↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Generating novel crystal structures with desired properties is crucial yet challenging. Current methods using crystallographic information files (CIFs) for language model processing suffer from the lack of unique and invariant sequence representations of 3D structures. This leads to inefficient and inaccurate crystal structure generation.
Mat2Seq overcomes this by introducing a novel tokenization method that transforms 3D crystal structures into unique 1D sequences while guaranteeing both SE(3) and periodic invariance. Experimental results demonstrate that this approach, when integrated with language models, significantly improves the accuracy and efficiency of crystal structure generation, outperforming existing methods in terms of matching rates and RMSE. This work makes significant contributions by providing a more robust and efficient approach to crystal structure prediction, thus advancing the field of materials science.
Key Takeaways#
Why does it matter?#
This paper is crucial for materials science researchers as it presents Mat2Seq, a novel method for invariant tokenization of crystalline materials, enabling the use of powerful language models for crystal structure generation and discovery. This opens new avenues for high-throughput materials discovery, addressing the computational cost limitations of traditional methods. The unique approach of Mat2Seq directly improves the efficiency and accuracy of LLM-based materials prediction.
Visual Insights#

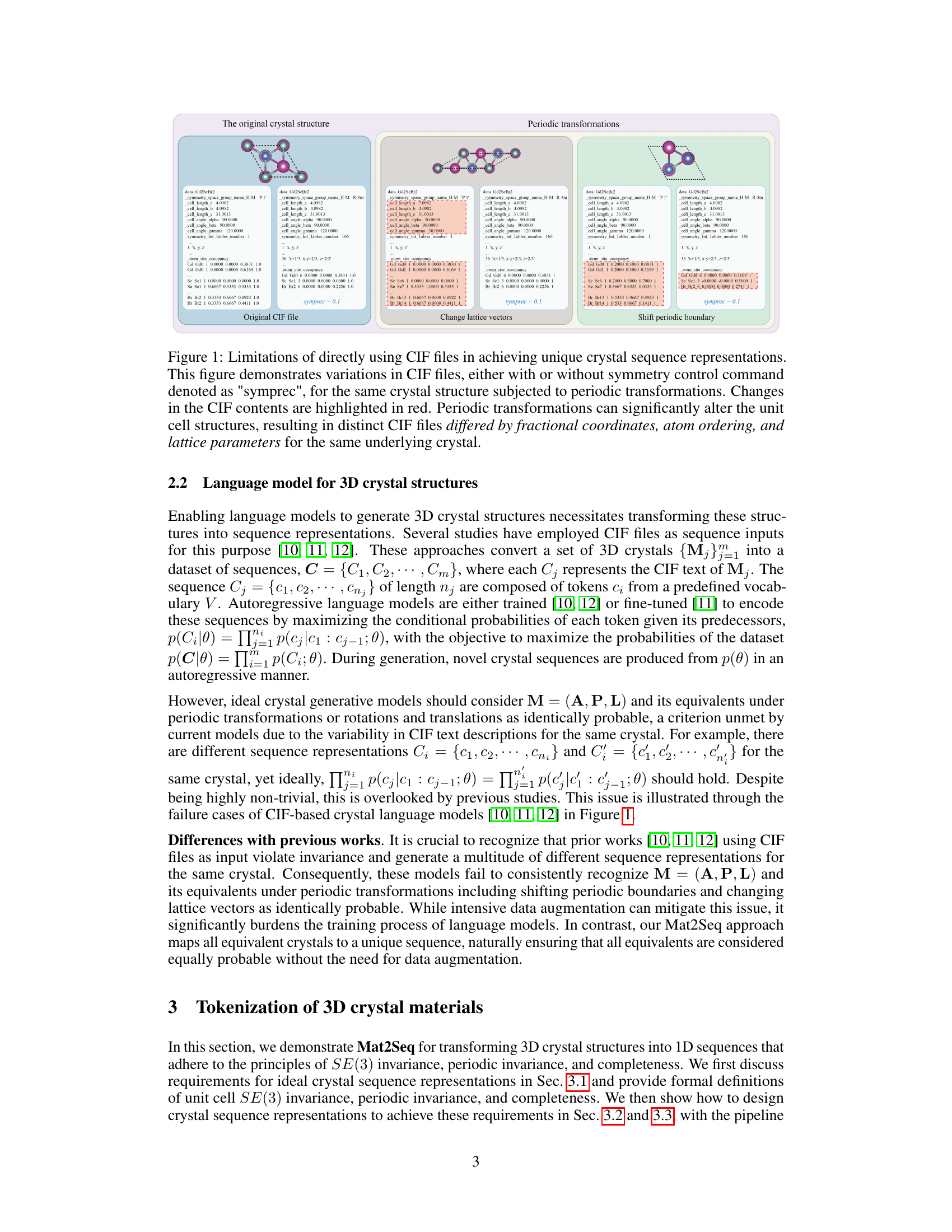
Figure 1 shows how different CIF files can represent the same crystal structure due to periodic transformations and variations in the way the unit cell is defined. The figure highlights the non-uniqueness problem in using CIF files directly for generating crystal structures with language models, as different CIF files may represent the same underlying crystal structure, leading to ambiguity in model training.
