↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Cryo-EM, crucial for determining biomolecular structures, faces challenges from noisy 2D images and multi-modal pose distributions, leading to slow and inaccurate 3D reconstructions. Existing deep learning approaches, using amortized inference, struggle with multi-modal distributions and slow convergence.
CryoSPIN overcomes these limitations using a two-stage approach: initial amortized inference using a multi-head encoder to explore multiple pose candidates, followed by direct pose optimization using stochastic gradient descent to refine poses locally. This semi-amortized method significantly accelerates convergence, improving reconstruction speed and accuracy. Experiments on both synthetic and real datasets demonstrated cryoSPIN’s superior performance over state-of-the-art methods, showing its potential for high-resolution, ab initio cryo-EM reconstruction.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in cryo-EM because it presents cryoSPIN, a novel semi-amortized method that significantly improves the speed and accuracy of ab initio 3D reconstruction. This addresses a critical bottleneck in the field, enabling faster analysis of complex biomolecular structures and accelerating the discovery of new insights. The introduction of a multi-head encoder and a direct pose optimization stage offers new avenues for enhancing pose inference techniques and improving the overall efficiency of cryo-EM workflows. The results also have implications for addressing conformational heterogeneity, a key challenge in the field.
Visual Insights#
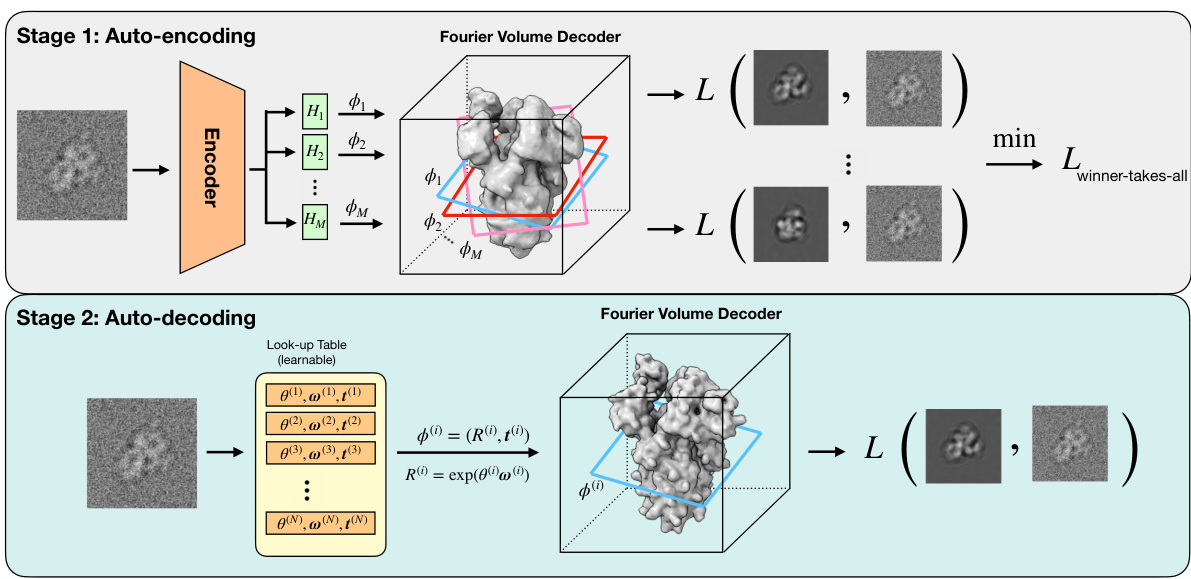
The figure illustrates the two-stage cryoSPIN architecture. Stage 1 (auto-encoding) uses a multi-head encoder to map an input image to multiple pose candidates. Projections are generated from these poses using a Fourier volume decoder, and the best pose is selected based on minimum reconstruction error. Stage 2 (auto-decoding) refines the selected pose using stochastic gradient descent and direct pose optimization, focusing on a single projection to reduce computational cost.
