↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Endoscopic video analysis faces challenges like complex camera movements and uneven lesion distribution. Current contrastive learning methods improve model discriminability but lack fine-grained information needed for pixel-level tasks. This paper introduces a novel Multi-view Masked Contrastive Representation Learning (M²CRL) framework to address these issues.
M²CRL employs a multi-view masking strategy with frame-aggregated attention guided tube mask for global views and random tube mask for local views. This, combined with contrastive learning, generates representations with both fine-grained perception and holistic discriminative capabilities. The framework was pre-trained on seven publicly available datasets and fine-tuned on three, outperforming state-of-the-art methods across classification, segmentation, and detection tasks. This work shows a promising direction for self-supervised pre-training in medical image analysis.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image analysis and computer vision due to its significant advancements in self-supervised learning for endoscopic video analysis. The multi-view masked contrastive representation learning (M²CRL) framework offers a novel approach to overcome limitations of existing methods and significantly improves performance in various downstream tasks (classification, segmentation, and detection). This research opens new avenues for exploring more effective and efficient self-supervised learning techniques for medical image applications and has the potential to improve the accuracy and efficiency of clinical diagnosis and healthcare services.
Visual Insights#
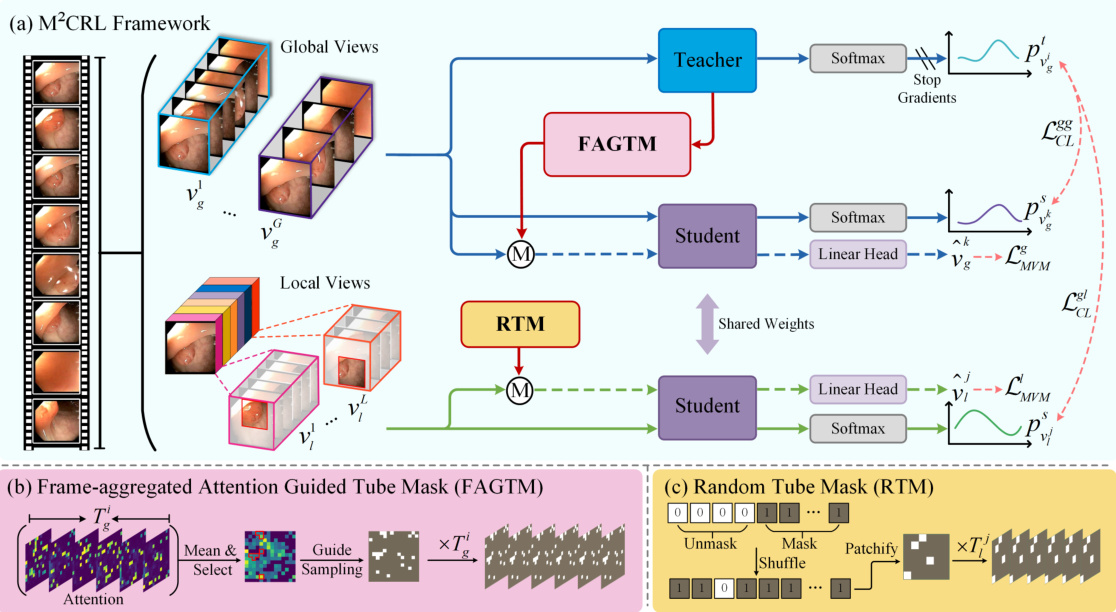
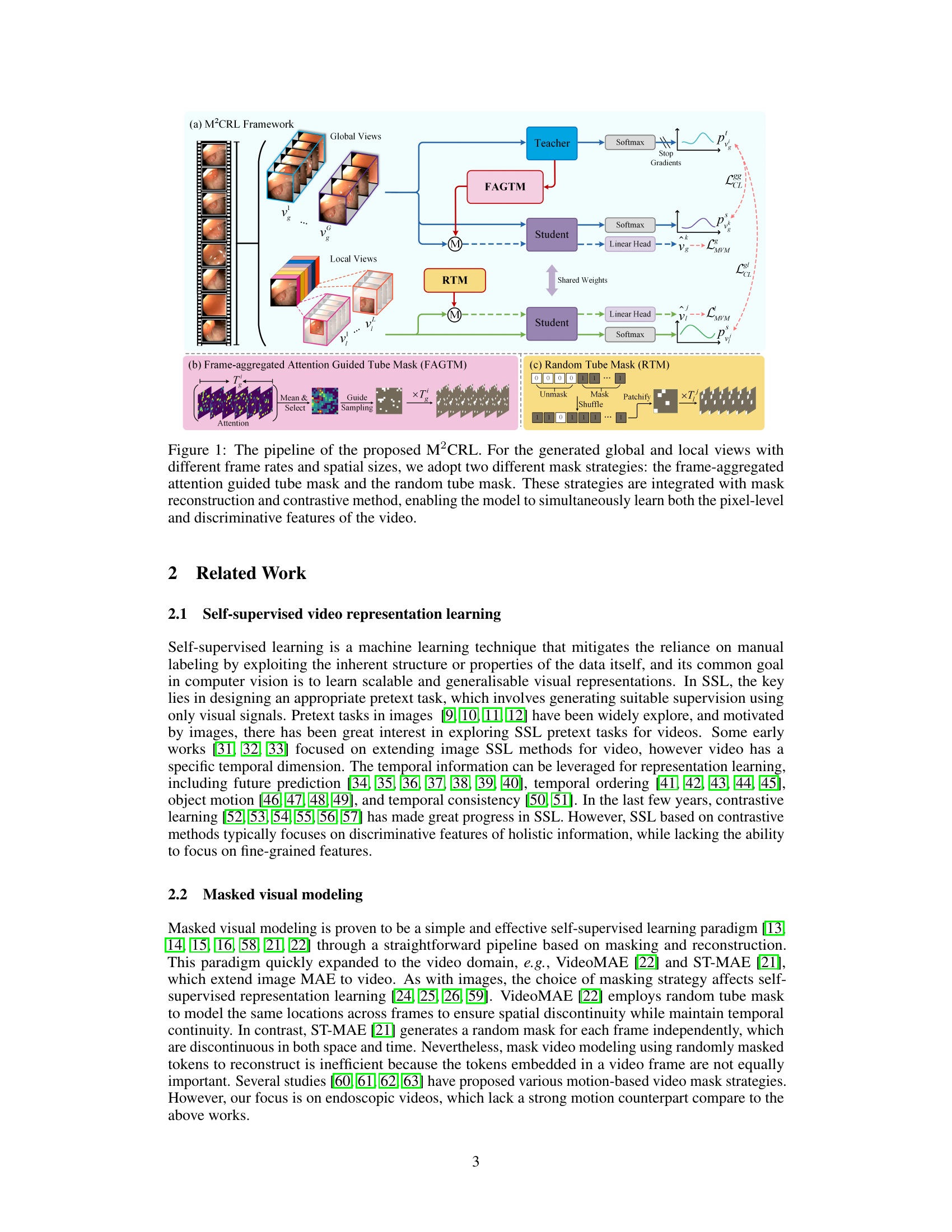
This figure illustrates the architecture of the proposed Multi-view Masked Contrastive Representation Learning (M²CRL) framework. It shows how global and local views of endoscopic videos are processed using different masking strategies (Frame-aggregated Attention Guided Tube Mask and Random Tube Mask) combined with contrastive learning and mask reconstruction. The goal is to learn both fine-grained pixel-level details and holistic discriminative features for improved performance on downstream tasks.