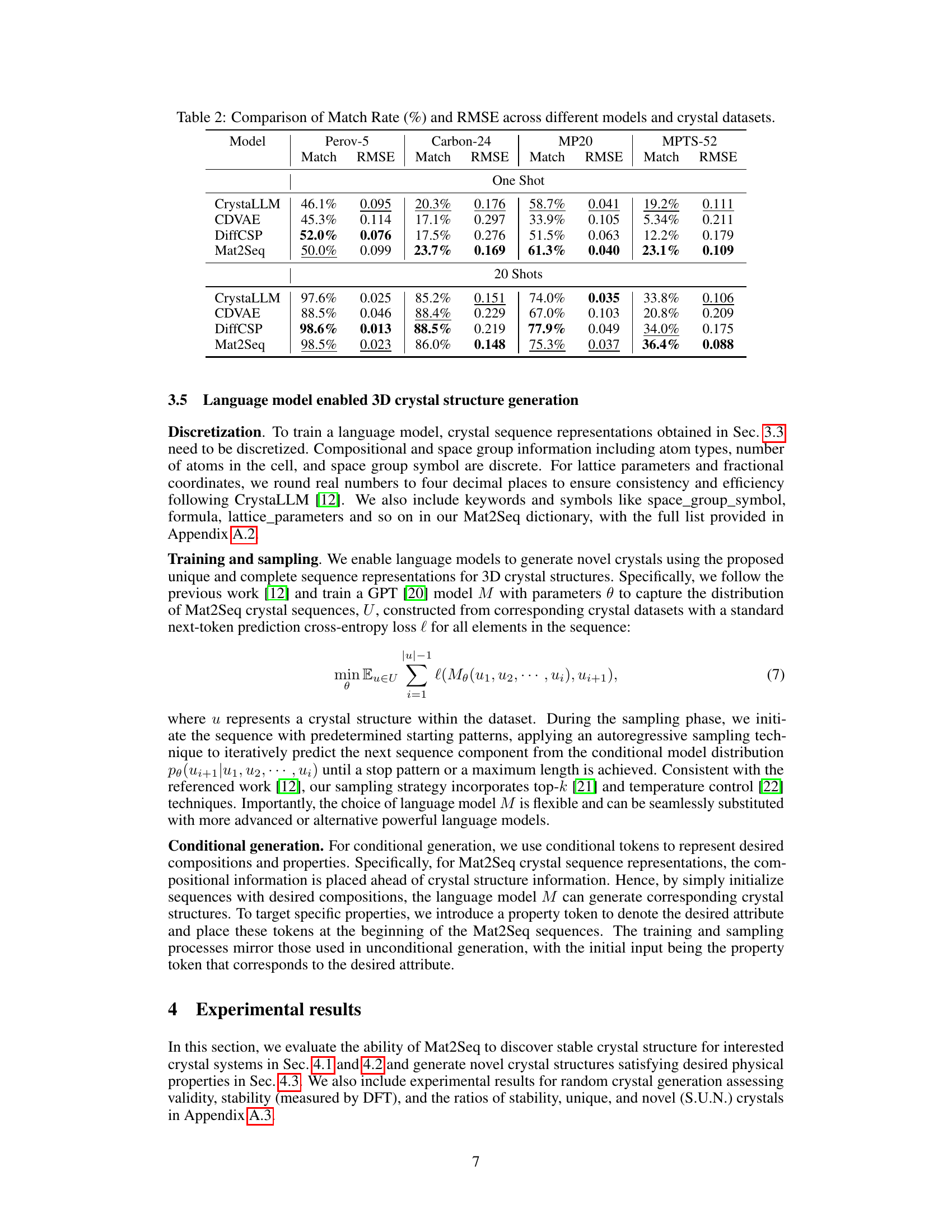
This table compares the performance of four different models (CrystaLLM, CDVAE, DiffCSP, and Mat2Seq) on four different crystal datasets (Perov-5, Carbon-24, MP20, and MPTS-52). The performance is measured using two metrics: Match Rate, which represents the percentage of generated crystal structures that match the ground truth structures, and RMSE, which represents the root mean squared error between the generated and ground truth structures. The table shows results for both one-shot and 20-shot generation settings. Mat2Seq generally shows better performance compared to the other methods.
