TL;DR#
Current methods for protein representation learning struggle with the diverse data modalities involved (sequence, structure, function) and the differences in data scale. Large language models are effective for text, but integrating this information with other protein data is challenging. Existing multi-modal methods typically ignore functional information or fail to address the data scale discrepancy.
This paper introduces ProtGO, a unified model that addresses these limitations. ProtGO uses a teacher network with a customized GNN and GO encoder to learn hybrid embeddings. This eliminates the need for additional function inputs and uses domain adaptation to guide training, improving performance without sample-wise alignment. ProtGO significantly outperforms state-of-the-art methods on several benchmarks, demonstrating the advantages of its unified approach.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in biomedicine and AI because it presents ProtGO, a novel unified framework for protein representation learning. This addresses a key challenge in the field by effectively integrating diverse data modalities (sequence, structure, function) for more accurate protein modeling. The improved accuracy in protein fold and function prediction has significant implications for drug discovery and related biomedical applications. This work also opens new avenues for research in multimodal learning and domain adaptation for protein data.
Visual Insights#

🔼 This figure illustrates the three core types of information crucial for understanding proteins: their amino acid sequence, their three-dimensional structure, and their biological functions (represented by Gene Ontology annotations). The sequence is shown as a linear chain of amino acids, the structure is a 3D model of the folded protein, and the functions are described using GO terms, which categorize proteins based on their molecular function, biological process, and cellular component.
read the caption
Figure 1: Protein sequence, structure, and function.
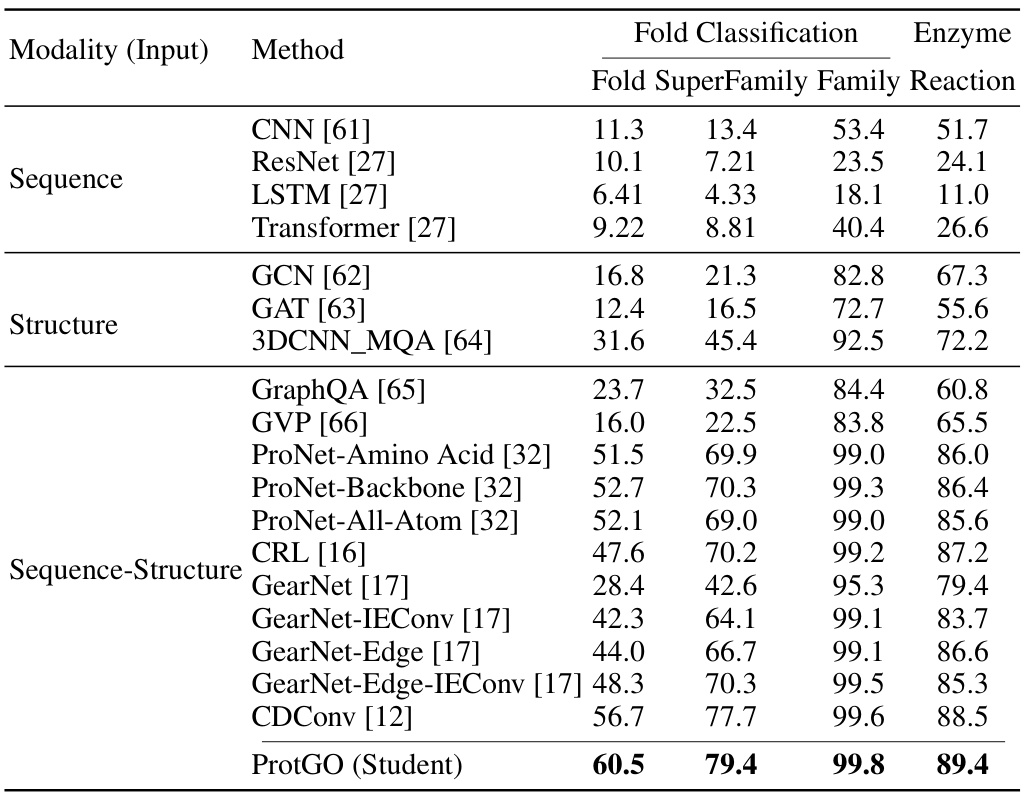
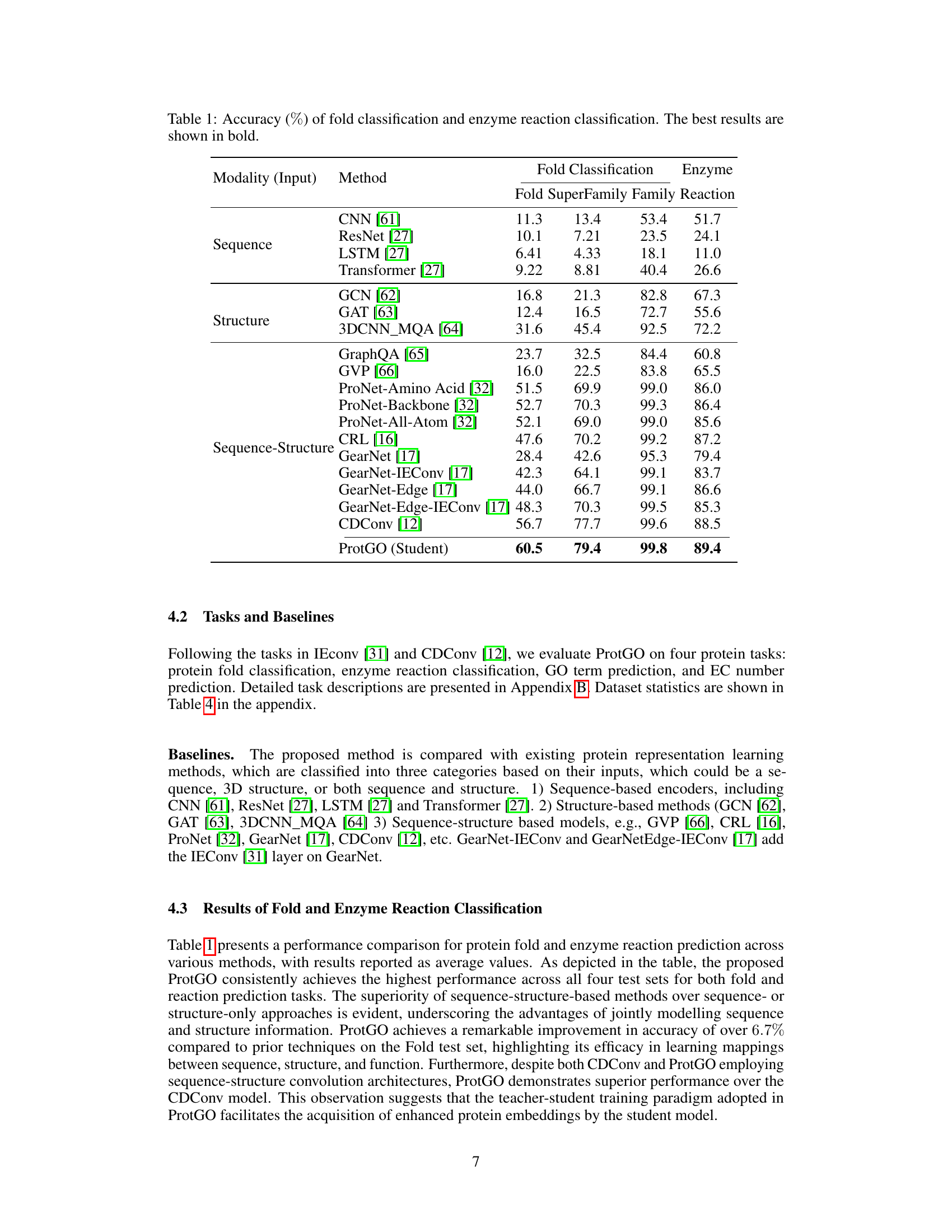
🔼 This table presents the accuracy results (%) for protein fold classification and enzyme reaction classification tasks. It compares the performance of ProtGO (the proposed method) against various other state-of-the-art methods categorized by the input modalities they use: sequence-only, structure-only, and sequence-structure. The results are broken down by fold classification (Fold, Superfamily, Family) and enzyme reaction classification. The best-performing method for each task and category is highlighted in bold, showcasing ProtGO’s superior performance across all categories.
read the caption
Table 1: Accuracy (%) of fold classification and enzyme reaction classification. The best results are shown in bold.