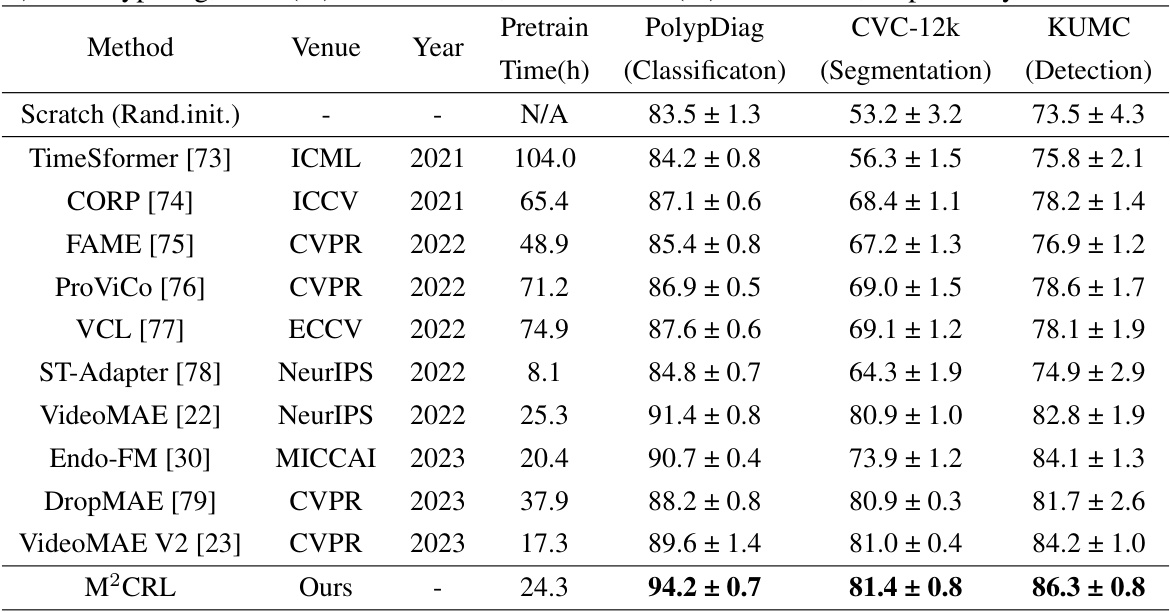
This table compares the performance of the proposed M²CRL method against other state-of-the-art (SOTA) self-supervised learning methods on three endoscopic video downstream tasks: Polyp classification (PolypDiag dataset), polyp segmentation (CVC-12k dataset), and polyp detection (KUMC dataset). The results are presented as F1 scores (percentage) for classification and detection tasks, and Dice scores (percentage) for the segmentation task. The table highlights the improvements achieved by M²CRL over existing methods.
In-depth insights#
Multi-view Masking#
The concept of “Multi-view Masking” in the context of a research paper likely refers to a technique that leverages multiple perspectives or representations of the same data for improved performance. This approach is particularly relevant in scenarios where a single view might be insufficient or prone to bias, such as in image or video analysis. By employing multiple views (e.g., local and global), the method might improve robustness, leading to more effective model training. The masking aspect likely involves strategically occluding portions of the data in each view, potentially enhancing feature learning by forcing the model to focus on specific details or relationships. The strategic masking patterns combined across multiple views would be key, creating a powerful data augmentation technique. The combination of multi-view and masking is likely designed to overcome limitations of traditional single-view approaches by allowing the model to capture complementary features and reduce dependence on any single view’s potential flaws. This method likely offers significant advantages for tasks like object detection and classification in image or video data where the richness of multiple viewpoints combined with masked data augmentation could lead to improved model generalization and robustness.
Contrastive Learning#
Contrastive learning, a self-supervised learning approach, is explored in the context of enhancing the discriminability of models for endoscopic video analysis. The core idea is to learn representations by pulling similar data points closer together and pushing dissimilar ones apart in a feature space. This technique is particularly valuable for endoscopic video analysis, which often suffers from challenges like complex camera movements and uneven lesion distributions. However, while contrastive learning effectively improves instance-level discriminability, it can be limited in its ability to capture fine-grained, pixel-level details crucial for tasks such as segmentation and detection. Therefore, the research often combines contrastive learning with other techniques like masked visual modeling to address this limitation. Masked visual modeling helps learn fine-grained features by reconstructing masked portions of the video, complementing the holistic discriminative abilities offered by contrastive learning. The combination aims to achieve a balance between holistic discriminative power and detailed, pixel-level perception. The effectiveness of this combined approach is often demonstrated empirically through downstream tasks like classification, segmentation, and detection on publicly available endoscopic video datasets.