In-depth insights#
Invariant Tokenization#
Invariant tokenization, in the context of crystalline materials, is a crucial preprocessing step for applying language models to material science. The core challenge lies in converting 3D crystal structures into a 1D sequence that is unique and invariant under various transformations (e.g., rotations, translations, and different unit cell choices). A naive approach of directly using CIF files fails because different CIF representations can describe the same crystal structure. Therefore, a robust invariant tokenization method must ensure that mathematically equivalent crystal structures are encoded into the same unique sequence, allowing the model to learn effectively from diverse representations of the same underlying structure. This is essential to prevent the model from overfitting to specific representations rather than learning the inherent properties of the material. Successful invariant tokenization will unlock the potential for language models to generate and discover novel crystalline materials with desired properties in a computationally efficient manner.
LLM Crystal Gen#
LLM Crystal Gen represents a significant advancement in materials science, leveraging the power of large language models (LLMs) to design and generate novel crystal structures. This approach moves beyond traditional trial-and-error methods and high-cost computational techniques. The core innovation likely involves representing 3D crystal structures as 1D sequences, a process crucial for LLM compatibility. This transformation must be carefully designed to ensure uniqueness and invariance, meaning different mathematical descriptions of the same crystal map to a single unique sequence. This is a key challenge, as the inherent periodicity and symmetry of crystals can lead to multiple valid representations. Successful LLM Crystal Gen hinges on efficient and complete tokenization to accurately capture the essential structural information. The resulting LLM should be capable of generating stable, valid crystal structures, potentially with desired properties specified as input conditions. This technology has the potential to accelerate materials discovery significantly, enabling faster identification of crystals with specific functionalities. The main challenges likely involve the design of the tokenization scheme and the training of robust LLMs capable of handling the complex mathematical properties of crystals.
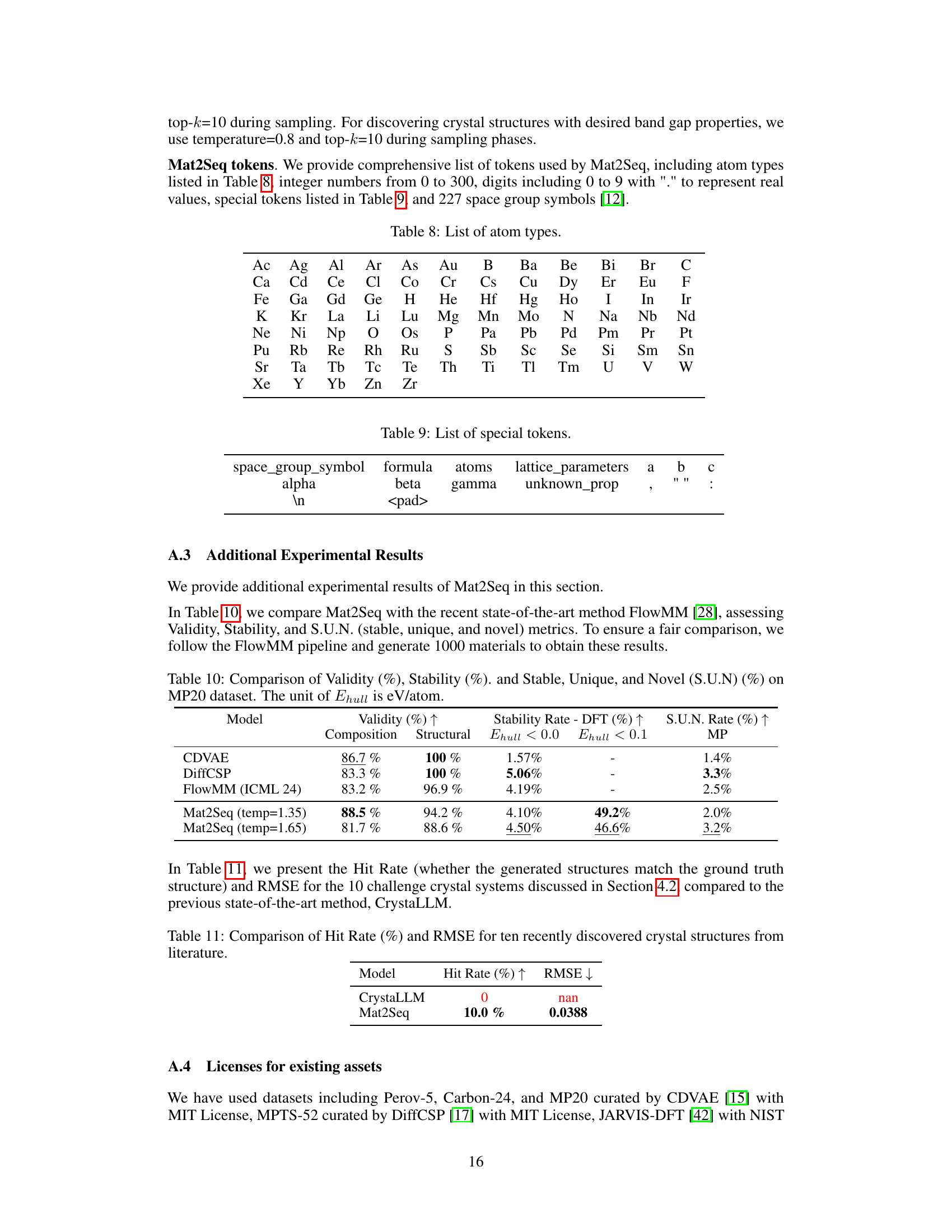
Mat2Seq Pipeline#