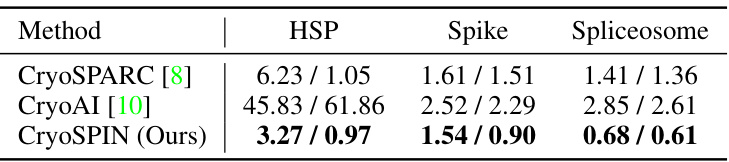
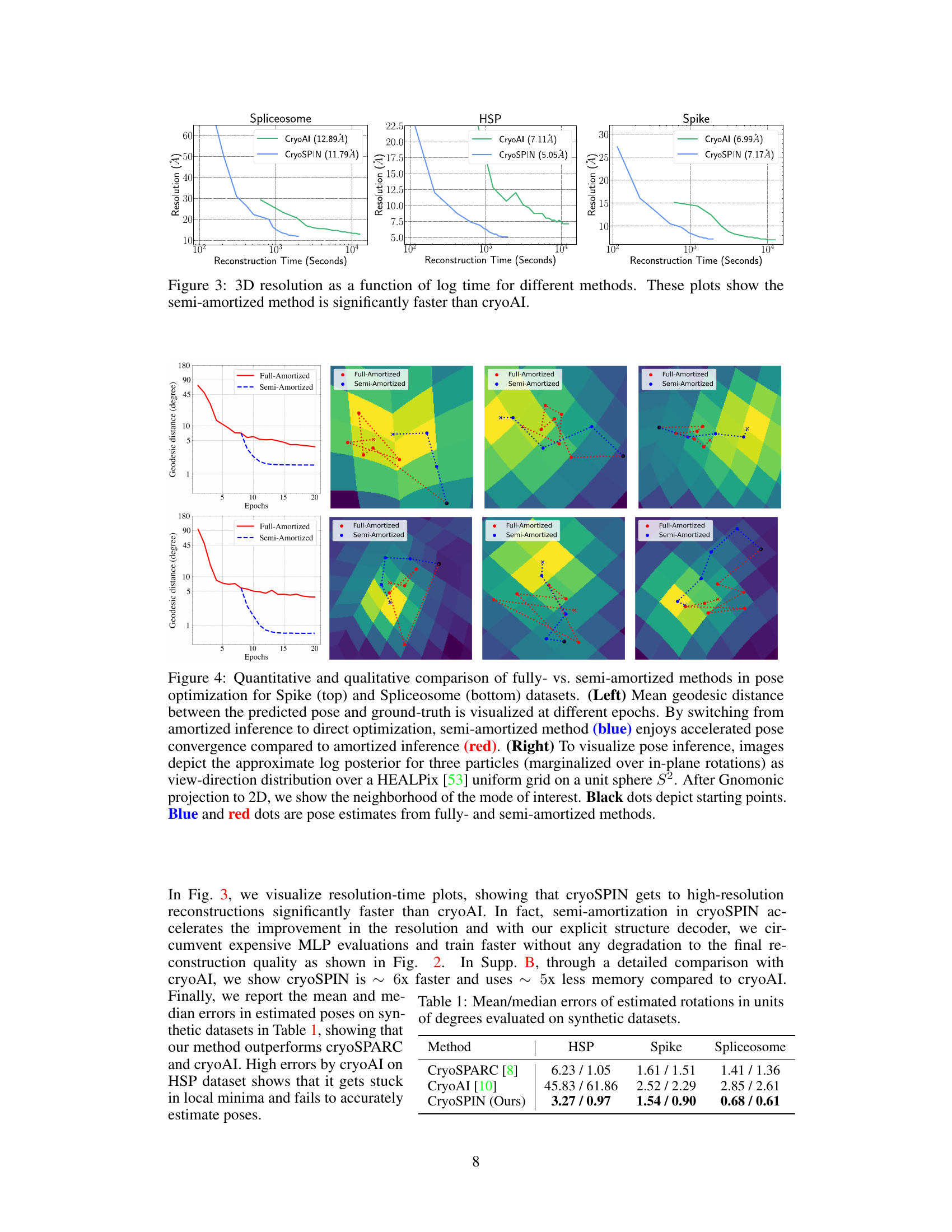
This table presents the mean and median errors in estimated rotations (in degrees) for three different methods: CryoSPARC, CryoAI, and CryoSPIN. The errors are calculated for three different synthetic datasets: HSP, Spike, and Spliceosome. CryoSPIN demonstrates significantly lower errors than the other two methods across all datasets, indicating improved accuracy in pose estimation.
In-depth insights#
Semi-Amortized CryoEM#
Semi-amortized cryoEM represents a significant advancement in cryo-electron microscopy (cryoEM) image processing. It cleverly combines the efficiency of amortized inference with the accuracy of direct optimization. Initial pose estimation leverages amortized inference, using a neural network to quickly predict likely particle poses from images. However, unlike fully amortized methods, cryoSPIN transitions to a direct optimization stage as the reconstruction progresses, refining poses with stochastic gradient descent. This hybrid approach addresses the limitations of purely amortized methods which can struggle with multi-modal pose distributions and slow convergence. The result is faster and more accurate pose estimation, leading to improved 3D structure reconstruction, especially beneficial in handling the inherent uncertainty and noise present in cryoEM data. A multi-head encoder architecture further enhances robustness by simultaneously predicting multiple possible poses, allowing the algorithm to explore and converge towards the optimal solution more effectively.
Multi-Head Pose#
The concept of “Multi-Head Pose” in cryo-EM reconstruction addresses the challenge of pose uncertainty inherent in single-particle cryo-EM. Standard methods often struggle with the highly multi-modal posterior distribution of particle orientations. A multi-head approach, as suggested by the name, uses multiple neural network “heads” to predict several pose candidates simultaneously. This contrasts with single-head methods, which only output a single pose estimate and are therefore susceptible to getting stuck in local minima. The key benefit is increased robustness and exploration of the pose space, potentially identifying poses missed by single-head approaches. The multiple hypotheses are then refined later, often using techniques like auto-decoding. This strategy combines the efficiency of amortized inference (multi-head) with the accuracy of direct pose optimization, improving the speed and quality of cryo-EM reconstruction significantly. The success hinges on the ability of the multi-head architecture to accurately capture and handle the inherent uncertainty in pose estimation.