In-depth insights#
ProtGO Overview#
ProtGO is a novel multimodal framework designed for unified protein representation learning. It leverages a teacher-student network architecture, where a teacher model integrates protein sequence, structure, and function data to generate rich embeddings. Importantly, ProtGO employs a domain adaptation technique to align the latent space distributions between the teacher and student models, enabling effective knowledge transfer even with limited functional annotations in the target domain. The student model, trained on sequence and structure data alone, benefits from the functional knowledge distilled from the teacher. This approach successfully addresses the data scarcity issue common in protein studies. The resulting unified representations are particularly advantageous for downstream tasks, demonstrating superior performance over existing unimodal and multimodal methods. The model’s efficiency and scalability make it well-suited for large-scale protein analysis.
Multimodal Fusion#
Effective multimodal fusion in protein representation learning is crucial for leveraging diverse data sources. ProtGO’s approach likely involves a strategic combination of methods to integrate protein sequence, structure, and functional annotations. This might include concatenating embedding vectors from separate unimodal models or employing attention mechanisms to weigh the importance of different modalities. Success hinges on careful alignment of data representations from different sources, possibly through domain adaptation techniques to ensure consistent feature spaces before fusion. The chosen fusion method must address inherent scale differences between modalities (e.g., the abundance of sequences compared to structures). A well-designed multimodal model should demonstrate improved predictive performance in downstream tasks such as function prediction and drug design, showcasing the synergistic effects of integrated information. ProtGO’s reported superior performance suggests a highly effective fusion strategy, though the specific details require further examination of the paper’s methodology.
Domain Adaptation#
Domain adaptation, in the context of the research paper, is crucial for bridging the gap between the teacher and student models. The discrepancy in data distributions between the source domain (teacher, with abundant functional annotations) and target domain (student, often lacking such annotations) presents a significant challenge. The paper effectively addresses this by focusing on aligning latent space distributions rather than individual samples. This approach using distribution approximation is robust and avoids