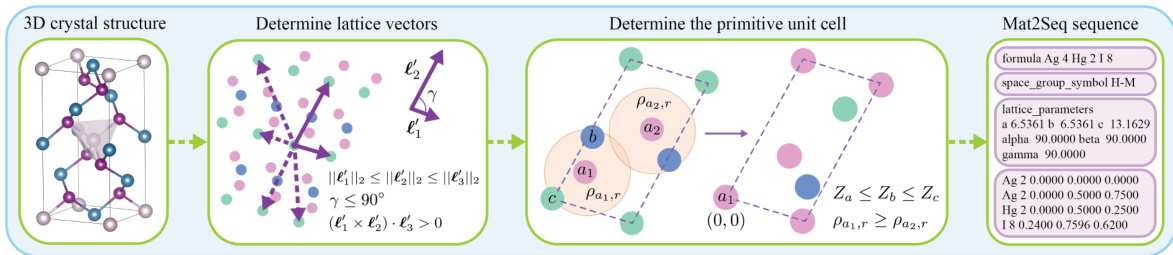
The Mat2Seq pipeline is a crucial contribution, tackling the challenge of converting 3D crystal structures into 1D sequences suitable for language model processing. Its novelty lies in guaranteeing SE(3) and periodic invariance, ensuring that mathematically equivalent descriptions of the same crystal yield identical 1D representations. This is achieved through a multi-step process. First, SO(3) equivariant unit cells are identified, employing Niggli cell reduction to find a unique, rotationally invariant cell. Next, the pipeline generates SE(3) invariant sequences, representing these cells in a way that’s invariant to both rotations and translations, achieving the desired uniqueness. The resulting sequences are thus complete, allowing for the reconstruction of the original 3D structure, crucial for successful LLM training and novel crystal structure generation. The pipeline’s rigorous approach addresses the limitations of prior methods that rely on CIF files, offering a significant improvement in the accuracy and efficiency of crystal structure prediction and discovery.
Uniqueness Proof#
A Uniqueness Proof in a research paper rigorously demonstrates that a proposed method or algorithm produces only one specific output for a given input, eliminating ambiguity. This is crucial for reproducibility and reliability, especially in fields like materials science where slight variations in input can lead to vastly different outcomes. The proof needs to consider all possible scenarios, accounting for any symmetries or equivalences in the input data. A robust proof would involve mathematical formalism, possibly leveraging group theory or other relevant mathematical structures to establish a bijective mapping between inputs and outputs. It should also explicitly address any potential edge cases or exceptions, demonstrating the method’s consistent behavior across a wide range of inputs. Strong uniqueness guarantees are paramount for applications requiring predictable and dependable results; the absence of a rigorous proof raises concerns about the generalizability and practical utility of the presented method. The strength of a uniqueness proof also depends on its clarity and accessibility – a mathematically sound but obscure proof is less valuable than a clear and concise one that facilitates understanding and verification by the wider scientific community. The quality of the proof directly impacts the credibility and trust placed in the research findings.