Auto-Decoding#
Auto-decoding, in the context of cryo-EM reconstruction, represents a crucial shift from the initial amortized inference stage. Instead of relying solely on a learned model to predict poses, auto-decoding employs a direct optimization approach, refining each particle’s pose individually. This refinement typically leverages stochastic gradient descent, allowing for greater flexibility and accuracy compared to the global, parametric limitations of amortized inference. The transition to auto-decoding is often triggered when the reconstruction’s resolution improves, signifying that the pose distribution shifts from multi-modal to predominantly unimodal. This makes direct optimization more efficient, avoiding the risk of the amortized model being trapped in suboptimal local minima. The advantages include faster convergence, enhanced accuracy, and the ability to handle more complex scenarios beyond the capacity of the earlier auto-encoding stage. The process becomes more focused on individual image details. This ultimately contributes to higher-quality reconstructions and faster processing times, making auto-decoding a key improvement in cryo-EM’s deep learning approaches.
Ab-initio Recon#
Ab-initio reconstruction in cryo-electron microscopy (cryo-EM) refers to the process of determining the 3D structure of a macromolecule solely from its 2D projection images, without relying on any prior structural information. This is a challenging computational problem because the images are noisy, the poses (orientations and positions) of the macromolecules are unknown, and the resulting posterior pose distribution is often multimodal. CryoSPIN, as described in the provided text, offers a novel semi-amortized approach to tackle these difficulties. It combines the efficiency of amortized inference, using a neural network to predict poses, with the accuracy of direct pose optimization via stochastic gradient descent. This two-stage method first utilizes a multi-head encoder to handle the multimodality of the early pose distribution and then switches to a more precise, per-image optimization once the structure is better estimated, leading to faster convergence and higher-resolution reconstructions. The use of an explicit volumetric decoder, unlike implicit methods, further enhances efficiency. This strategy appears particularly effective in dealing with complex, noisy datasets where traditional methods often struggle. The results demonstrate that cryoSPIN surpasses existing methods in terms of both speed and reconstruction quality, particularly where the pose posterior distribution presents multiple modes. The combination of amortized and deterministic methods is a key innovation for improving the efficiency and accuracy of ab-initio cryo-EM reconstruction.