overfitting to specific samples. The method is especially valuable because functional annotation data is limited, thereby improving the generalizability of the student model for broader downstream applications. This is accomplished by minimizing the Kullback-Leibler (KL) divergence, a measure of the difference between two probability distributions. The teacher network guides the training of the student using domain adaptation techniques to ensure optimal knowledge transfer without direct sample alignment, ultimately leading to a more accurate and adaptable protein representation learning model.
ProtGO Evaluation#
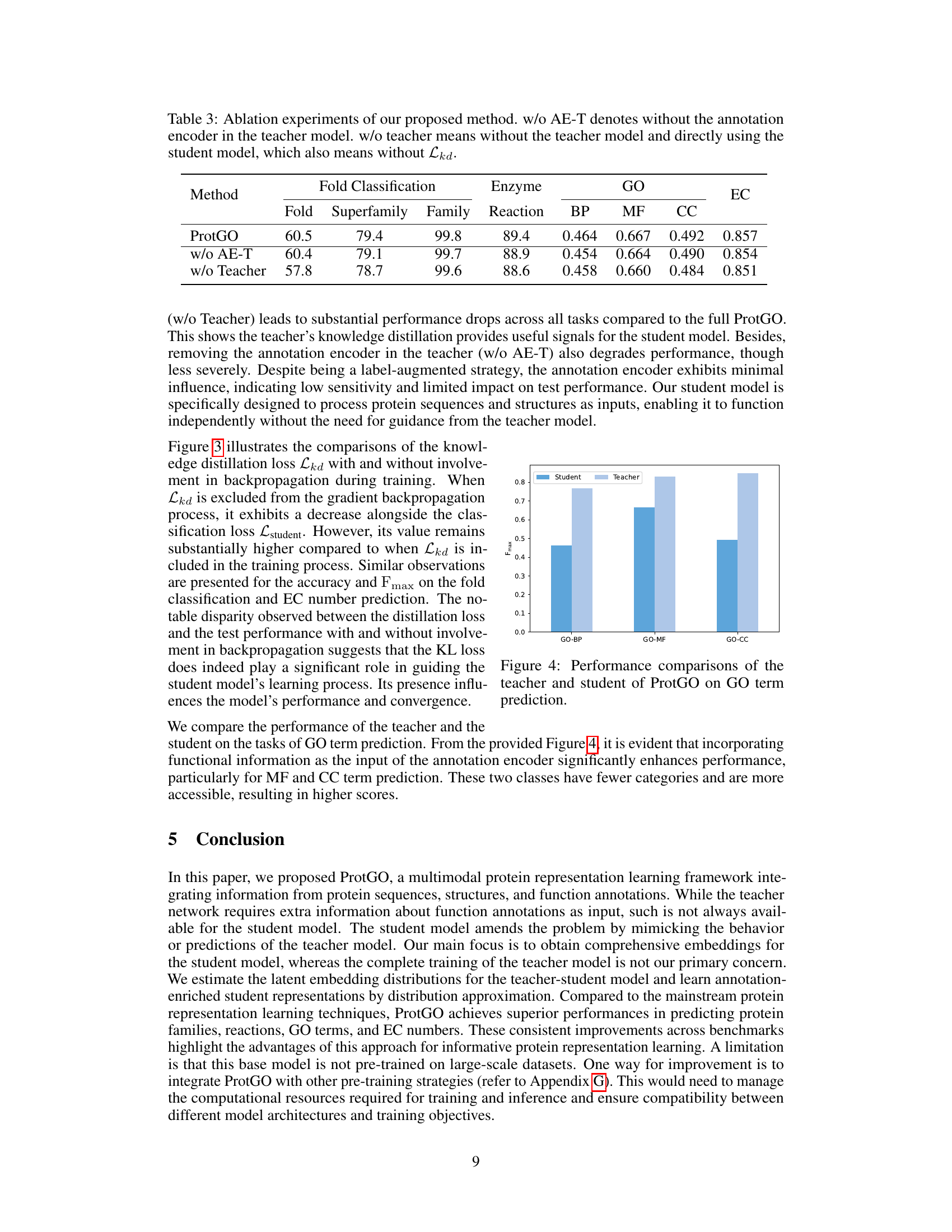
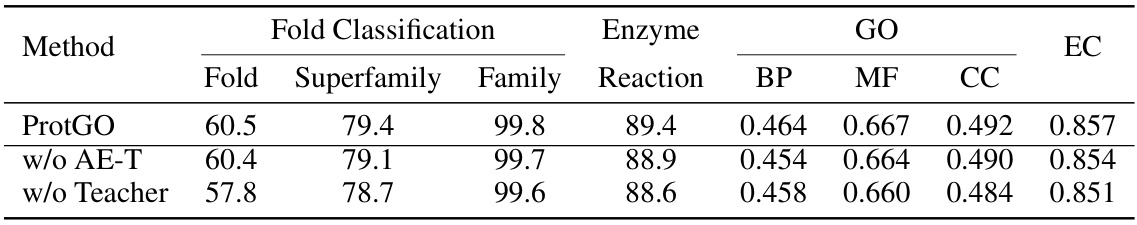
A hypothetical ‘ProtGO Evaluation’ section would likely detail the rigorous benchmarking of the ProtGO model against state-of-the-art baselines across multiple protein-related tasks. Key metrics such as accuracy, precision, recall, and F1-score would be reported, potentially broken down by task (fold prediction, enzyme reaction classification, GO term prediction, EC number prediction) and input modality (sequence, structure, both). The evaluation would emphasize multimodal performance, highlighting ProtGO’s advantage in integrating sequence, structure, and function information compared to unimodal or limited-modality methods. A discussion of statistical significance (p-values, confidence intervals) would be crucial to validate the observed improvements. Finally, the evaluation would likely incorporate an ablation study, systematically removing components of ProtGO (e.g., the teacher model, the annotation encoder) to demonstrate the contribution of each module to overall performance. This detailed analysis would ultimately establish ProtGO’s efficacy and robustness for unified protein representation learning.
Future Directions#
Future research could explore several promising avenues. Improving ProtGO’s scalability to handle even larger datasets and more complex protein structures is crucial. Incorporating additional data modalities, such as protein-protein interaction networks or experimental data like binding affinities, could further enhance the model’s predictive capabilities. Investigating alternative knowledge distillation techniques to optimize knowledge transfer and potentially reduce the computational overhead would be beneficial. Furthermore, applying ProtGO to a wider range of downstream tasks, including protein design and engineering, would be valuable. Finally, developing more robust methods for handling data imbalance and uncertainty in the available functional annotations will be key to improving the model’s generalizability and reliability.