Future Directions#
Future research could explore extending Mat2Seq to handle more complex crystal structures, including those with defects or disorder, which are common in real-world materials. Improving the efficiency of the Mat2Seq algorithm is also crucial, particularly for larger crystals or high-throughput screening applications. Investigating the use of more advanced language models, such as those with greater capacity and more sophisticated architectures, could further enhance the performance and capabilities of the LLM-based crystal structure generation. Exploring novel sequence representations that capture additional material properties beyond compositional and structural information, like electronic and magnetic properties, would broaden the scope of the technology. Finally, integrating Mat2Seq with other experimental and computational tools to accelerate the crystal discovery process would be a valuable future direction.
More visual insights#
More on figures
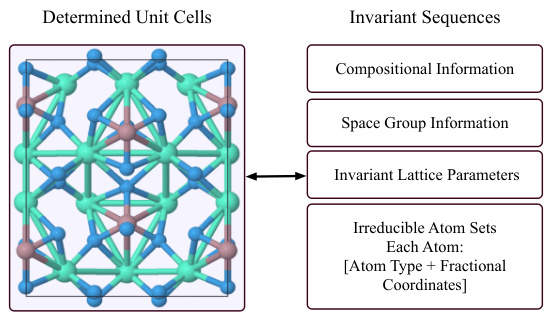
This figure illustrates the Mat2Seq pipeline’s final step, converting the determined SO(3) equivariant unit cells into SE(3) invariant sequences. It details the components of the generated sequence: compositional information (chemical formula), space group information (symmetry), invariant lattice parameters (unit cell dimensions and angles), and irreducible atom sets. Each atom within the irreducible set is further represented by its atom type and fractional coordinates within the unit cell. This unique sequence representation ensures SE(3) and periodic invariance for the crystal structure.
This figure shows how different CIF files can represent the same crystal structure due to the flexibility in describing crystal structures with CIF. The variations highlighted in red demonstrate how periodic transformations (shifting the unit cell or changing lattice vectors) lead to different CIF files with varying fractional coordinates, atom ordering, and lattice parameters despite representing the same underlying crystal structure. This lack of unique representation is a limitation for using CIF files directly in language models for crystal structure generation.
More on tables

This table compares the success rates of different methods in achieving unique crystal sequence representations. Mat2Seq demonstrates a 100% success rate, significantly outperforming previous methods (CIF and CIF with symmetry) which had success rates of 0% and 30%, respectively. This highlights Mat2Seq’s ability to generate unique and consistent representations for identical crystal structures, regardless of their mathematical description.
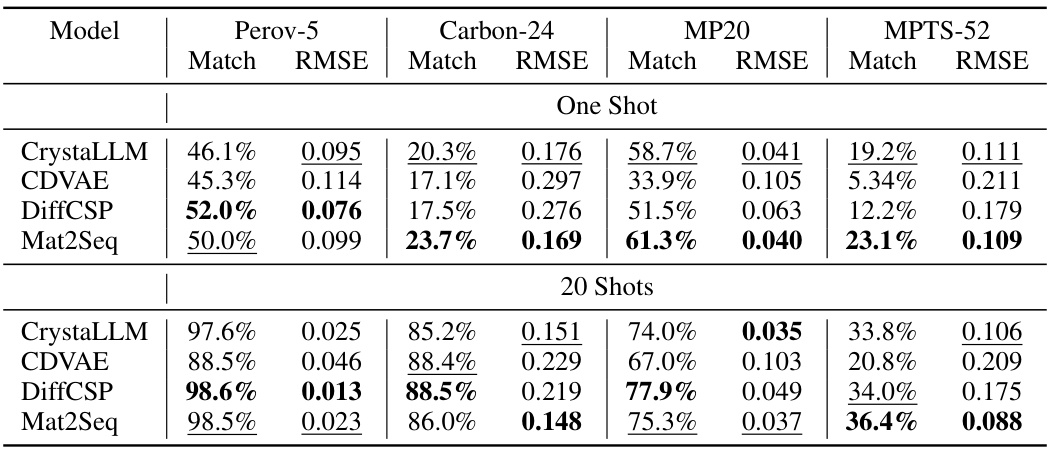
This table compares the performance of four different models (CrystaLLM, CDVAE, DiffCSP, and Mat2Seq) on four crystal datasets (Perov-5, Carbon-24, MP20, and MPTS-52) in terms of Match Rate and RMSE. The results are shown for both one-shot and 20-shot generation settings. Match Rate indicates the percentage of generated crystal structures that match the ground truth structures. RMSE represents the root mean squared error, measuring the difference between generated and ground truth crystal structures.