CryoSPIN Limits#
CryoSPIN, while a significant advancement in ab-initio cryo-EM reconstruction, possesses inherent limitations. Its reliance on a semi-amortized approach introduces a trade-off. While speeding up pose optimization, it might not fully capture the complexity of multi-modal pose distributions, potentially hindering accuracy, especially in the early reconstruction stages. The method’s success depends on the accuracy of the encoder’s initial pose estimations, and limitations in this aspect could propagate through the refinement process. Furthermore, the assumption of rigid structures is a simplification, neglecting the inherent flexibility of many biomolecules, which could affect the overall accuracy and resolution of the final reconstruction. The reliance on an explicit volumetric decoder, while speeding computation, could potentially limit its ability to capture fine-grained structural details compared to implicit methods. Lastly, while capable of handling multi-modal pose distributions better than previous fully amortized methods, CryoSPIN may still struggle with highly symmetric structures where pose uncertainty remains high even at later stages, causing the optimization to converge to local minima. Addressing these limitations through incorporating flexibility, developing more robust pose estimation techniques, and exploring different decoder architectures could lead to further advancements.
More visual insights#
More on figures
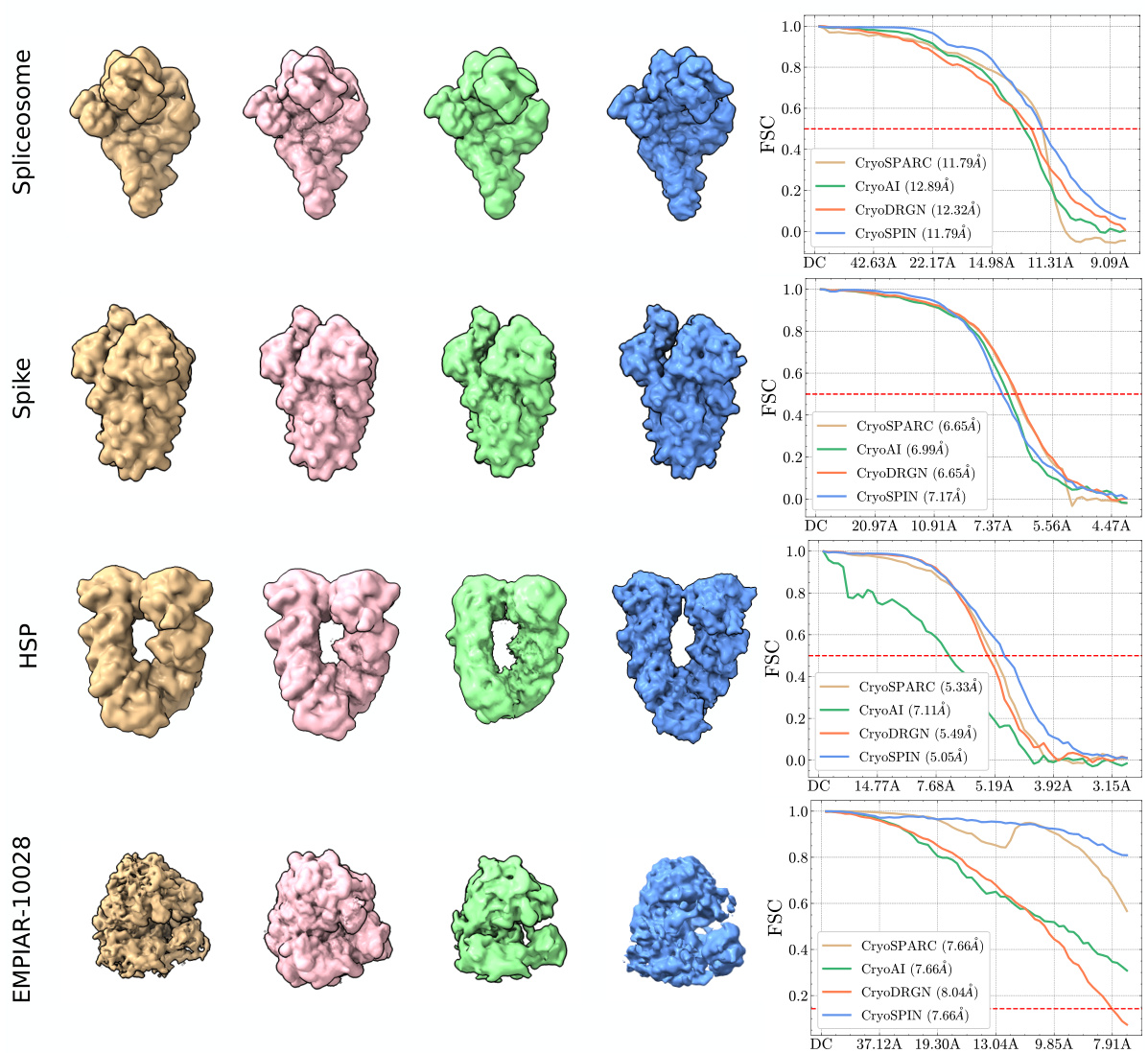
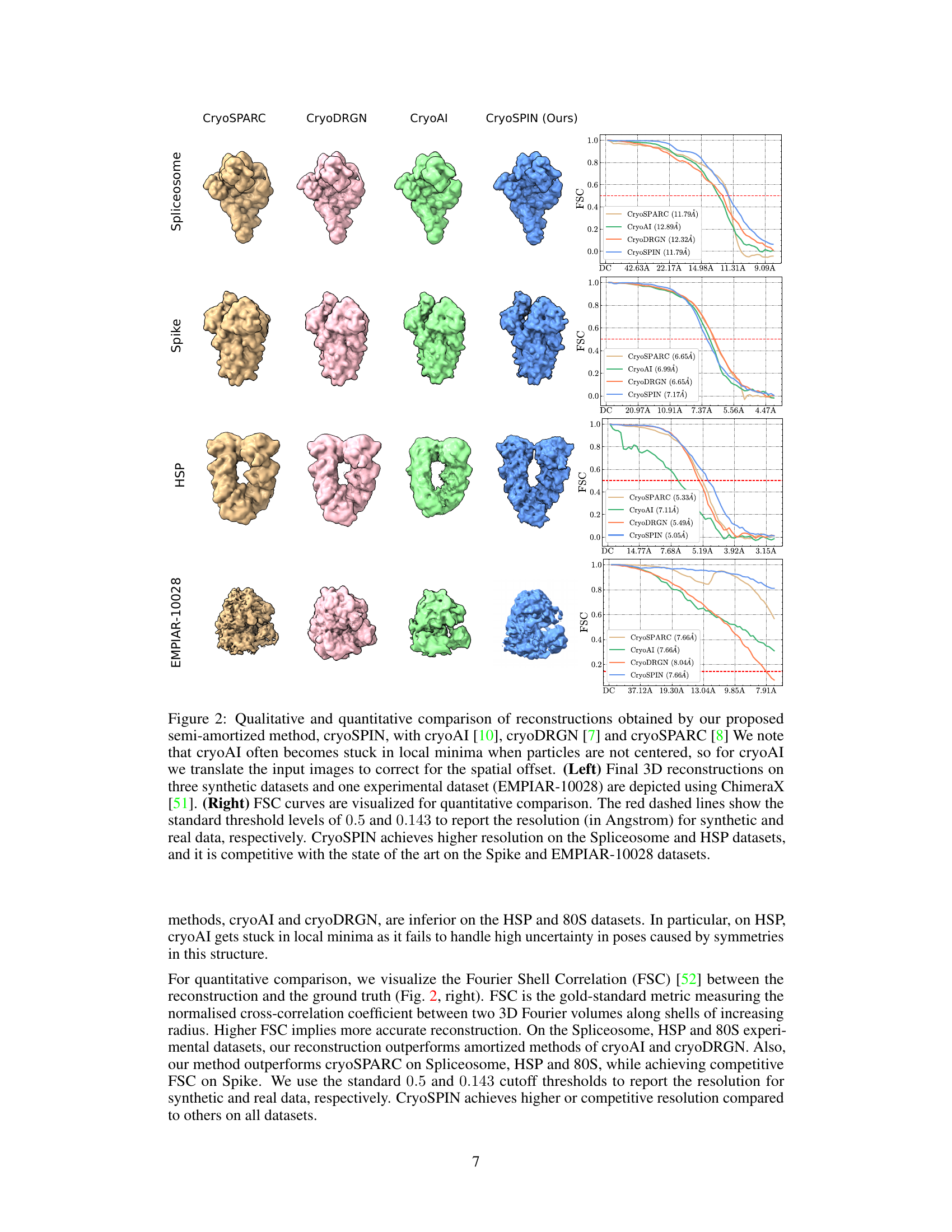
This figure compares the 3D reconstruction results and Fourier Shell Correlation (FSC) curves of cryoSPIN with three other methods (cryoAI, cryoDRGN, and cryoSPARC) on four datasets (Spliceosome, Spike, HSP, and EMPIAR-10028). The left panel shows 3D visualizations of the reconstructed structures, highlighting the differences in resolution and accuracy. The right panel presents the FSC curves, a quantitative measure of reconstruction resolution, showing that cryoSPIN often achieves comparable or higher resolution compared to the other methods, especially on the Spliceosome and HSP datasets. CryoAI’s performance is notably affected when particles aren’t centered.
