TL;DR#
Medical image analysis often struggles with aligning image and text data, especially due to limited labeled datasets. This data reliance results in models that don’t generalize well to new data. This paper tackles this issue by proposing a novel approach.
The proposed Eye-gaze Guided Multi-modal Alignment (EGMA) framework uses radiologists’ eye-gaze data as auxiliary information to improve the alignment process. EGMA shows significant performance improvements in image classification and image-text retrieval on multiple medical datasets, demonstrating superior generalization compared to existing methods. This highlights the value of incorporating eye-gaze data in medical multi-modal learning.
Key Takeaways#
Why does it matter?#
This paper is crucial because it introduces a novel framework, EGMA, that leverages eye-gaze data to improve the alignment of medical images and text. This addresses a critical challenge in medical multi-modal learning, leading to better model generalization and improved performance on downstream tasks. The approach is particularly relevant given the scarcity of labeled medical data and opens exciting avenues for future research in medical image analysis and diagnosis.
Visual Insights#
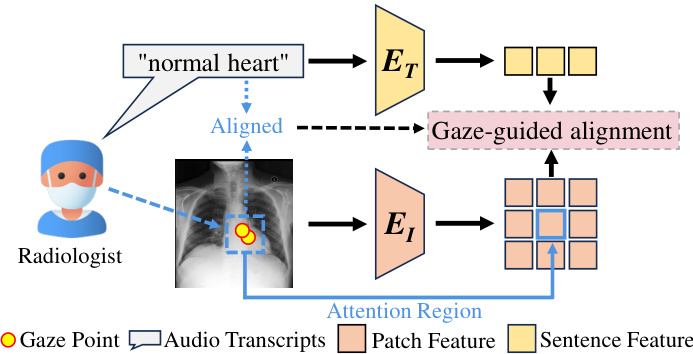
🔼 This figure illustrates how radiologists’ eye gaze data acts as an auxiliary tool in aligning multimodal medical data. The diagram shows a radiologist looking at a chest X-ray, with their gaze focused on a specific region of the image. Simultaneously, the radiologist provides a diagnostic text description (’normal heart’). The dashed line connecting the gaze and the text represents the alignment guided by the eye gaze. The process is further explained with visual representation of gaze points, audio transcripts, attention regions, patch features, and sentence features, which are all processed to enhance the alignment of image and text data.
read the caption
Figure 1: The guiding role of radiologists' eye-gaze data. The text provided by radiologists during diagnosis aligns naturally with the attention regions.
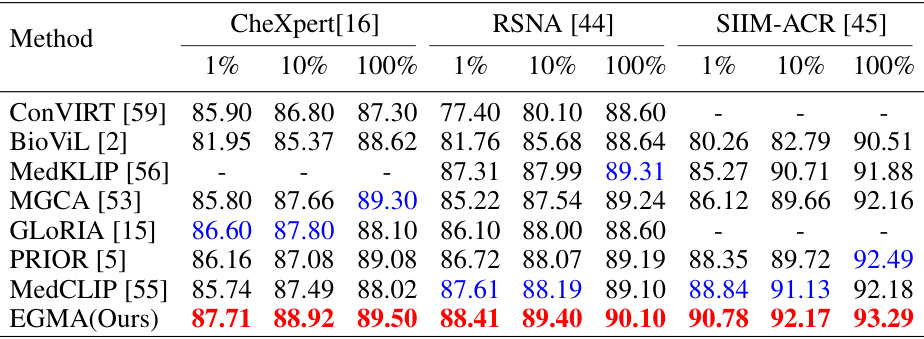
🔼 This table presents a comparison of the Area Under the Receiver Operating Characteristic Curve (AUROC) scores achieved by different state-of-the-art (SOTA) models and the proposed Eye-gaze Guided Multi-modal Alignment (EGMA) model on three chest X-ray datasets (CheXpert, RSNA, and SIIM-ACR). The comparison is done using three different training set sizes (1%, 10%, and 100%), illustrating the performance impact of the size of the training data. The best and second-best performing models are highlighted.
read the caption
Table 1: Comparison results of supervised classification task with other SOTA models on CheXpert, RSNA, and SIIM-ACR datasets. Area under ROC curve (AUROC) is reported with different portions of training data: 1%, 10%, 100%. Red and blue denote the best and second-best results.
In-depth insights#
Gaze-guided Alignment#
The concept of ‘Gaze-guided Alignment’ in the context of multimodal learning for medical image analysis is innovative and impactful. It leverages the inherent knowledge embedded in radiologists’ eye-gaze patterns during diagnosis to improve the alignment of visual and textual features. This approach is particularly valuable because it tackles the inherent challenge of implicitly aligned features in existing medical multimodal models. By explicitly incorporating gaze data as an auxiliary signal, the model can learn stronger and more generalizable relationships, leading to improved performance on downstream tasks such as image classification and retrieval. The utilization of eye-gaze data is also efficient compared to manual annotation, making it a scalable solution. This method is particularly useful in scenarios with limited labeled medical data, where explicit alignment guidance is crucial. The success of this method highlights the power of integrating human expertise into machine learning models for enhanced accuracy and generalizability.
Med-VLP Advances#
Med-VLP (Medical Vision-Language Pre-training) has significantly advanced medical image analysis by integrating vision and language models. Early Med-VLP approaches focused on simple fusion of visual and textual features, often relying heavily on large, well-annotated datasets. However, the scarcity of labeled medical data poses a major challenge. Recent advances leverage techniques like contrastive learning (e.g., MedCLIP) and self-supervised learning to improve model performance with less annotation, though scaling remains an issue. Incorporating auxiliary information, such as radiologist eye-gaze data (as explored in the provided paper), offers a promising pathway to enhance alignment between modalities and boost generalization. This approach leverages inherent expert knowledge within the eye-gaze patterns, making training more efficient. Future directions include exploring more sophisticated multi-modal alignment strategies and investigating ways to better handle the inherent complexity and variability within medical data. The use of weak supervision and transfer learning will likely play a significant role in Med-VLP’s ongoing evolution.
EGMA Framework#
The EGMA framework, as described in the research paper, is a novel approach for medical multi-modal alignment that leverages the power of radiologists’ eye-gaze data. The core innovation lies in integrating this auxiliary data to enhance the alignment of medical visual and textual features. Unlike previous methods relying solely on implicit data relationships, EGMA explicitly incorporates the radiologists’ attention patterns, improving cross-modality feature alignment. This framework demonstrates superior performance and generalization across various medical datasets, suggesting that incorporating eye-gaze data is highly beneficial for multi-modal learning in the medical domain. Furthermore, the results indicate that even a limited amount of eye-gaze data yields notable improvements, making EGMA a feasible and efficient method for medical image-text understanding. The framework’s robust performance and enhanced generalization capabilities highlight the significant potential of incorporating auxiliary information, such as eye-gaze data, to advance multi-modal learning in medical applications.
