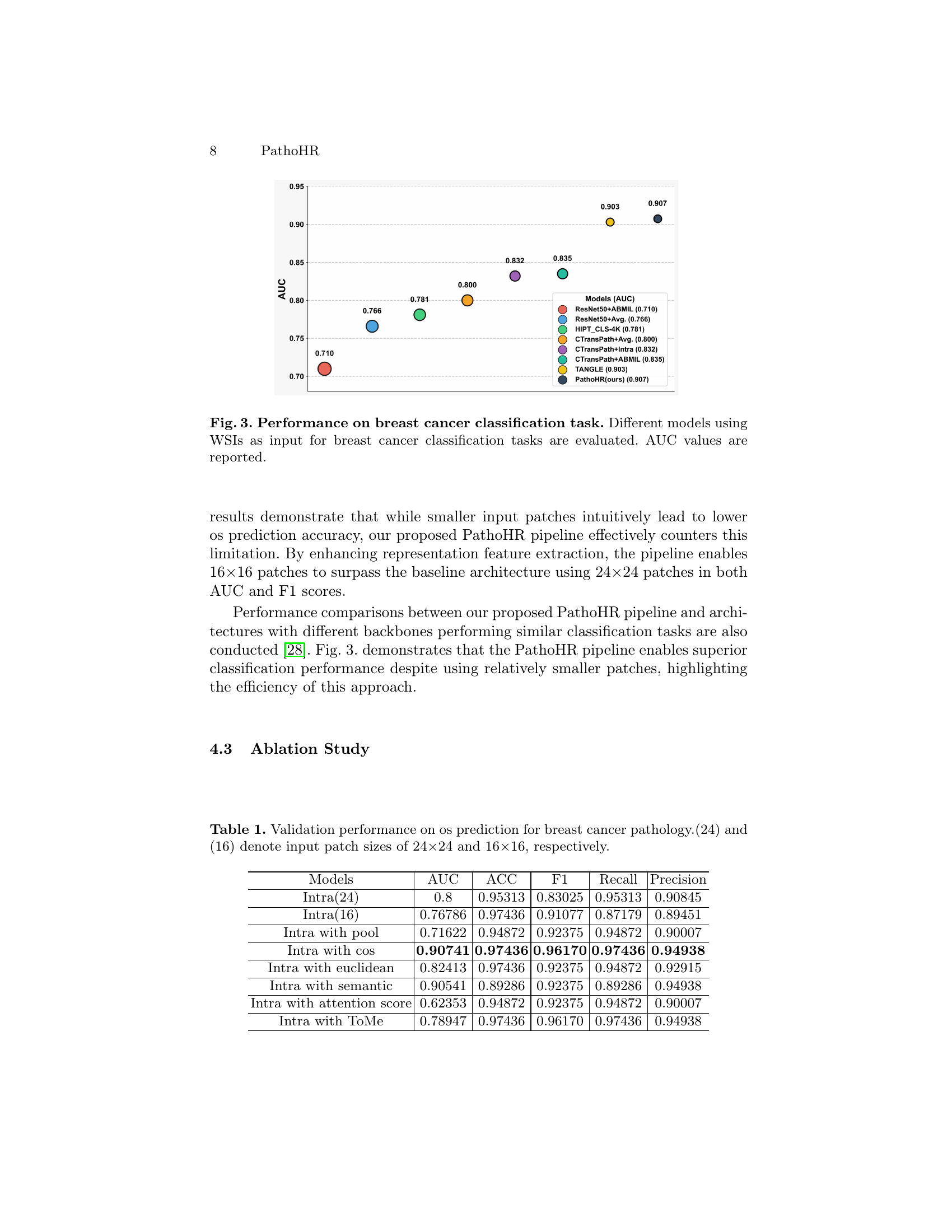
🔼 Figure 3 presents a comparative analysis of breast cancer classification performance using Whole Slide Images (WSIs) as input. Multiple models, including several variations of the proposed PathoHR pipeline and other established methods (ResNet50+ABMIL, ResNet50+Avg., HIPT_CLS-4K, CTransPath+Avg., CTransPath+Intra, CTransPath+ABMIL, and TANGLE), were evaluated on this task. The figure displays a bar chart representing the Area Under the Curve (AUC) values for each model, providing a visual comparison of their performance in classifying breast cancer types. The AUC serves as a metric to evaluate the model’s ability to distinguish between different classes of breast cancer, with higher AUC values indicating better performance. The chart allows for a direct comparison of the efficacy of various models in handling this classification challenge using WSIs.
read the caption
Figure 3: Performance on breast cancer classification task. Different models using WSIs as input for breast cancer classification tasks are evaluated. AUC values are reported.
Full paper#