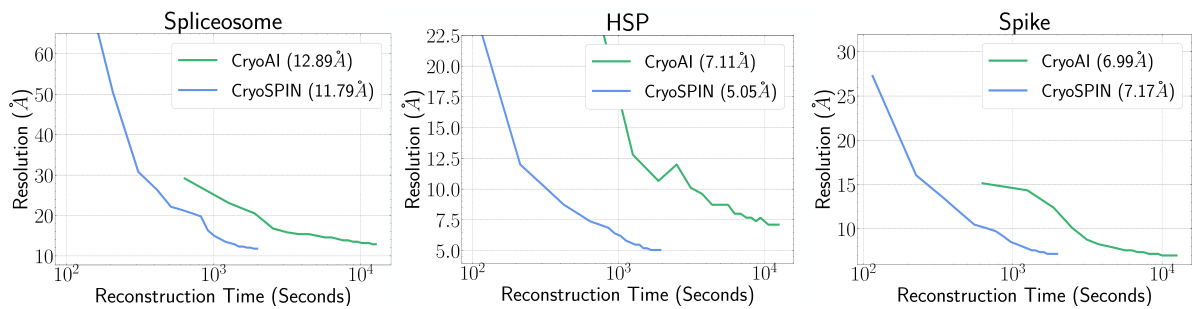
This figure compares the 3D reconstruction resolution achieved by cryoSPIN and cryoAI over time. The x-axis represents reconstruction time in seconds (log scale), while the y-axis shows the achieved resolution in Angstroms. Separate plots are provided for three different datasets: Spliceosome, HSP, and Spike. The plots demonstrate that cryoSPIN consistently reaches higher resolution significantly faster than cryoAI across all three datasets.
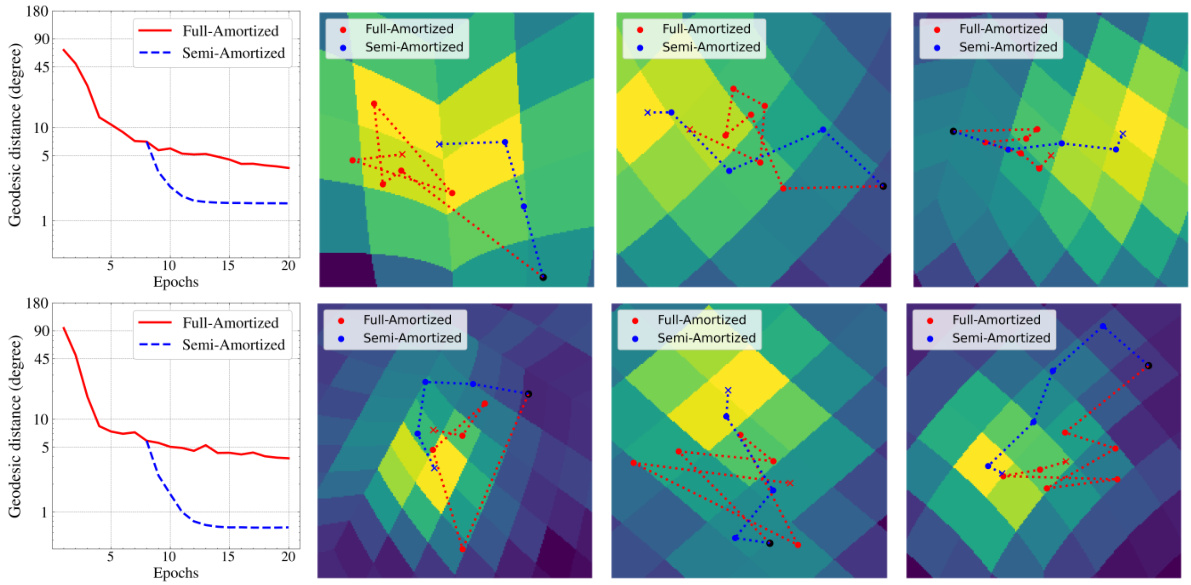
This figure compares the performance of fully-amortized and semi-amortized methods in pose optimization. The left plots show the mean geodesic distance between predicted and ground truth poses over epochs. The right plots visualize the pose inference process by depicting the approximate log posterior for three particles, showing how semi-amortized method converges faster to the optimal point.