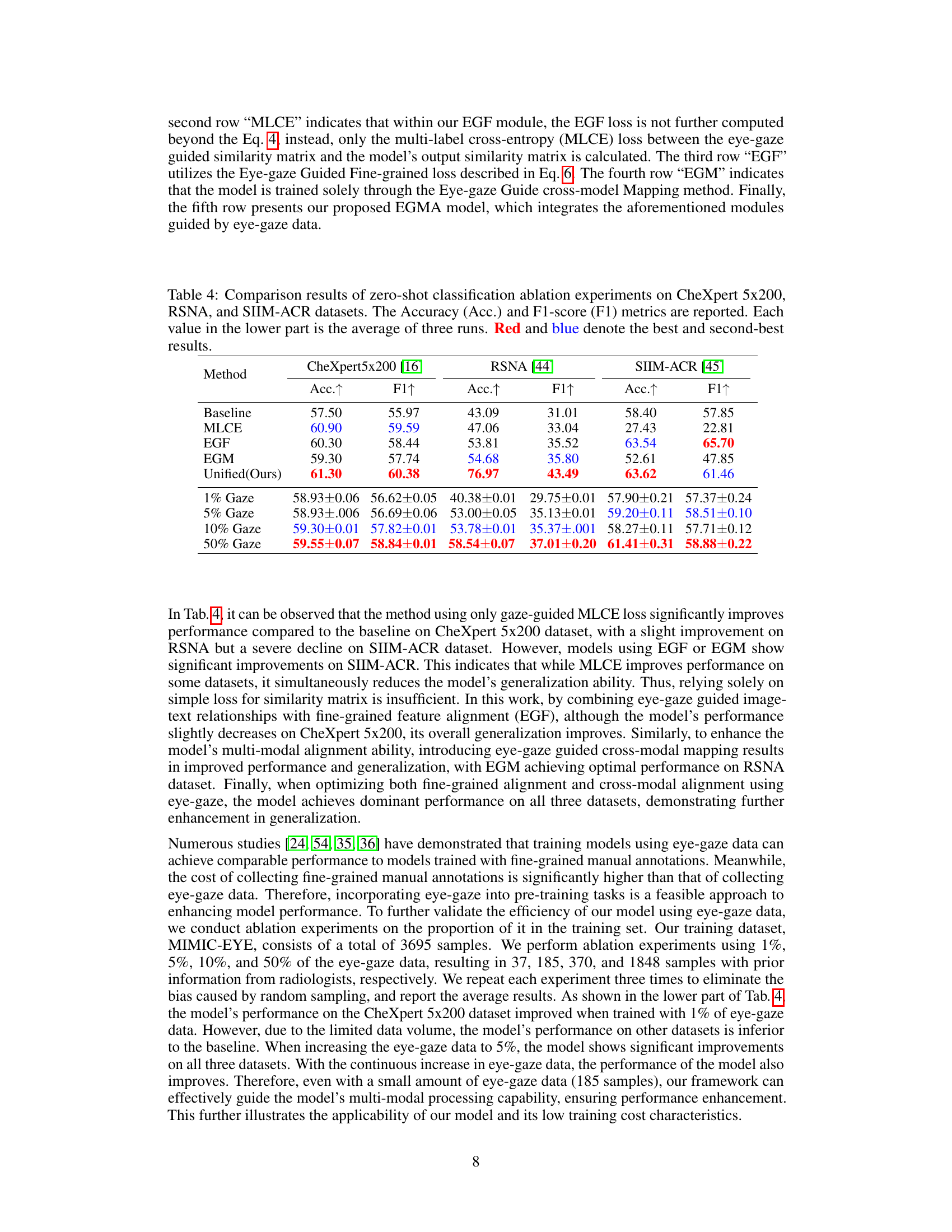
Ablation Experiments#
Ablation experiments systematically remove components of a model to assess their individual contributions. In this context, it would involve removing or disabling parts of the proposed Eye-gaze Guided Multi-modal Alignment (EGMA) framework. Key areas of ablation would include the eye-gaze guided fine-grained alignment module and the eye-gaze guided cross-modality mapping module. By evaluating performance after removing each part, researchers can determine their relative importance in achieving state-of-the-art results. The ablation study should show that both modules significantly contribute to performance, demonstrating the effectiveness of integrating eye-gaze data. The results would offer valuable insights into the EGMA architecture and might suggest areas for further optimization or simplification. Furthermore, varying the amount of eye-gaze data used could reveal whether it’s crucial to have a large amount for effective alignment or if smaller amounts offer a good balance between performance and cost-effectiveness. A well-designed ablation study is crucial for establishing the model’s robustness and understanding the interplay between different components.
Future Directions#
Future directions for research in eye-gaze guided multi-modal medical image analysis are promising. Improving data collection methods is crucial, moving beyond existing datasets to capture more diverse scenarios and handle the challenges of real-time data acquisition during clinical practice. This includes exploring alternative annotation strategies that may be less time-consuming than full bounding box annotations. Developing more robust models that are less sensitive