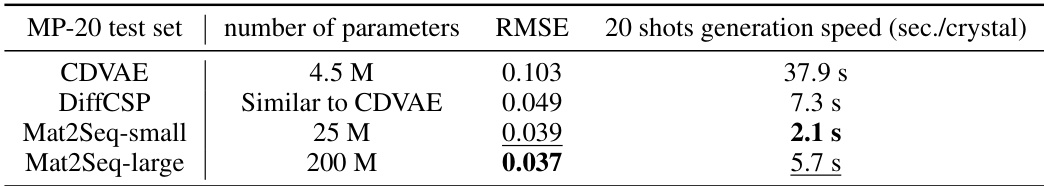
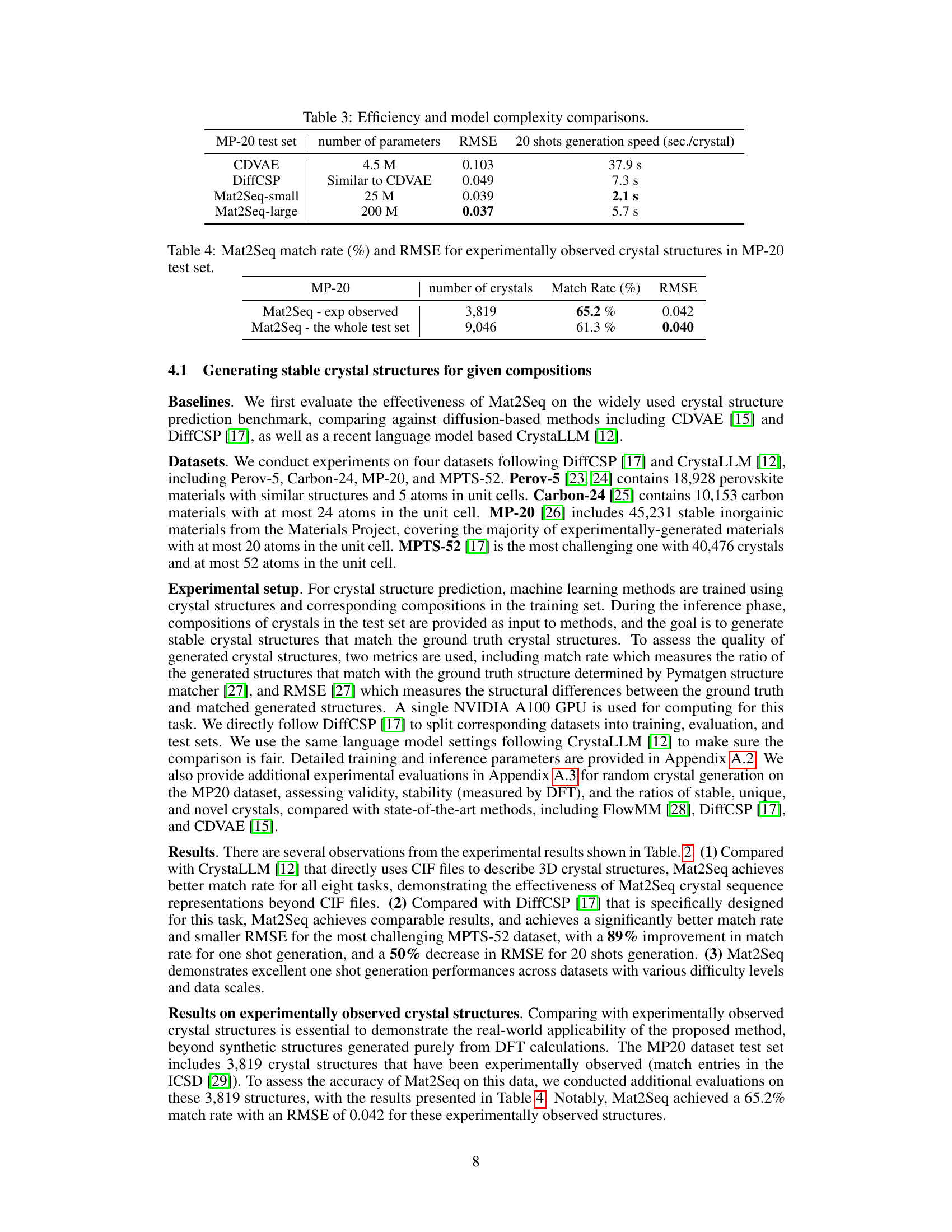
This table compares the efficiency and model complexity of different methods for crystal structure generation, including CDVAE, DiffCSP, and Mat2Seq (both small and large versions). The metrics compared are the number of model parameters, the root mean squared error (RMSE) of the generated structures compared to the ground truth, and the time taken to generate structures using 20 shots.