More visual insights#
More on figures
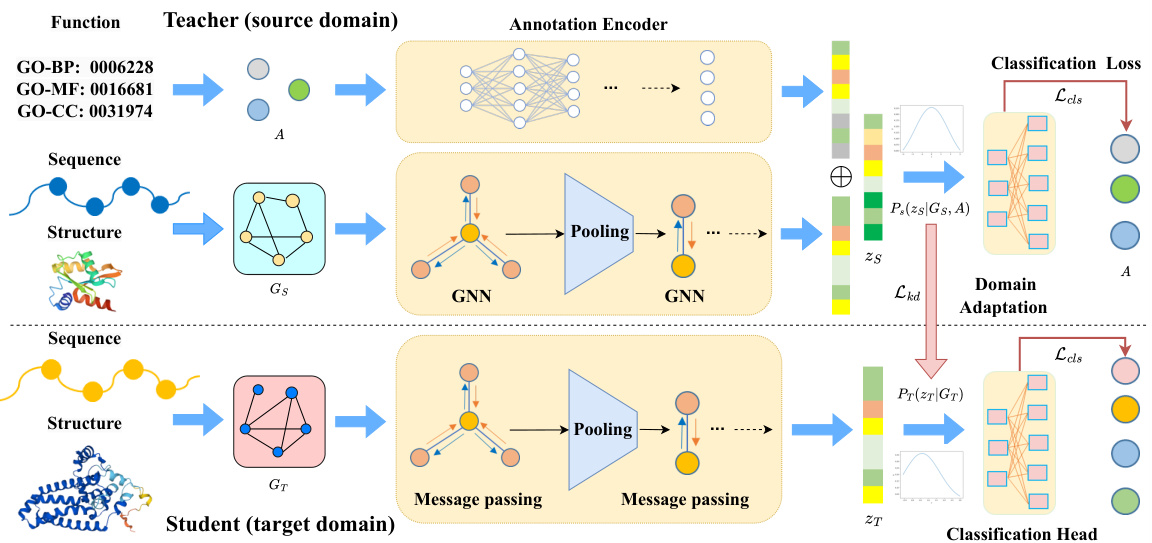
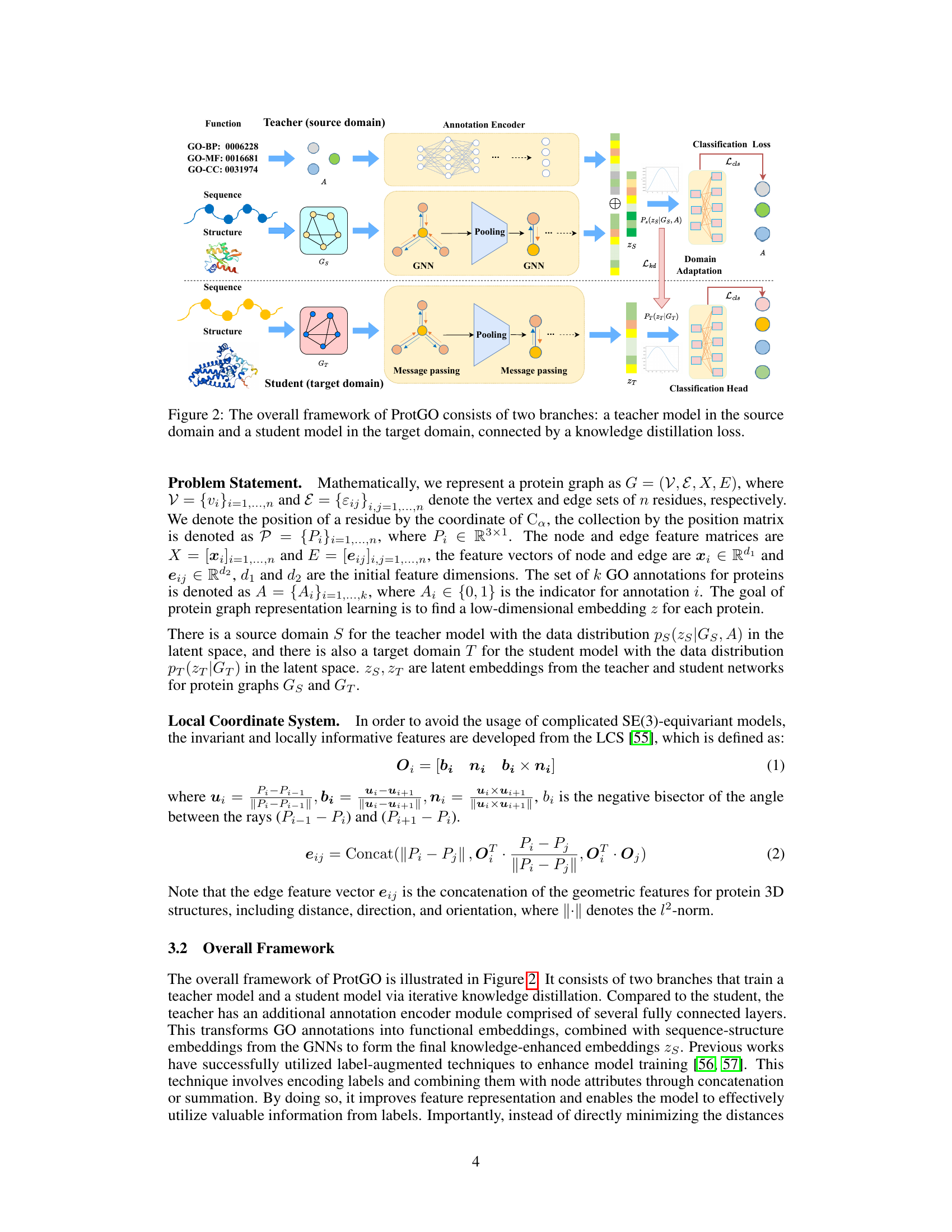
🔼 The figure illustrates the architecture of ProtGO, a multimodal protein representation learning framework. It consists of two branches: a teacher model trained on data including sequence, structure, and function information, and a student model trained primarily on sequence and structure data. The teacher model utilizes a customized graph neural network (GNN) and an annotation encoder to generate hybrid embeddings. Knowledge distillation, specifically minimizing the Kullback-Leibler (KL) divergence between the teacher and student latent space distributions, guides the training of the student model. This approach facilitates the transfer of functional knowledge from the teacher to the student, even when functional annotations are limited for the student’s training data. Domain adaptation is employed to ensure that the student model can generalize well to new data.