to noise in eye-gaze data and can better handle variations in radiologist behavior is vital. This would likely involve incorporating techniques to better filter noise and potentially using data augmentation strategies that address the inherent variability of eye-gaze. Investigating the potential of eye-gaze data in other medical imaging modalities beyond chest x-rays will expand this technology’s impact. Combining eye-gaze with other auxiliary data, such as physiological signals or clinical notes, could yield even more powerful multi-modal learning frameworks. Finally, careful consideration must be given to addressing ethical and privacy concerns associated with the use of eye-gaze data, particularly in sensitive contexts such as medical diagnosis.
More visual insights#
More on figures
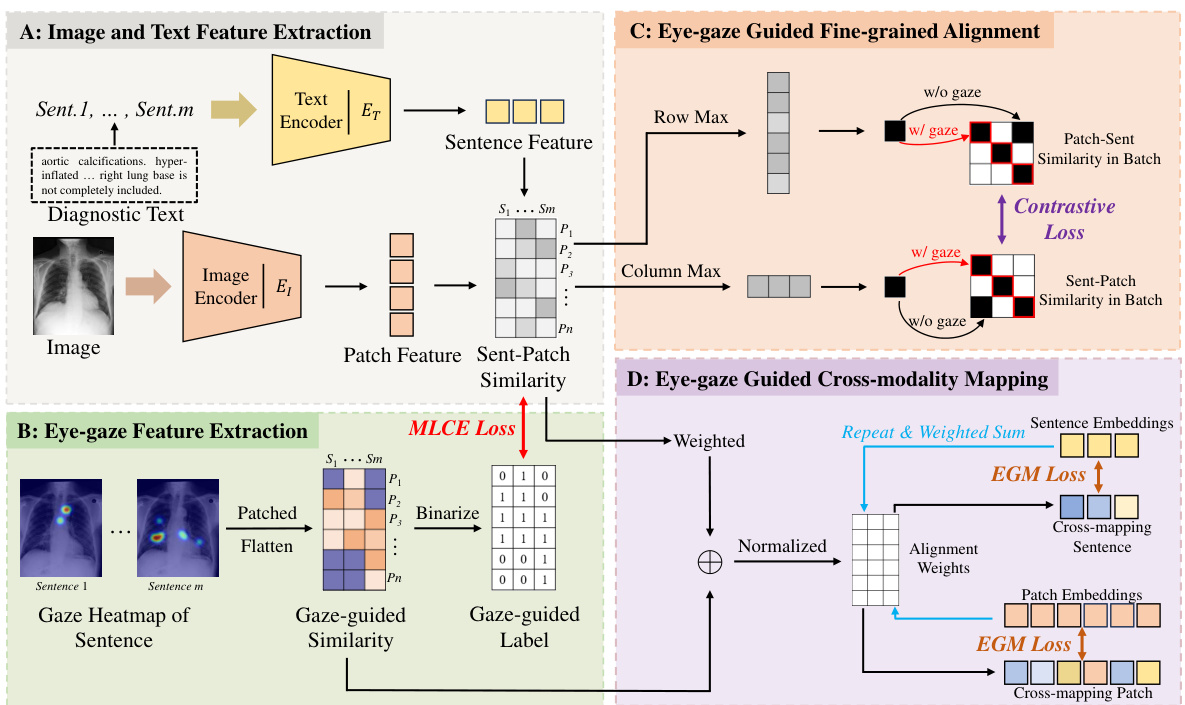
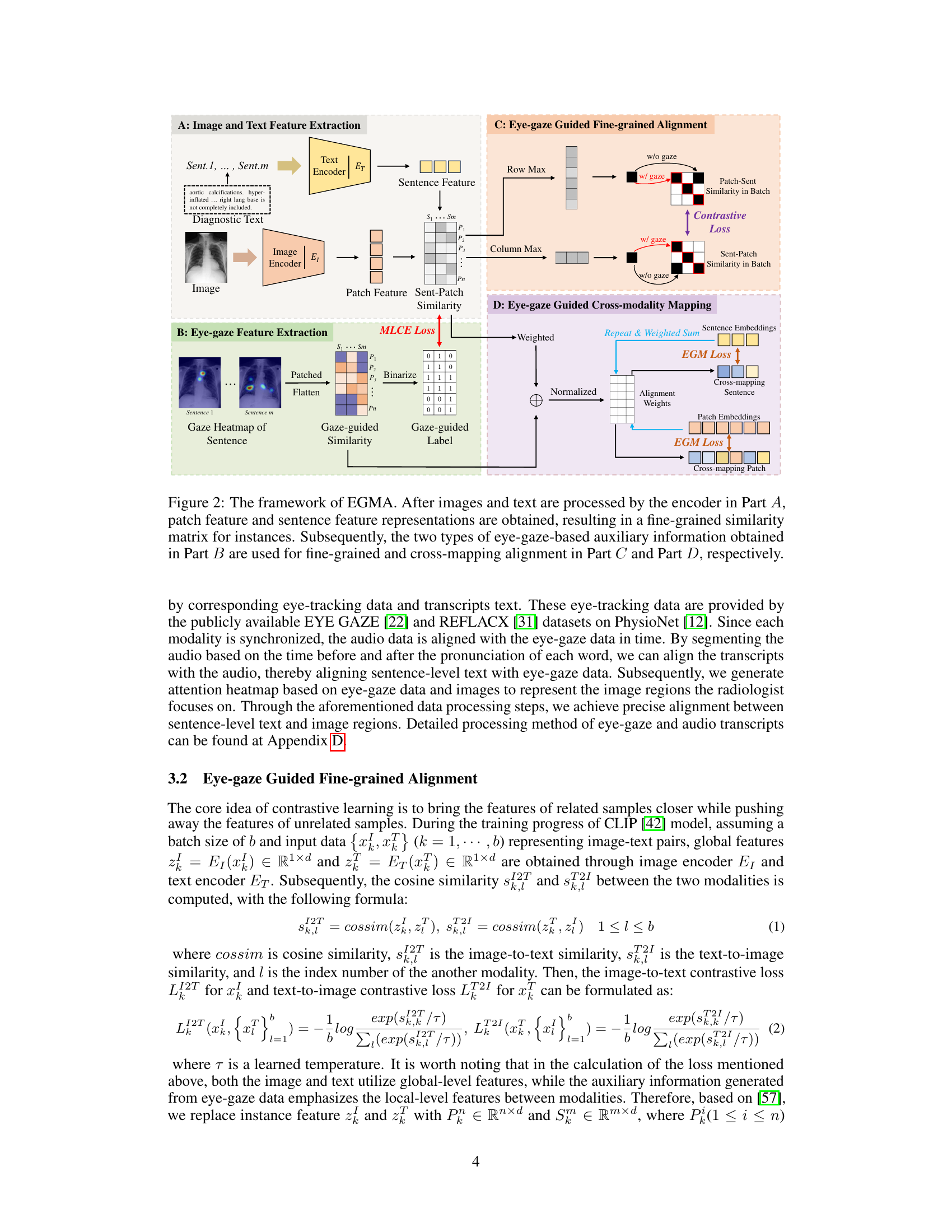
🔼 This figure illustrates the EGMA framework’s four main components. Part A shows image and text feature extraction, creating an instance-level similarity matrix. Part B extracts eye-gaze features, creating attention heatmaps that indicate radiologist focus areas. Part C refines the alignment using eye-gaze data, employing fine-grained contrastive loss. Finally, Part D leverages eye-gaze data to guide cross-modality mapping, improving text-image alignment.
read the caption
Figure 2: The framework of EGMA. After images and text are processed by the encoder in Part A, patch feature and sentence feature representations are obtained, resulting in a fine-grained similarity matrix for instances. Subsequently, the two types of eye-gaze-based auxiliary information obtained in Part B are used for fine-grained and cross-mapping alignment in Part C and Part D, respectively.