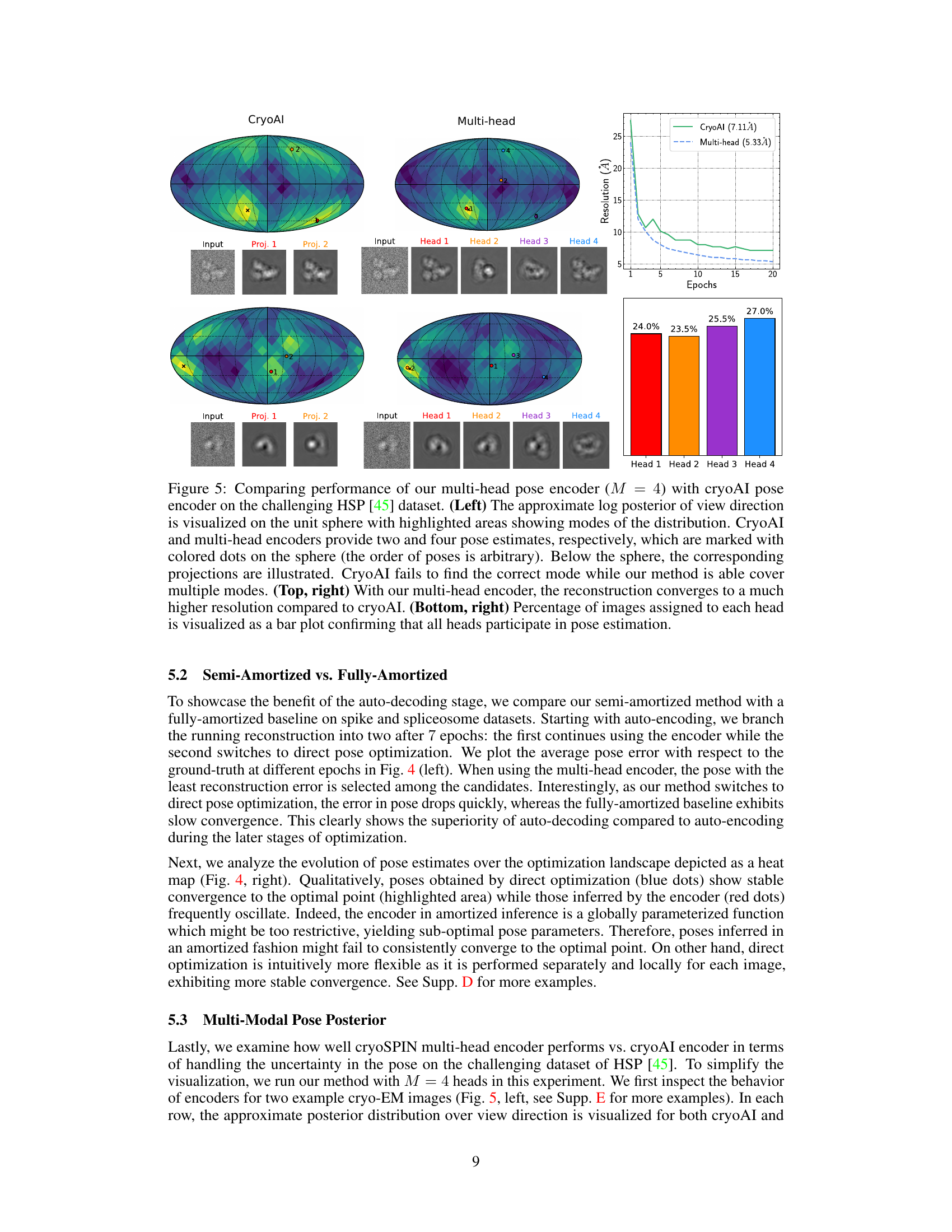
This figure compares the performance of the proposed multi-head pose encoder against cryoAI’s pose encoder on a challenging dataset (HSP). The left panel visually shows the posterior distribution of view directions on a unit sphere, highlighting the superior ability of the multi-head encoder to identify multiple modes. The top-right panel demonstrates the faster convergence and higher resolution achieved by the multi-head encoder. The bottom-right panel shows the even distribution of image assignments across different heads, indicating that all heads contribute effectively to the pose estimation process.