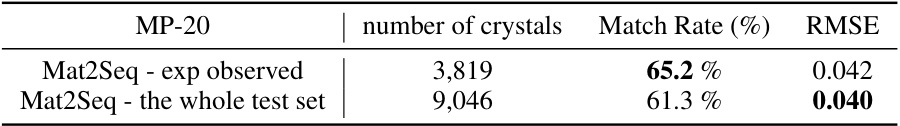
This table presents the performance of the Mat2Seq model on a subset of the MP-20 test set containing only experimentally observed crystal structures. It shows the match rate (percentage of generated structures that match existing known structures) and the root mean squared error (RMSE) which measures the structural difference between generated and experimental structures. Two rows are provided; one showing performance on experimentally observed structures, and another on the entire MP-20 test set for comparison.

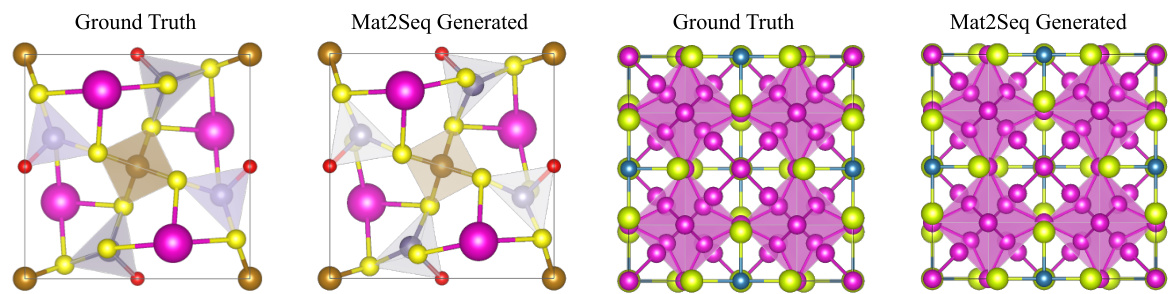
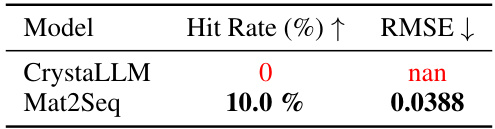
This table compares the ability of CrystaLLM and the proposed Mat2Seq method to generate recently discovered crystal structures from literature. The ‘Validity’ metric, along with its standard deviation, represents the percentage of successfully generated structures that are considered valid based on some criteria (not explicitly detailed in the provided text). The results suggest that Mat2Seq shows improved validity compared to CrystaLLM.