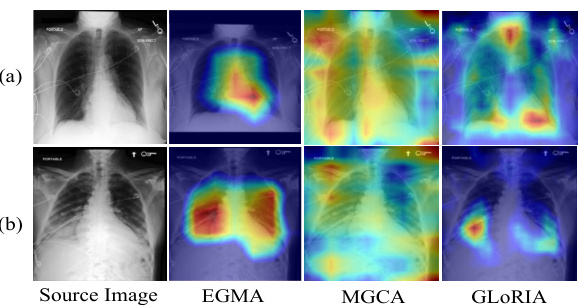
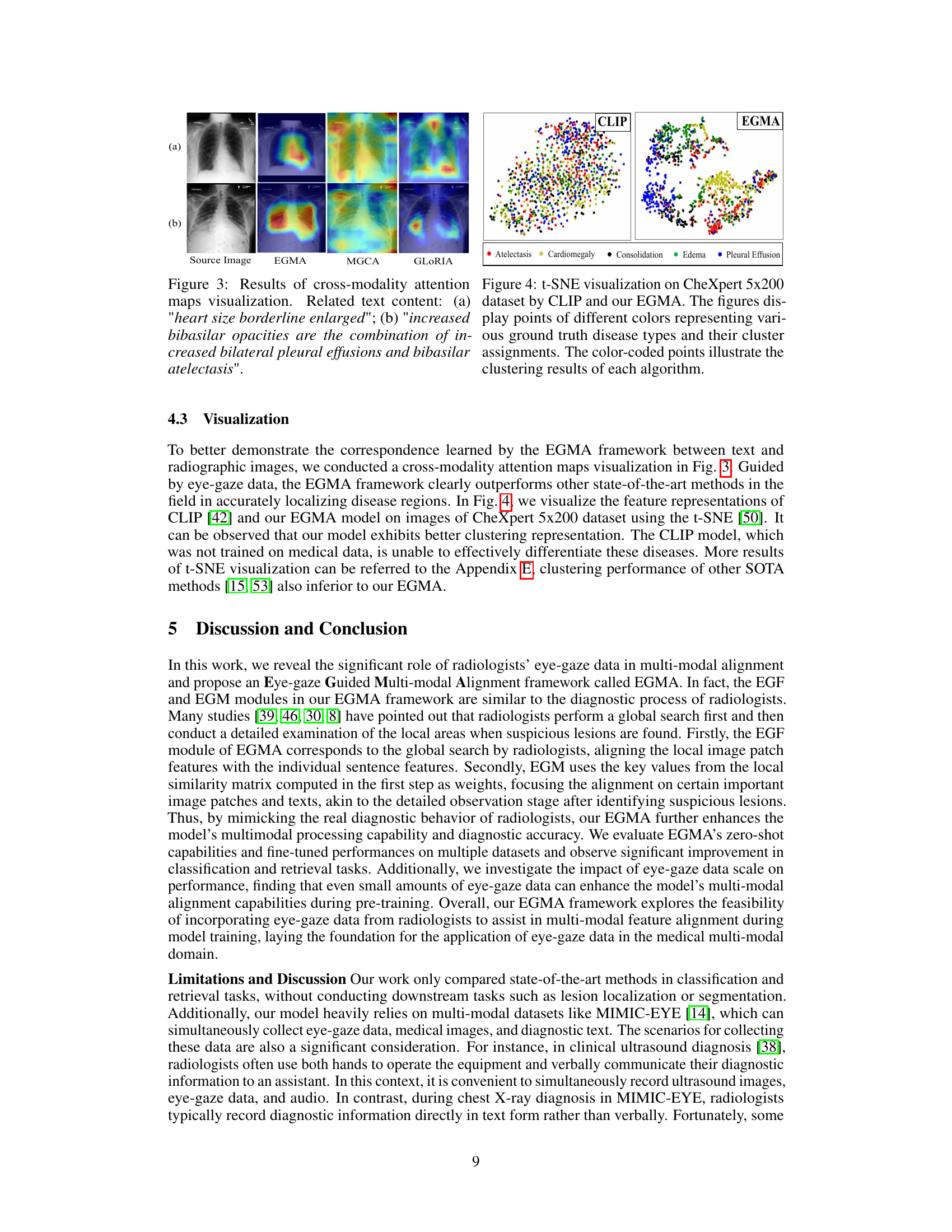
🔼 This figure visualizes the cross-modality attention maps generated by the EGMA model, showcasing its ability to accurately pinpoint disease regions in chest X-ray images. It compares EGMA’s performance with other methods (MGCA and GLORIA) and shows how EGMA better aligns text descriptions with relevant image regions, highlighting the effectiveness of incorporating eye-gaze data in multi-modal alignment. Two examples are provided: one showing the attention map generated for the phrase ‘heart size borderline enlarged’, and another for the more complex phrase ‘increased bibasilar opacities are the combination of increased bilateral pleural effusions and bibasilar atelectasis’.
read the caption
Figure 3: Results of cross-modality attention maps visualization. Related text content: (a) 'heart size borderline enlarged'; (b) 'increased bibasilar opacities are the combination of increased bilateral pleural effusions and bibasilar atelectasis'.
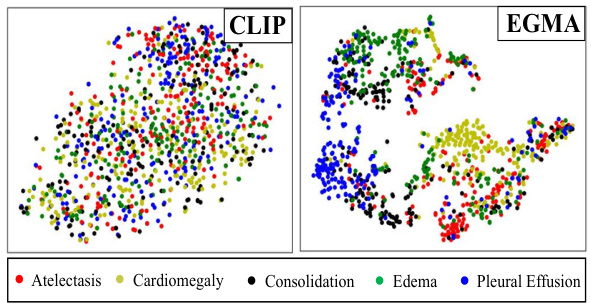
🔼 This figure shows a t-SNE visualization comparing the feature representations learned by CLIP and EGMA on the CheXpert 5x200 dataset. Each point represents an image, and the color indicates the ground truth disease label. The visualization helps to understand how well each model separates the different disease classes in the feature space. EGMA shows a better clustering and separation of the disease classes than CLIP, indicating improved representation learning.
read the caption
Figure 4: t-SNE visualization on CheXpert 5x200 dataset by CLIP and our EGMA. The figures display points of different colors representing various ground truth disease types and their cluster assignments. The color-coded points illustrate the clustering results of each algorithm.