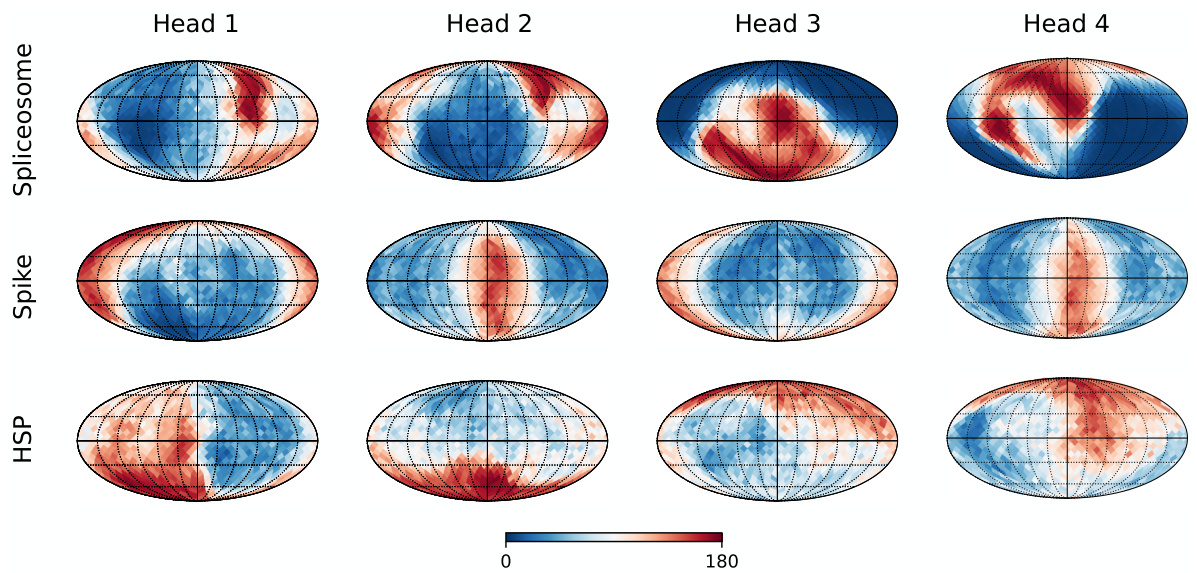
This figure visualizes the performance of each head in the multi-head pose encoder on three datasets: Spliceosome, Spike, and HSP. The sphere represents the view direction space, where each cell represents a particular view direction. The color in each cell shows the average rotation error for that view direction, with blue representing low error and red representing high error. This visualization demonstrates how different heads specialize in predicting poses for different view directions, showcasing the effectiveness of the multi-head approach in handling pose ambiguity.

This figure compares the performance of fully and semi-amortized methods in pose optimization. The left panel shows the mean geodesic distance to the ground