Endoscopy App.#
An endoscopy application, or “Endoscopy App,” would represent a significant advancement in healthcare, potentially revolutionizing how medical professionals perform and interpret endoscopic procedures. Key features could include high-resolution image and video capture, real-time image processing for enhanced visualization (e.g., chromoendoscopy simulation), AI-powered lesion detection and classification to assist with diagnosis, and integration with electronic health records (EHRs) for seamless data management. User-friendliness would be paramount, requiring intuitive interfaces and tools for easy navigation and operation, even for less experienced endoscopists. Data security and privacy would be critical considerations, requiring robust measures to protect patient information. The app’s success would hinge on the accuracy and reliability of its AI capabilities, requiring rigorous testing and validation. Furthermore, integration with existing hospital systems would be essential for adoption, ensuring that the app seamlessly integrates with workflow and data storage infrastructure. Finally, the “Endoscopy App” would likely need continuous updates and improvements, incorporating advancements in AI technology and user feedback to enhance its capabilities and value.
Ablation Studies#
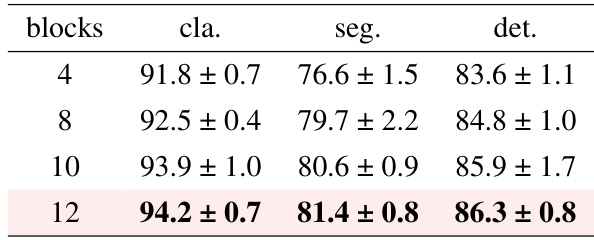
Ablation studies systematically assess the contribution of individual components within a model. In the context of a research paper, they are crucial for understanding the impact of specific design choices. A well-designed ablation study isolates variables, removing or altering one at a time to observe the effect on overall performance. This allows researchers to determine which parts are essential, redundant, or even detrimental. The results help refine the model, optimize its efficiency, and build a stronger understanding of the underlying mechanisms driving its success. For instance, an ablation study might involve removing a particular module, altering hyperparameters, changing input data, or changing the optimization strategy. By carefully analyzing the results from each ablation, researchers can gain insights into which features are most impactful and why. This approach not only validates the core design, but also identifies weaknesses and suggests avenues for future improvement, showcasing a rigorous methodology.
Future Works#
Future work in this research area could explore several promising directions. Extending the model to incorporate multimodal data, such as audio and text streams alongside video, could significantly enrich the model’s understanding of endoscopic procedures and improve diagnostic accuracy. Addressing the computational cost of pre-training is crucial for wider applicability; investigating more efficient training methods or exploring model compression techniques would be valuable. Investigating different masking strategies beyond the current multi-view approach could further optimize the model’s ability to learn fine-grained spatial and temporal features. Finally, thorough evaluation on a broader range of endoscopic datasets is needed to demonstrate the generalizability of the model’s performance across diverse clinical scenarios and imaging modalities. A focus on improving robustness and handling unseen data is vital for practical clinical implementation.
More visual insights#
More on figures

This figure shows a qualitative comparison of the segmentation and detection results obtained using the proposed M²CRL method and other state-of-the-art self-supervised pre-training methods on the CVC-12k and KUMC datasets. The left side displays segmentation results, highlighting the model’s ability to accurately segment polyp regions, even in challenging scenarios with overlapping or small polyps. The right side shows detection results, demonstrating the method’s effectiveness in identifying and localizing polyps with varying sizes and contrasts. The figure visually demonstrates the superior performance of M²CRL in both tasks, especially in capturing fine-grained details and handling challenging scenarios.

This figure shows a qualitative comparison of the segmentation and detection results obtained using different methods on two benchmark datasets for endoscopic video analysis. The left half displays the segmentation results on the CVC-12k dataset, while the right half shows detection results on the KUMC dataset. Each column represents a different method, allowing for a visual comparison of how well each method performs in segmenting polyps and detecting them within endoscopic videos. The original images and ground truth annotations are also included for reference.