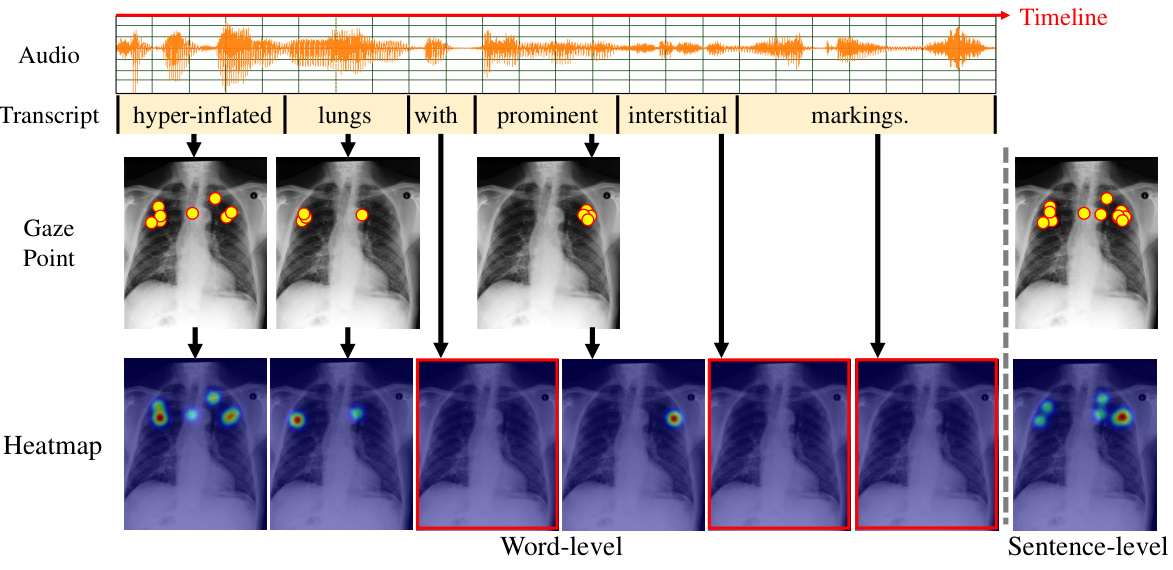
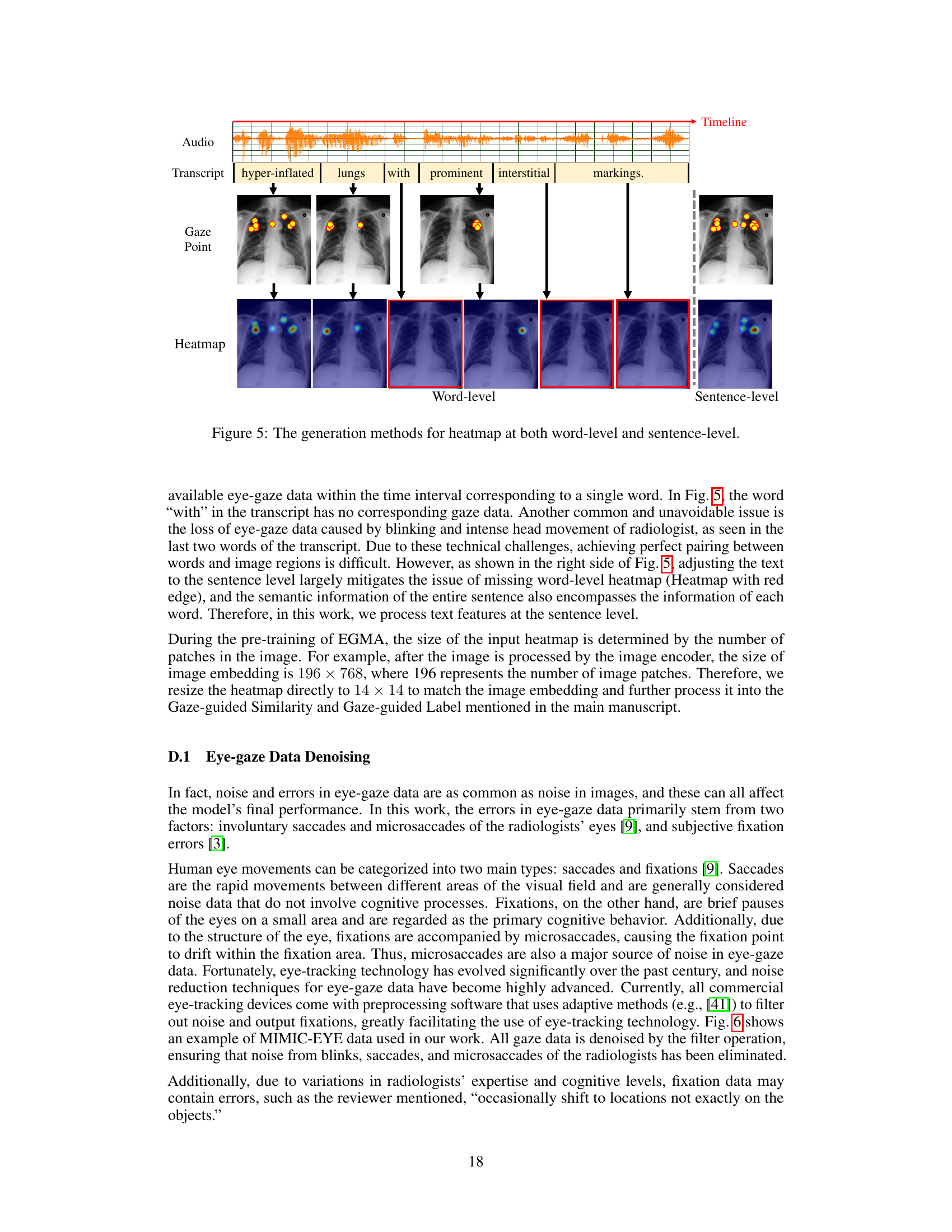
🔼 This figure illustrates the process of generating attention heatmaps from radiologists’ eye-gaze data. The top shows the audio recording of the radiologist, its transcription and the timeline. Each word in the transcription is aligned with the corresponding eye-gaze data points marked on the chest X-ray image. The middle panel shows the word-level gaze data, with each image corresponding to one word in the transcription. The bottom panel shows the resulting word-level and sentence-level heatmaps, which are used to align the textual and visual features. The red boxes indicate how word-level heatmaps are aggregated into sentence-level heatmaps.
read the caption
Figure 5: The generation methods for heatmap at both word-level and sentence-level.
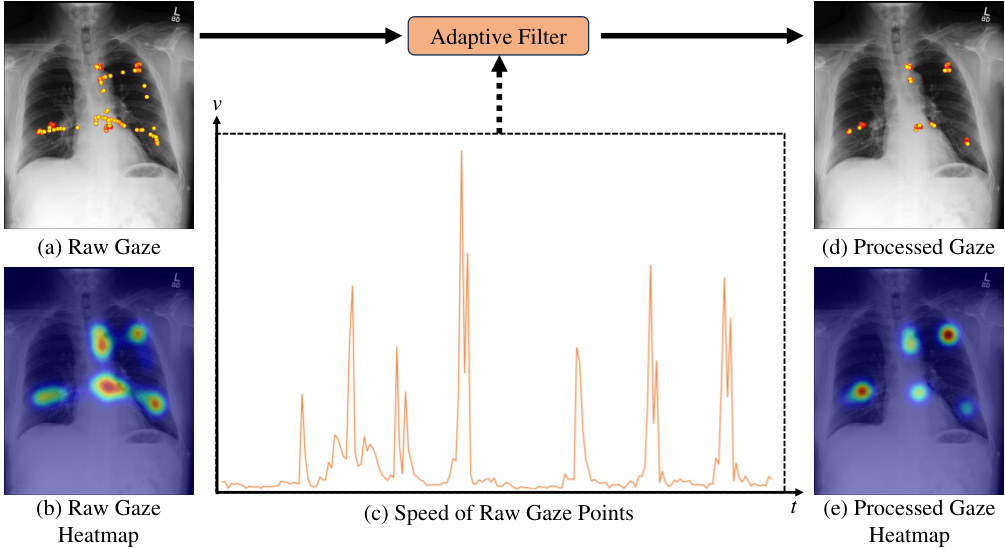
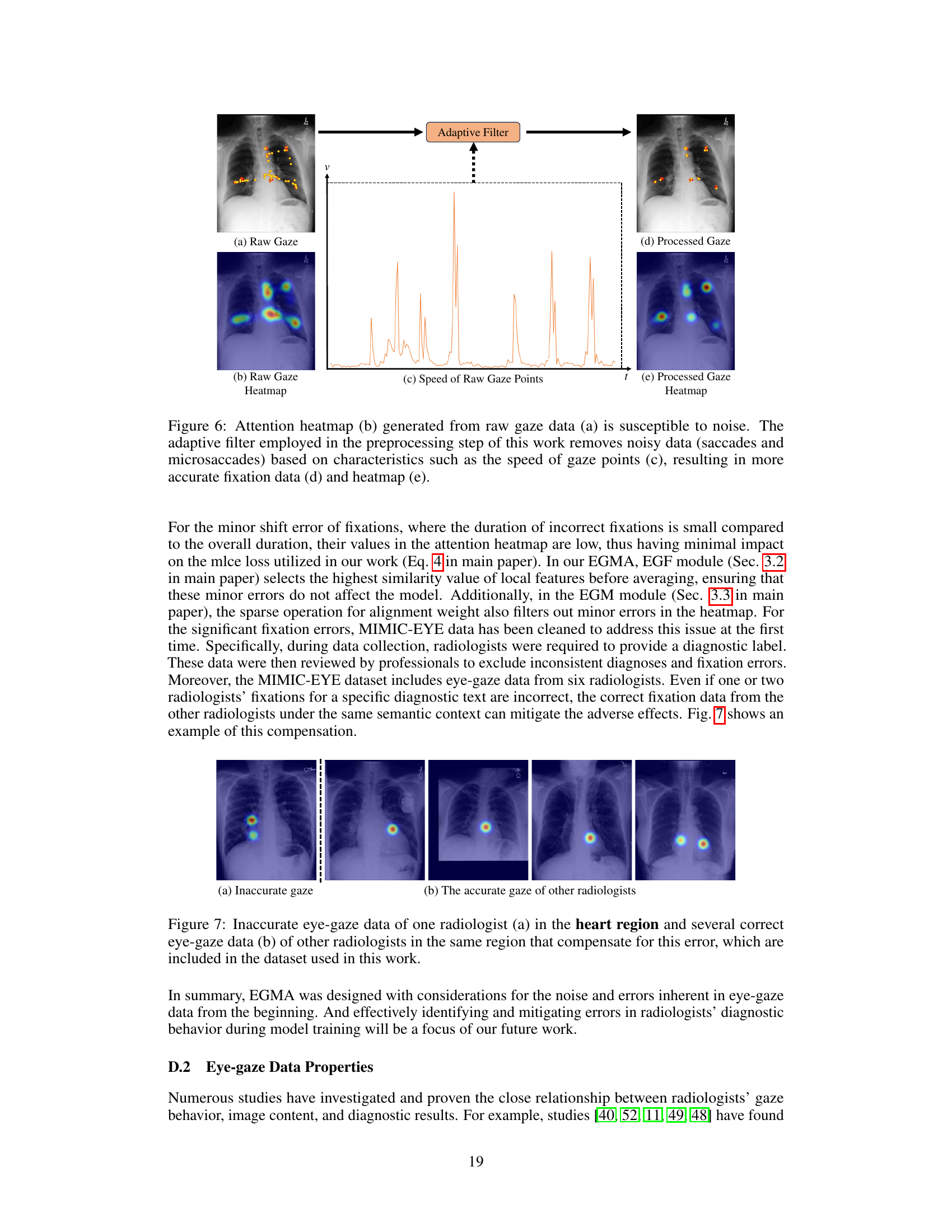
🔼 This figure demonstrates the denoising process applied to raw eye-gaze data. Panel (a) shows the raw gaze data overlaid on an image, highlighting the noisy nature of the raw data, which includes many saccades and microsaccades. Panel (b) displays the heatmap generated from this raw gaze data. Panel (c) shows the speed of gaze points over time, illustrating high speed during saccades and lower speed during fixations. An adaptive filter is applied to the raw gaze data, resulting in the processed gaze data and heatmap shown in (d) and (e), respectively. This filtering process significantly reduces noise and improves the quality of the gaze data for downstream analysis.