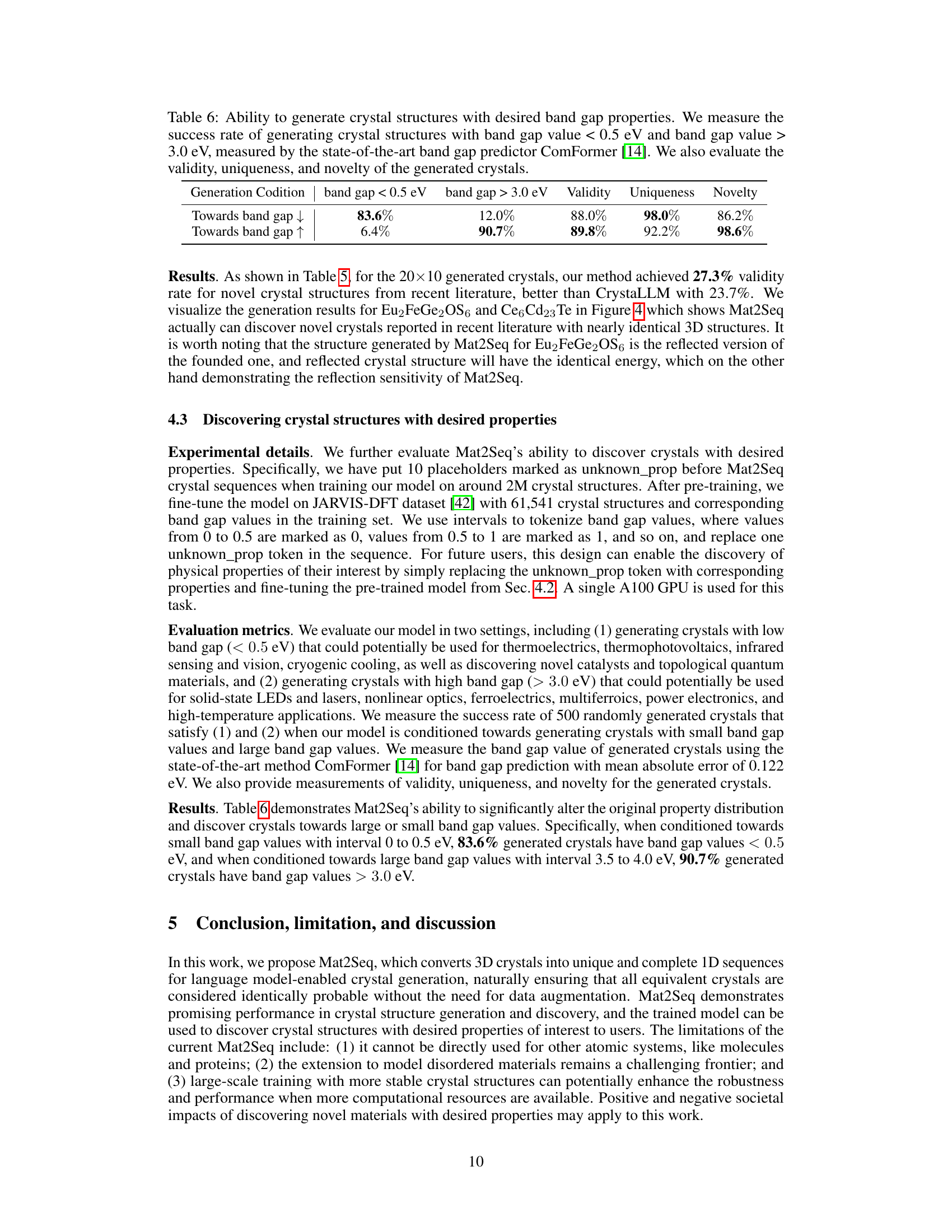
This table presents the results of experiments aiming to generate crystals with specific band gap properties using the Mat2Seq model. It shows the success rates of generating crystals with band gaps below 0.5 eV and above 3.0 eV, assessed using the ComFormer model. Further, it evaluates the validity, uniqueness, and novelty of the generated crystals.

This table lists the hyperparameters used to train the Mat2Seq model for crystal structure prediction on different datasets. It includes the window size, batch size, learning rate, number of training iterations, and dropout rate for each dataset. The datasets used are Perov-5, Carbon-24, MP20, MPTS52, CrystaLLM 2.3M, and JARVIS bandgap.

This table compares the success rate of achieving unique crystal sequence representations for different methods, including Mat2Seq and prior methods that use CIF files. The uniqueness of crystal sequence representation is crucial for successful LLM-based discovery of novel crystalline materials. Mat2Seq significantly outperforms previous methods with 100% success, highlighting its effectiveness in creating unique representations for identical crystals under various transformations.

This table lists the special tokens used in the Mat2Seq model. These tokens represent various pieces of information about the crystal structure, including the space group symbol, chemical formula, number of atoms, lattice parameters (a, b, c, alpha, beta, gamma), and a placeholder for unknown properties.

This table compares the performance of several models (CDVAE, DiffCSP, FlowMM, and Mat2Seq) on the MP20 dataset for crystal structure generation. It evaluates the generated structures based on their validity (how well they match the ground truth), stability (assessed using DFT calculations), and S.U.N rate (percentage of generated structures that are stable, unique, and novel). Two different temperatures were used for Mat2Seq, resulting in slightly different outcomes.
This table compares the success rate of the Mat2Seq method with previous methods (CrystaLLM, CIF with and without symmetry control) in achieving unique crystal sequence representations. The uniqueness is evaluated by checking if different CIF files representing the same crystal structure (subjected to periodic transformations) lead to the same sequence representation. A success rate of 100% indicates that Mat2Seq successfully generated unique sequences for all equivalent crystal structures.
Full paper#