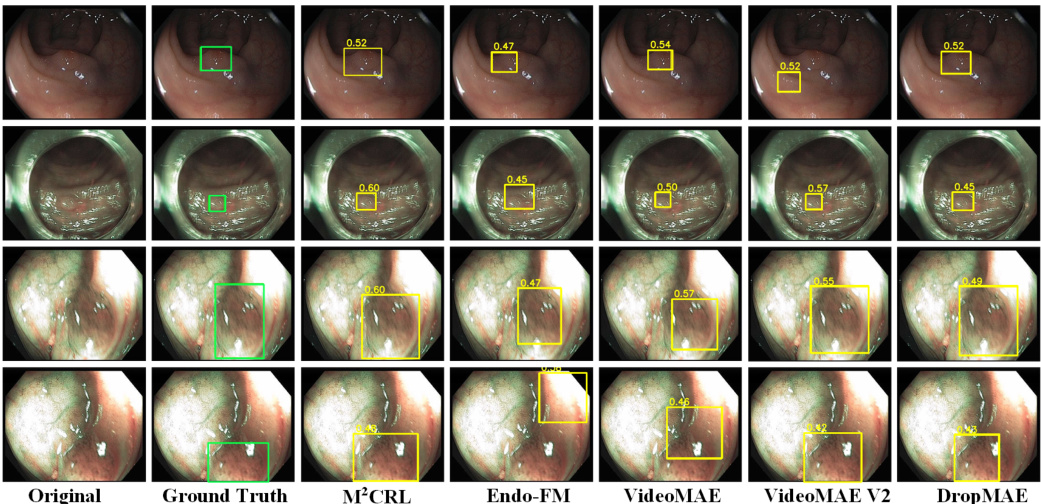
This figure shows a qualitative comparison of the segmentation and detection results obtained using different methods on the CVC-12k and KUMC datasets. The left side displays the segmentation results, showcasing how various methods segment polyps in endoscopic images from the CVC-12k dataset. The right side shows the detection performance on images from the KUMC dataset, demonstrating the ability of each method to accurately locate polyps. The comparison helps to visualize the differences in accuracy and precision of various methods in identifying polyps.

This figure visualizes the frame-aggregated attention guided tube masking strategy used in the paper. It shows how the model aggregates attention maps across multiple frames to identify regions of high importance. These regions are then used to guide the sampling of visible patches, ensuring that the most relevant information is used for masked reconstruction and contrastive learning.
More on tables
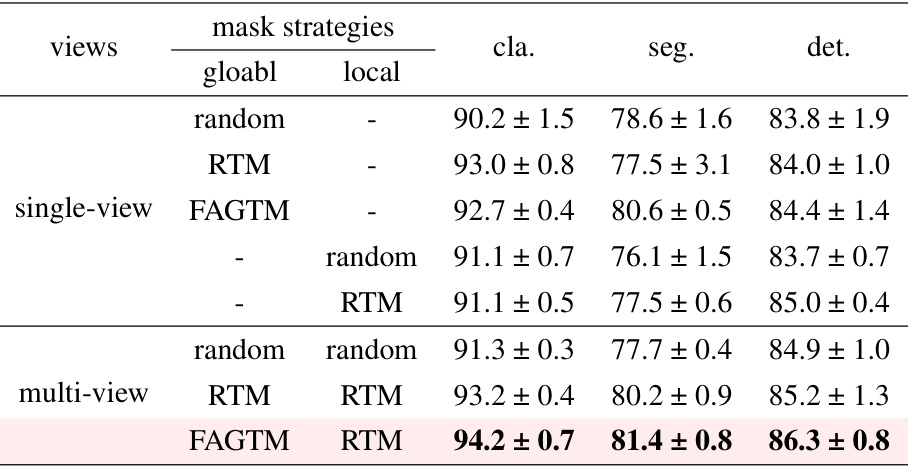
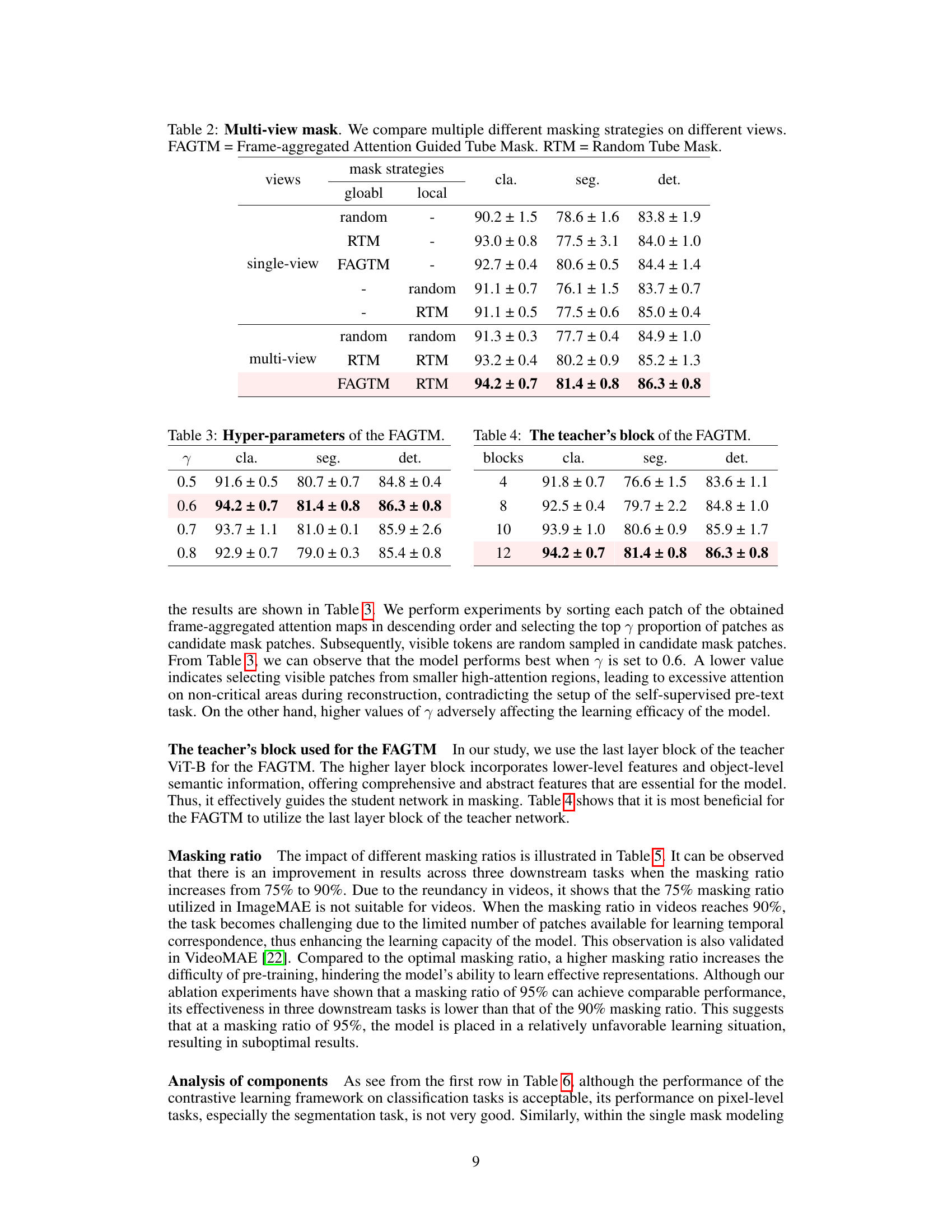
This table presents ablation study results comparing different masking strategies applied to global and local views within a multi-view masked contrastive representation learning framework. It shows the performance (classification, segmentation, and detection F1 scores) achieved using various combinations of masking methods (random, RTM, FAGTM) applied independently to the global and local views, highlighting the effectiveness of the proposed multi-view masking strategy.

This ablation study investigates the effect of different hyperparameter values (γ) of the Frame-aggregated Attention Guided Tube Mask (FAGTM) on the model’s performance across three downstream tasks: classification, segmentation, and detection. The results show that a γ value of 0.6 yields the best overall performance.
This table compares the performance of the proposed M²CRL method against other state-of-the-art (SOTA) self-supervised learning methods on three downstream tasks: Polyp classification (PolypDiag dataset), polyp segmentation (CVC-12k dataset), and polyp detection (KUMC dataset). The table shows the F1 score for classification and detection tasks, and the Dice score for the segmentation task. It highlights the improvements achieved by M²CRL compared to other methods.

This table compares the performance of the proposed M²CRL model with other state-of-the-art (SOTA) self-supervised learning methods on three endoscopic video analysis tasks: Polyp classification (PolypDiag dataset), polyp segmentation (CVC-12k dataset), and polyp detection (KUMC dataset). The results are presented in terms of F1 score (for classification and detection) and Dice score (for segmentation), showing the improvement achieved by M²CRL over existing methods.