read the caption
Figure 6: Attention heatmap (b) generated from raw gaze data (a) is susceptible to noise. The adaptive filter employed in the preprocessing step of this work removes noisy data (saccades and microsaccades) based on characteristics such as the speed of gaze points (c), resulting in more accurate fixation data (d) and heatmap (e).
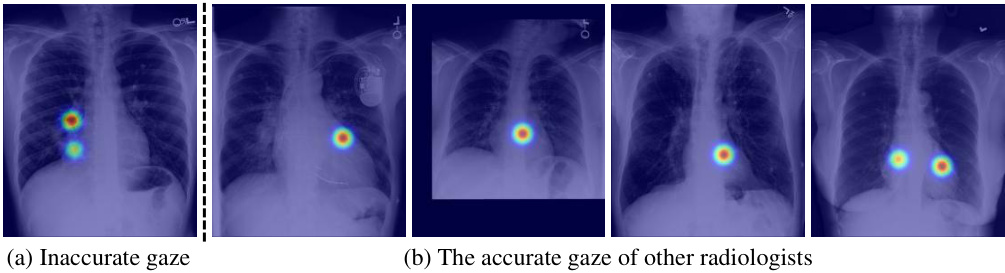
🔼 This figure demonstrates how the proposed method handles inaccurate eye gaze data. In (a), one radiologist’s gaze is not accurately focused on the relevant region of the heart. However, (b) shows that data from other radiologists in the dataset correctly identify the heart region, compensating for the inaccurate data point and improving overall accuracy.
read the caption
Figure 7: Inaccurate eye-gaze data of one radiologist (a) in the heart region and several correct eye-gaze data (b) of other radiologists in the same region that compensate for this error, which are included in the dataset used in this work.
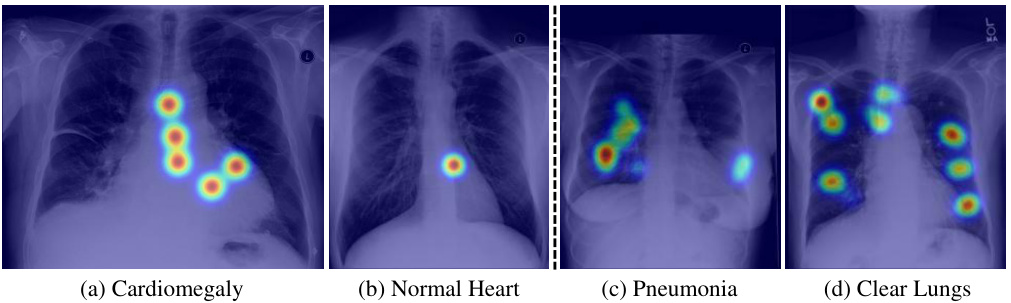
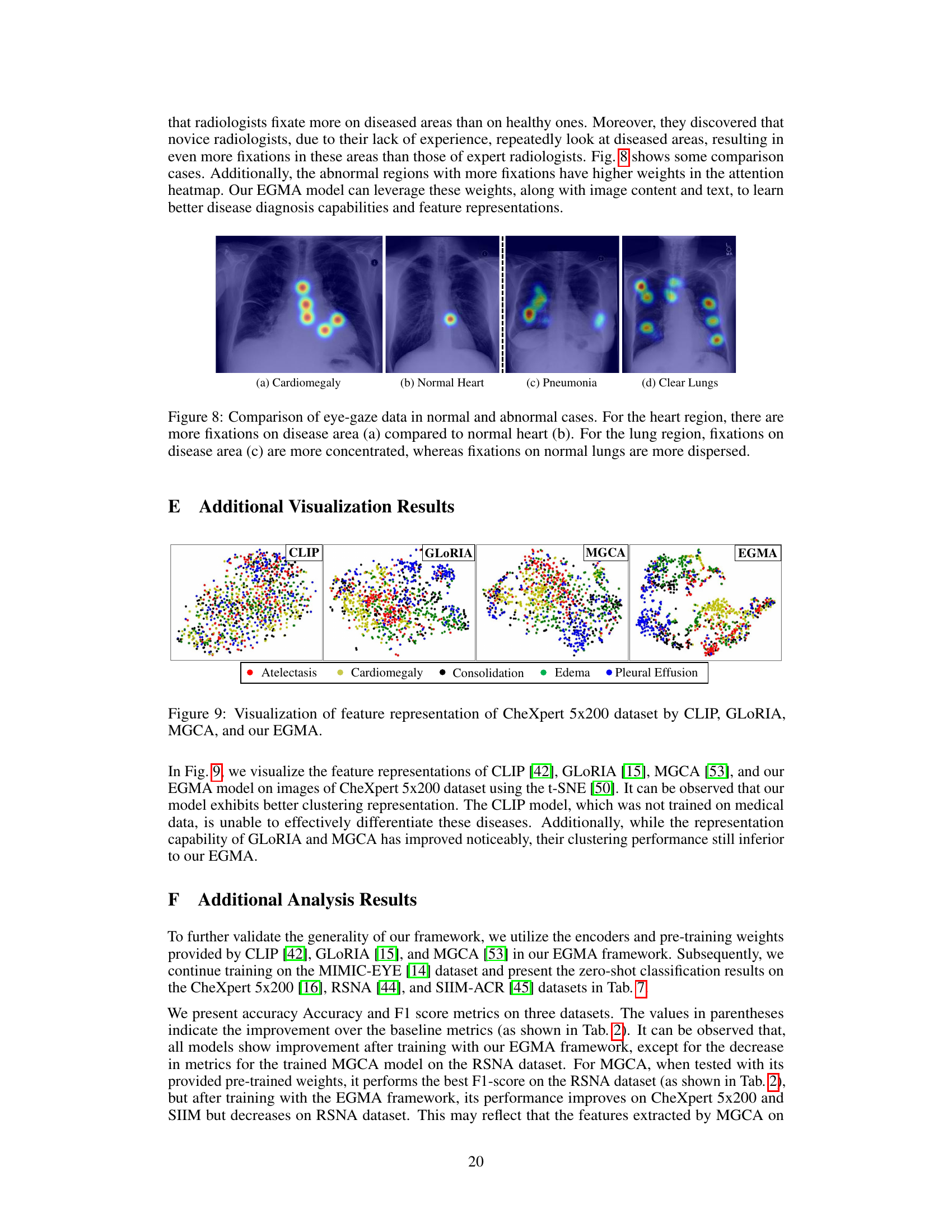
🔼 This figure visually demonstrates the differences in radiologists’ eye-gaze patterns between normal and abnormal chest X-rays. The heatmaps overlaid on the images show the concentration of gaze points. In cases of cardiomegaly (a) and pneumonia (c), there’s a noticeably higher concentration of gaze in the affected areas (heart and lungs, respectively) compared to normal (b) and clear lungs (d). This highlights how radiologists naturally focus their attention on the regions of interest.