read the caption
Figure 2: The overall framework of ProtGO consists of two branches: a teacher model in the source domain and a student model in the target domain, connected by a knowledge distillation loss.
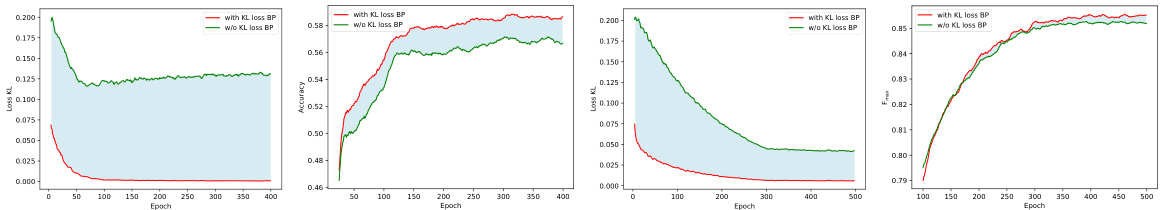
🔼 This figure shows the training loss curves and test performance for fold classification and EC number prediction tasks. Two versions are compared: one where the KL divergence loss (Lkd) is used for domain adaptation, and another where it is not. The results indicate the effectiveness of incorporating Lkd in the training process for improving performance.
read the caption
Figure 3: The KL training loss curves (a), (c) and test performance (b), (d) on the tasks of fold classification and EC number prediction. The red curve denotes that Lkd conducts its function, while the green curve denotes we calculated the value of Lkd, but it is not involved in the process of the gradient backpropagation (BP).