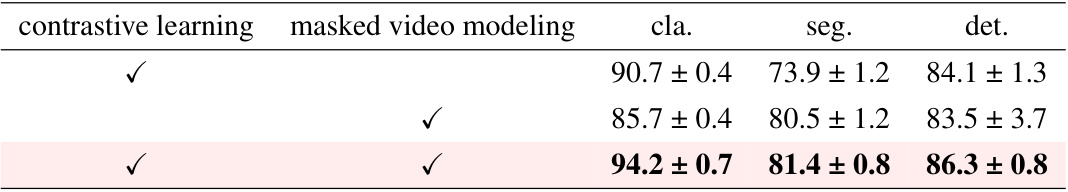
This table presents an ablation study comparing the performance of the proposed M²CRL method against models using only contrastive learning or only masked video modeling. The results, shown for classification, segmentation, and detection tasks, demonstrate that combining both techniques yields superior performance compared to using either method alone. The scores indicate F1-score for classification and detection tasks and Dice score for segmentation.

This table compares the performance of the proposed M²CRL method with other state-of-the-art (SOTA) self-supervised learning methods on three downstream tasks: Polyp classification (PolypDiag dataset), polyp segmentation (CVC-12k dataset), and polyp detection (KUMC dataset). The results are presented as F1 scores for classification and detection, and Dice scores for segmentation. The table highlights the improvements achieved by M²CRL in comparison to other methods.
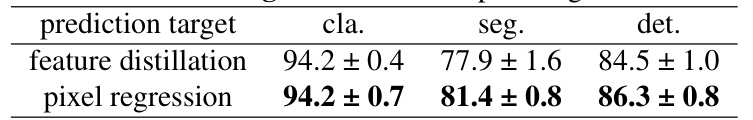
This table presents the ablation study on different prediction targets used in the M²CRL model. The results are shown for three downstream tasks: classification, segmentation and detection. Pixel regression as the prediction target outperforms feature distillation, indicating the advantage of using pixel-level reconstruction for improving the performance of downstream tasks.

This table presents the ablation study results on different loss functions used in the masked modeling component of the proposed M2CRL model. The table shows that different loss functions have minimal impact on the final results across classification, segmentation, and detection tasks, indicating robustness of the model to the choice of loss function.
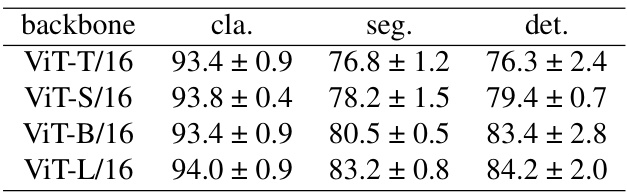
This table presents the results of ablation studies conducted on different Vision Transformer (ViT) architectures to evaluate their impact on the performance of the proposed M²CRL framework across three downstream tasks: classification, segmentation, and detection. The results show that larger models (ViT-L/16) generally perform better, but the improvement diminishes as the model size increases, potentially due to overfitting. The ViT-B/16 architecture shows a good balance between performance and computational cost.

The table compares the performance of the proposed M²CRL method with other state-of-the-art (SOTA) self-supervised learning methods on three downstream tasks: Polyp classification (PolypDiag dataset), polyp segmentation (CVC-12k dataset), and polyp detection (KUMC dataset). For each method, it reports the training time (in hours), and the F1 score (for classification and detection) and Dice score (for segmentation). The results show that M²CRL outperforms other methods by a significant margin.
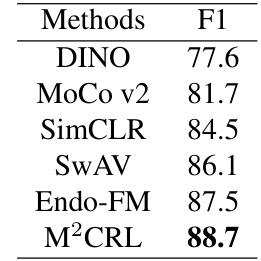
This table compares the performance of the proposed M²CRL method against other state-of-the-art (SOTA) self-supervised learning methods on three downstream tasks: classification (PolypDiag dataset), segmentation (CVC-12k dataset), and detection (KUMC dataset). It shows the F1 score for classification and detection, and Dice score for segmentation, highlighting the improvement achieved by M²CRL.
Full paper#