read the caption
Figure 8: Comparison of eye-gaze data in normal and abnormal cases. For the heart region, there are more fixations on disease area (a) compared to normal heart (b). For the lung region, fixations on disease area (c) are more concentrated, whereas fixations on normal lungs are more dispersed.
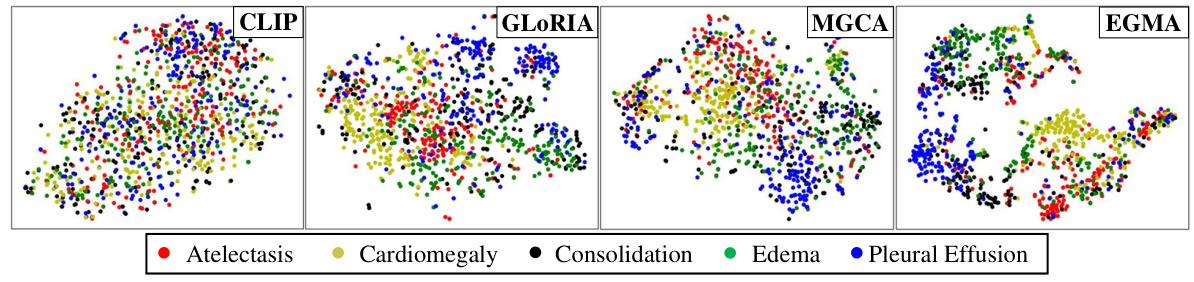
🔼 This figure visualizes the feature representations of the CLIP and EGMA models on the CheXpert 5x200 dataset using t-SNE. Different colors represent different disease categories. The visualization shows that EGMA achieves better clustering of the data points than CLIP, indicating its superior ability to differentiate between diseases.
read the caption
Figure 4: t-SNE visualization on CheXpert 5x200 dataset by CLIP and our EGMA. The figures display points of different colors representing various ground truth disease types and their cluster assignments. The color-coded points illustrate the clustering results of each algorithm.
More on tables
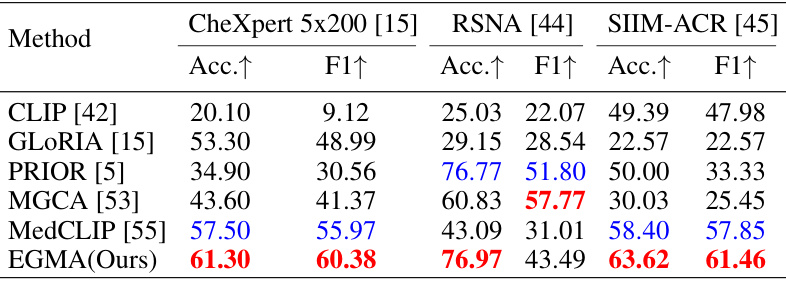
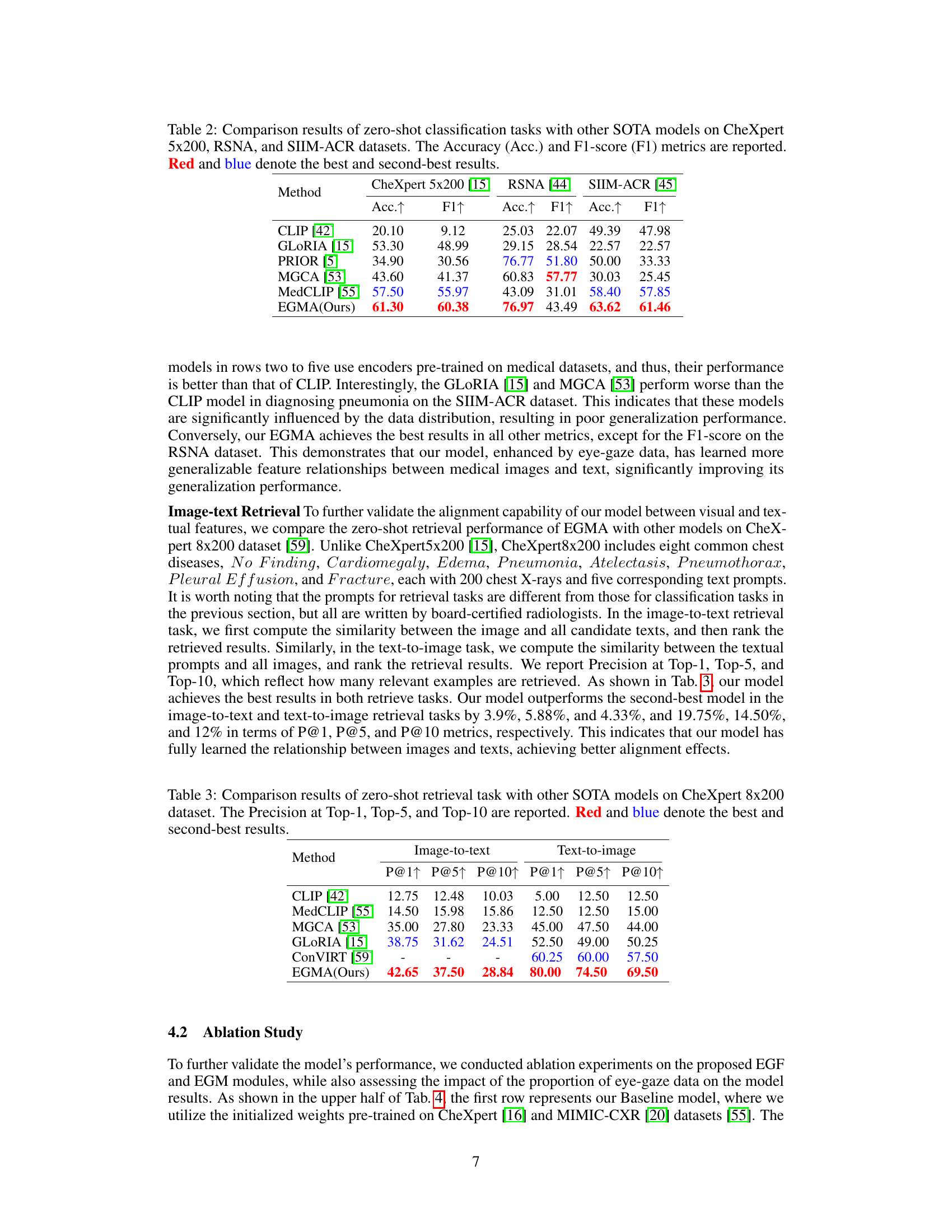
🔼 This table compares the performance of the proposed EGMA model against other state-of-the-art models on three different datasets (CheXpert 5x200, RSNA, and SIIM-ACR) using zero-shot classification. The results are presented as Accuracy and F1-score, showing that EGMA outperforms other models in most cases.