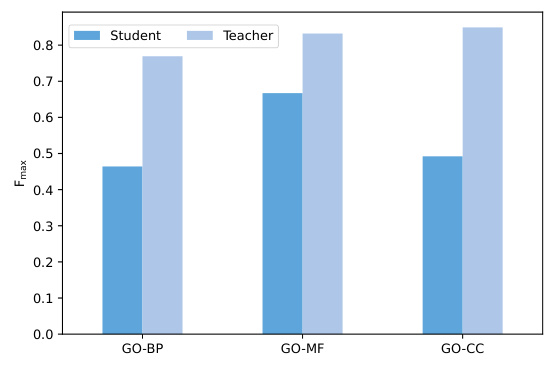
🔼 This figure shows the training and test performance of the ProtGO model on fold classification and EC number prediction tasks. It compares the performance when the KL divergence loss (Lkd) is included in the training process (red curves) versus when it is excluded (green curves). The results demonstrate the impact of the KL loss on the model’s learning and generalization ability.
read the caption
Figure 3: The KL training loss curves (a), (c) and test performance (b), (d) on the tasks of fold classification and EC number prediction. The red curve denotes that Lkd conducts its function, while the green curve denotes we calculated the value of Lkd, but it is not involved in the process of the gradient backpropagation (BP).
More on tables
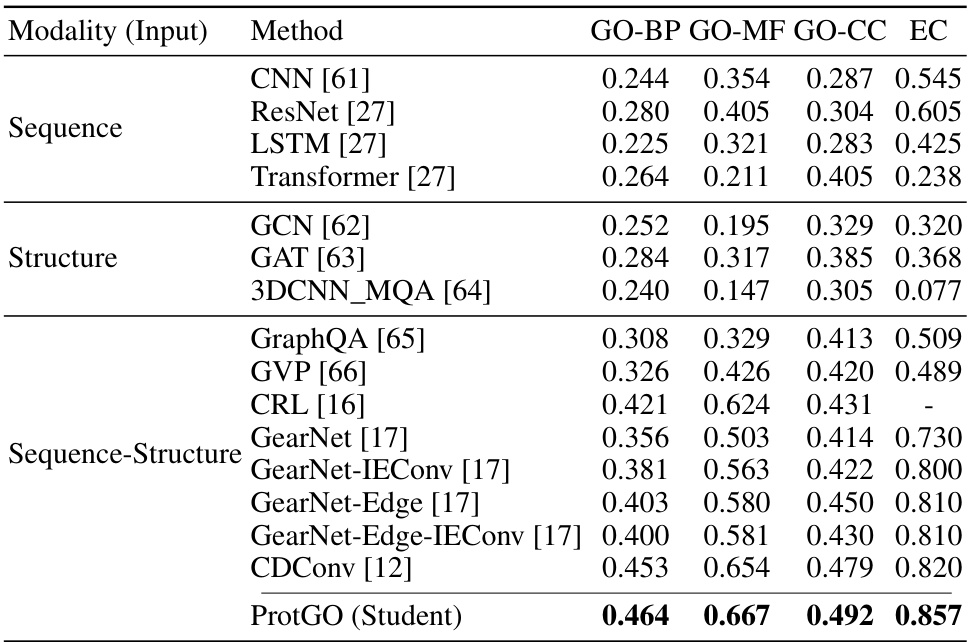
🔼 This table presents the F-measure (Fmax) results for two protein function prediction tasks: Gene Ontology (GO) term prediction and Enzyme Commission (EC) number prediction. It compares the performance of various methods across different modalities (sequence-only, structure-only, and sequence-structure). The best performing method for each task and each GO category (GO-BP, GO-MF, GO-CC) is highlighted in bold. The table helps illustrate the improvement in prediction accuracy achieved by considering multiple protein data modalities.
read the caption
Table 2: Fmax of GO term prediction and EC number prediction. The best results are shown in bold.
🔼 This table presents the accuracy results for protein fold and enzyme reaction classification. It compares the performance of ProtGO against various other methods categorized by the type of input used (sequence only, structure only, or both sequence and structure). The accuracy is reported for different levels of granularity in fold classification (Fold, Superfamily, Family) and for enzyme reactions. The best-performing method for each task is highlighted in bold. The table demonstrates ProtGO’s superior performance, highlighting its ability to integrate multimodal data effectively.
read the caption
Table 1: Accuracy (%) of fold classification and enzyme reaction classification. The best results are shown in bold.

🔼 This table presents the accuracy results for protein fold and enzyme reaction prediction, comparing various methods. The accuracy is calculated as a percentage. Different modalities of input data (sequence only, structure only, and sequence and structure combined) are compared across multiple methods. The best performing method in each category is highlighted in bold.
read the caption
Table 1: Accuracy (%) of fold classification and enzyme reaction classification. The best results are shown in bold.