truth over training epochs, demonstrating faster convergence for the semi-amortized approach. The right panel visualizes the pose posterior distribution for three particles across training epochs, highlighting the more stable convergence of the semi-amortized method.
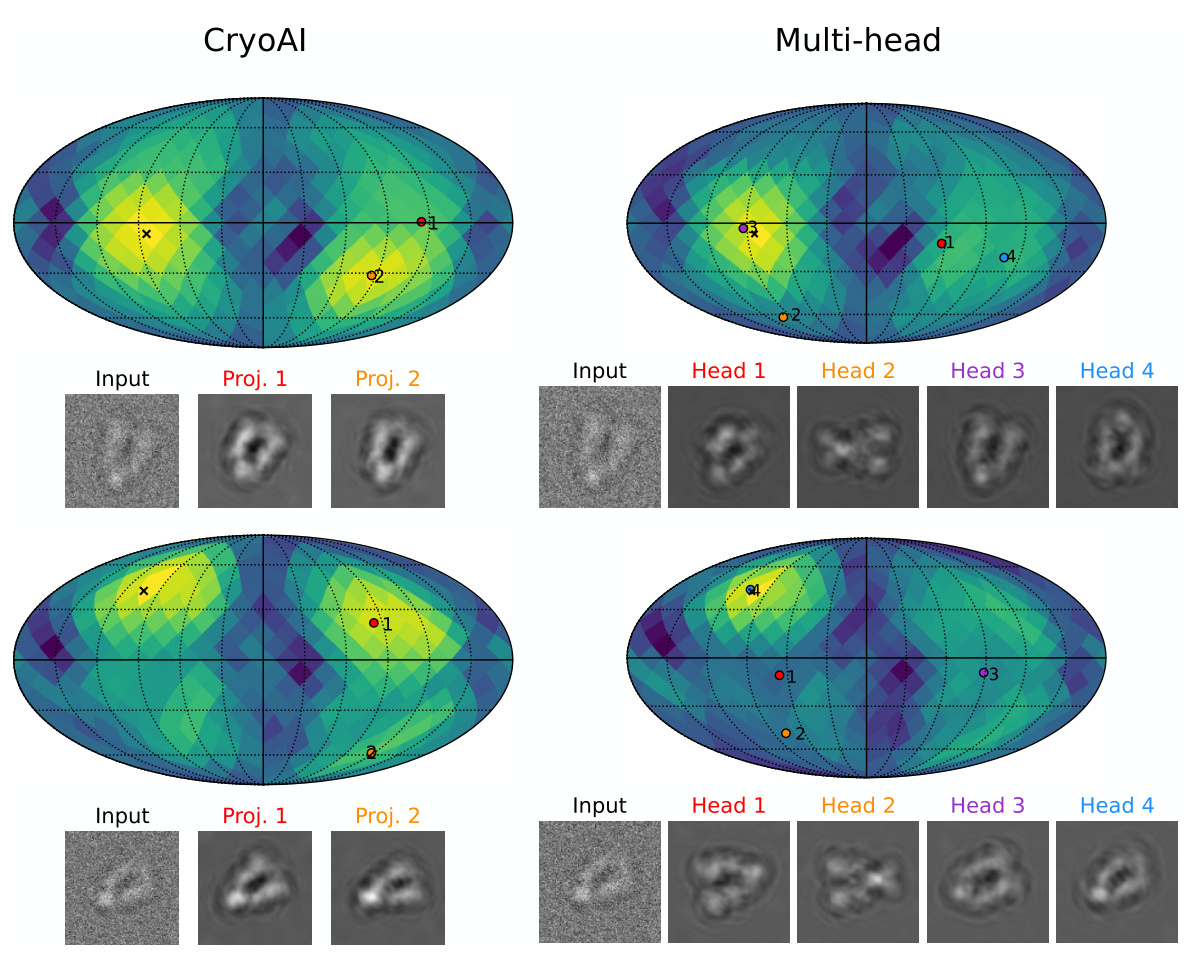
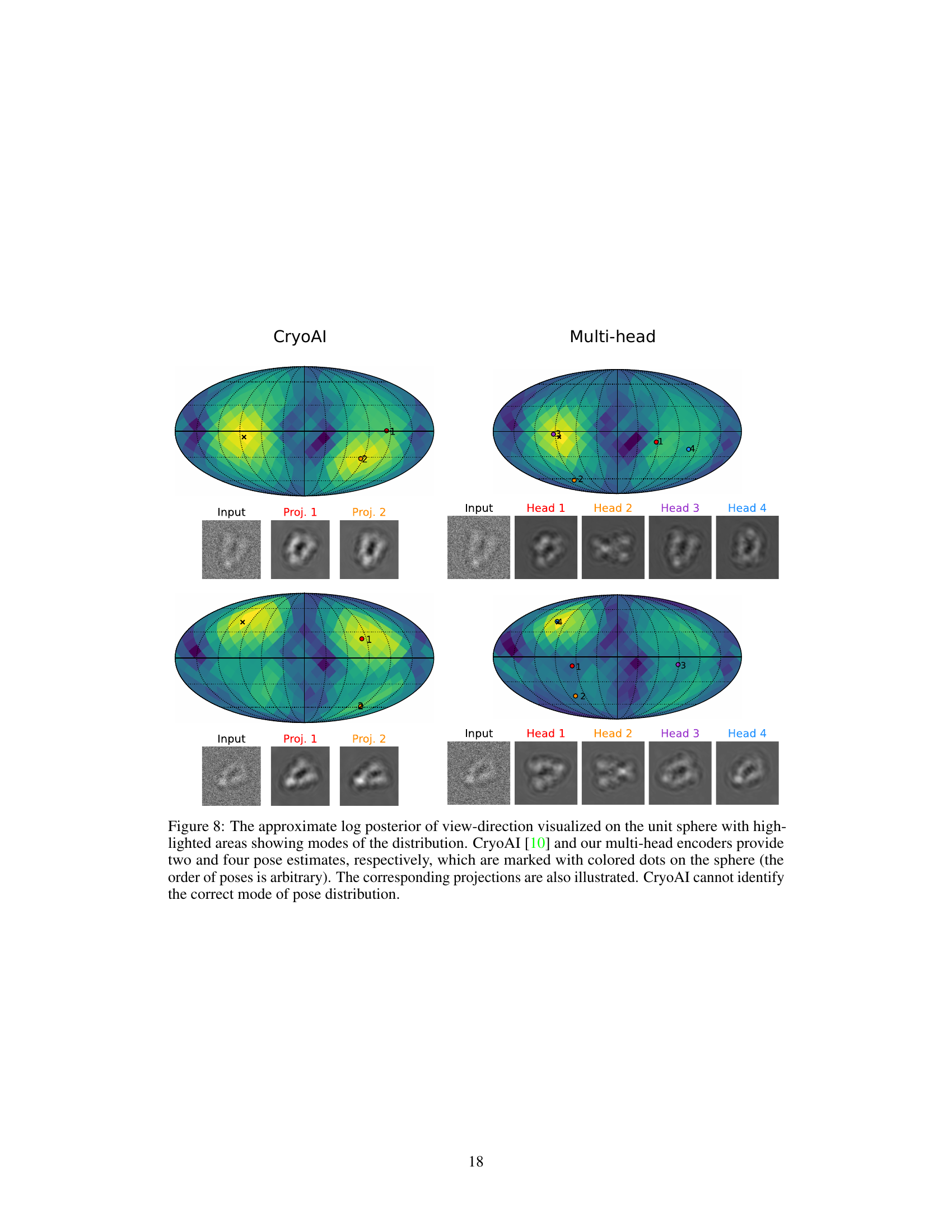
This figure compares the performance of the proposed multi-head pose encoder with cryoAI’s pose encoder on a challenging dataset. The multi-head encoder shows better performance in identifying the correct pose mode and achieving higher reconstruction resolution. The visualization uses a unit sphere to represent the distribution of view directions and shows the projections corresponding to the predicted poses.
Full paper#