read the caption
Table 2: Comparison results of zero-shot classification tasks with other SOTA models on CheXpert 5x200, RSNA, and SIIM-ACR datasets. The Accuracy (Acc.) and F1-score (F1) metrics are reported. Red and blue denote the best and second-best results.
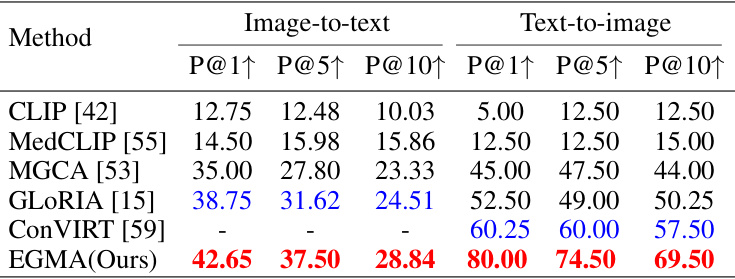
🔼 This table presents a comparison of the zero-shot image-to-text and text-to-image retrieval performance of the proposed EGMA model against several state-of-the-art methods on the CheXpert 8x200 dataset. The metrics used are Precision@1, Precision@5, and Precision@10, indicating the percentage of correctly retrieved items within the top 1, 5, and 10 results, respectively. The best and second-best results are highlighted.
read the caption
Table 3: Comparison results of zero-shot retrieval task with other SOTA models on CheXpert 8x200 dataset. The Precision at Top-1, Top-5, and Top-10 are reported. Red and blue denote the best and second-best results.
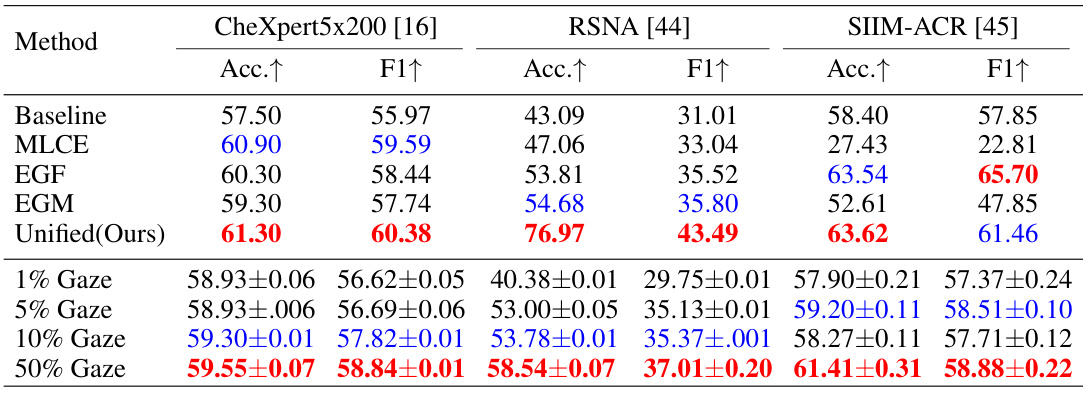
🔼 This table presents the results of ablation experiments performed on the proposed EGMA model for zero-shot classification. It compares the performance of the full model against versions with different components removed (MLCE, EGF, EGM). It also shows the effect of varying the amount of eye-gaze data used for training (1%, 5%, 10%, 50%). The metrics used are accuracy and F1-score, across three different datasets (CheXpert 5x200, RSNA, and SIIM-ACR).
read the caption
Table 4: Comparison results of zero-shot classification ablation experiments on CheXpert 5x200, RSNA, and SIIM-ACR datasets. The Accuracy (Acc.) and F1-score (F1) metrics are reported. Each value in the lower part is the average of three runs. Red and blue denote the best and second-best results.
🔼 This table lists examples of the possible sub-types, severities, and locations of three common chest diseases (Atelectasis, Consolidation, and Pleural Effusion) as provided by a radiologist in the CheXpert 5x200 dataset. This information shows the level of detail in the text descriptions used in the image-text matching task, illustrating the complexity of the relationships between image features and textual descriptions.
read the caption
Table 5: Examples of possible sub-types, severities, and locations provided by the radiologist in CheXpert 5x200 dataset.
🔼 This table presents a comparison of the Area Under the Receiver Operating Characteristic Curve (AUROC) scores achieved by different state-of-the-art (SOTA) models and the proposed EGMA model on three different chest X-ray datasets (CheXpert, RSNA, and SIIM-ACR) for a supervised image classification task. The performance is evaluated using 1%, 10%, and 100% of the training data to assess the impact of data size and model generalization. The best and second-best performing models are highlighted.
read the caption
Table 1: Comparison results of supervised classification task with other SOTA models on CheXpert, RSNA, and SIIM-ACR datasets. Area under ROC curve (AUROC) is reported with different portions of training data: 1%, 10%, 100%. Red and blue denote the best and second-best results.
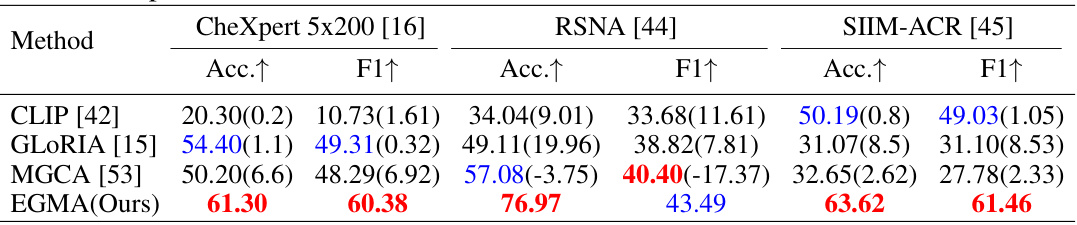
🔼 This table compares the performance of different state-of-the-art models on the zero-shot classification task after fine-tuning them using the proposed EGMA framework. The models include CLIP, GLORIA, and MGCA. The results are shown for three different datasets: CheXpert 5x200, RSNA, and SIIM-ACR. The table shows the accuracy and F1 scores for each model and dataset. The values in parentheses indicate the improvement achieved by each model after being fine-tuned with the EGMA framework. The best and second-best results are highlighted in red and blue, respectively.
read the caption
Table 7: Comparison results of zero-shot classification after continue pre-training using the backbones of other SOTA models in our EGMA framework. Red and blue denote the best and second-best results. The values in (parentheses) represents the improvement over the baseline metrics in Table 1 of main manuscript.
Full paper#