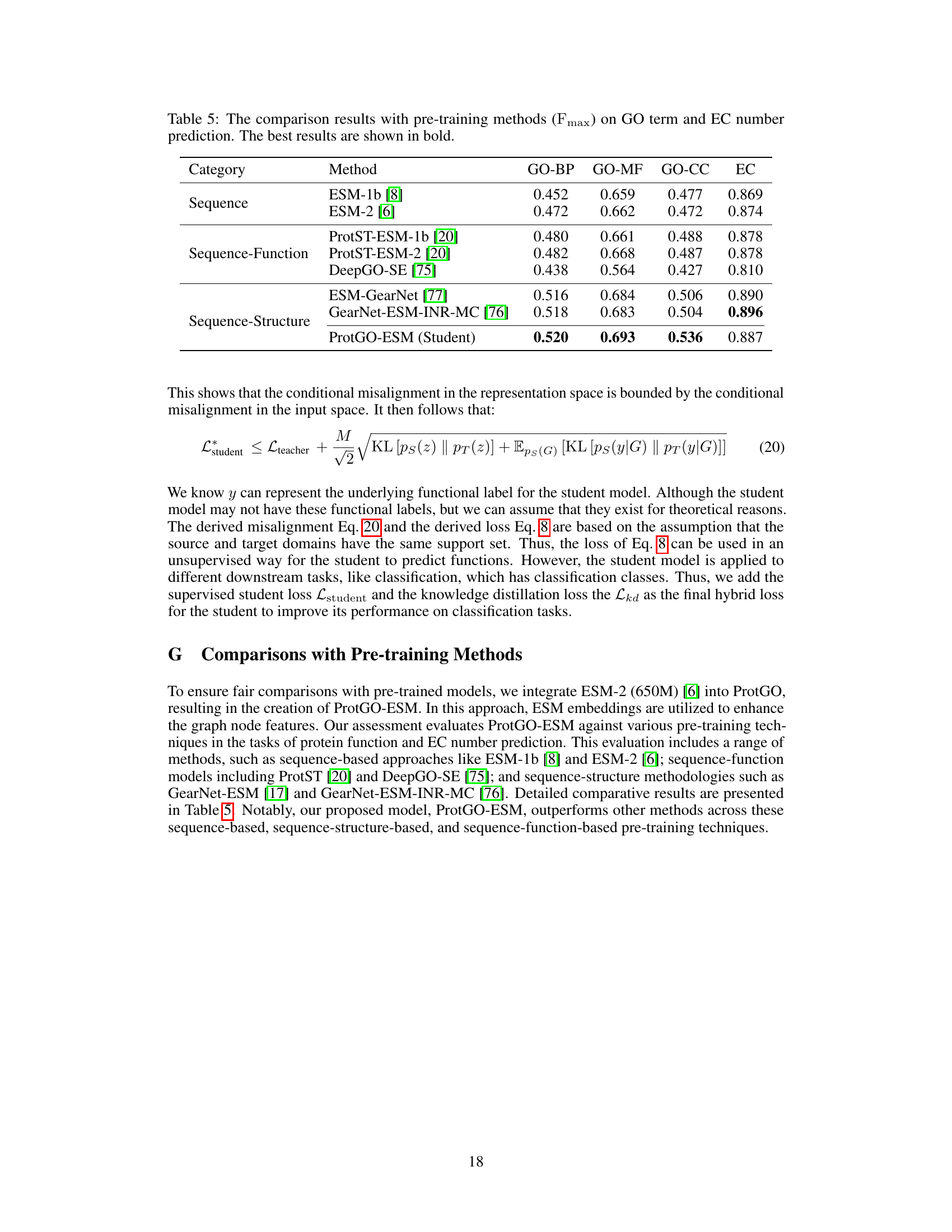
🔼 This table compares the performance of ProtGO-ESM (a version of ProtGO that incorporates ESM embeddings) against other pre-training methods on GO term and EC number prediction tasks. The methods are categorized into sequence-only, sequence-function, and sequence-structure approaches. The Fmax score (a combined measure of precision and recall) is used to evaluate performance. ProtGO-ESM shows competitive results compared to existing pre-training methods.
read the caption
Table 5: The comparison results with pre-training methods (Fmax) on GO term and EC number prediction. The best results are shown in bold.
Full paper#