↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many Graph Neural Network (GNN) models struggle with link prediction when data exhibits heterophily—the tendency of connected nodes to have different features. Existing methods often rely on the assumption of homophily. This research directly addresses this limitation. The paper lacks class labels, making the problem even more challenging.
This research introduces a theoretical framework that defines homophilic and heterophilic link prediction. It proposes innovative GNN encoders and decoders tailored for these tasks, focusing on the separation of ego- and neighbor-embedding to handle varying feature similarity. The study validates these designs on six new real-world datasets, ranging from homophilic to heterophilic structures, showing improved performance on various tasks.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers working with graph neural networks (GNNs) and link prediction. It addresses the challenge of heterophily, a common issue where connected nodes have dissimilar features, significantly impacting GNN performance. The paper provides a theoretical framework, new benchmark datasets, and innovative model designs which directly helps to improve the accuracy and generalizability of GNNs in various applications. This work opens doors to further research on enhancing GNN robustness and adaptability to diverse network structures.
Visual Insights#
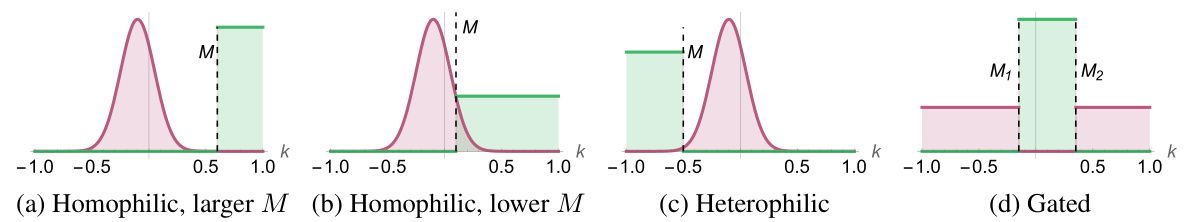
This figure categorizes link prediction tasks based on the distribution of feature similarity scores. It shows four scenarios: (a) Homophilic with a large separation between positive and negative similarity scores, (b) Homophilic with a smaller separation, (c) Heterophilic, where the negative scores are larger than positive, and (d) Gated, where the distributions overlap significantly. The key is whether positive similarity scores are above or below a threshold (or thresholds).
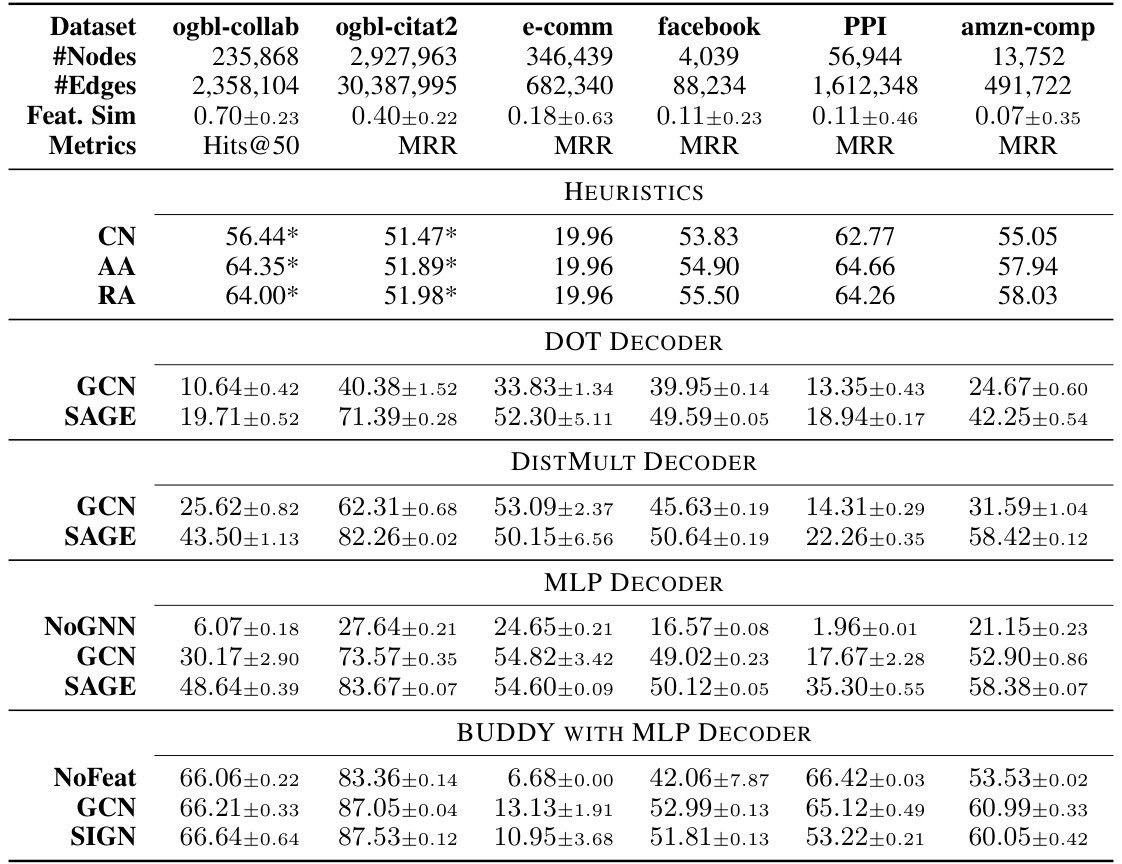
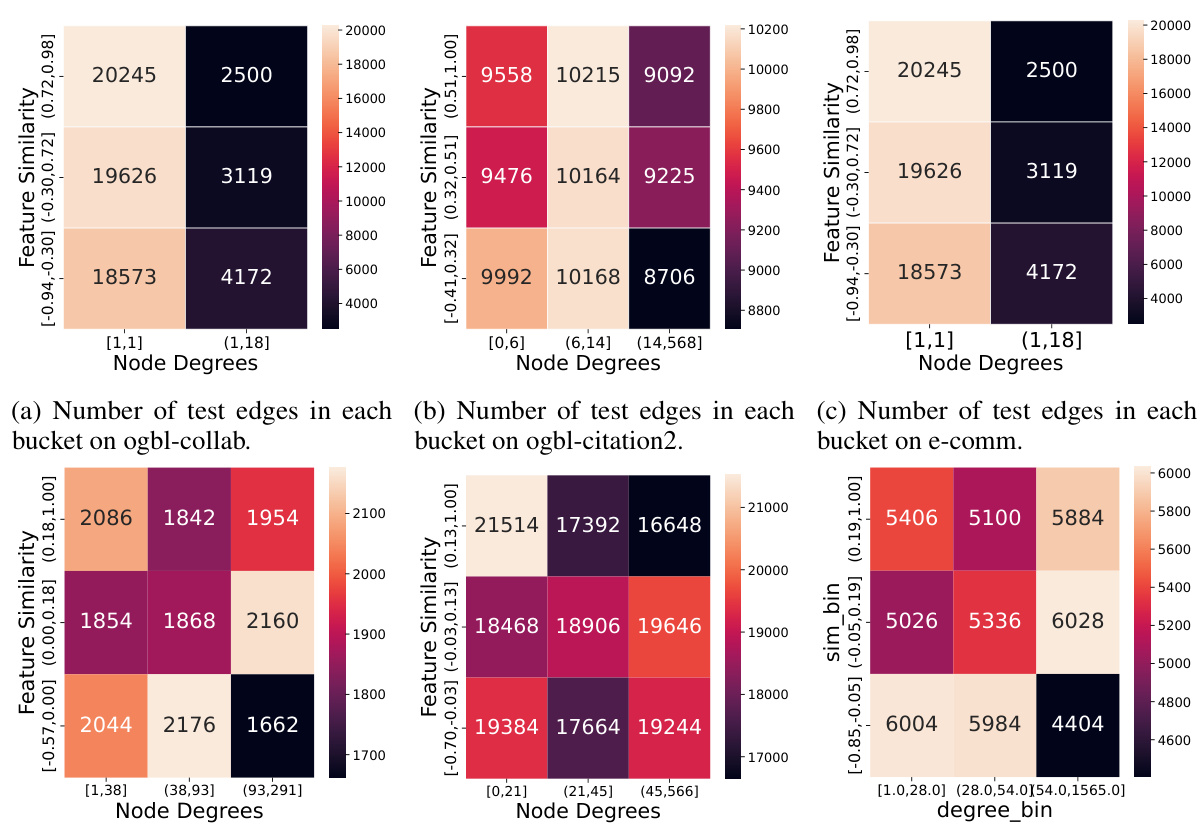
This table presents the results of applying various link prediction methods to six real-world datasets. The datasets vary significantly in size (number of nodes and edges) and feature similarity. The methods compared include heuristics (CN, AA, RA), GNN-based approaches using different encoders (GCN, GraphSAGE, NoGNN) and decoders (DOT, DistMult, MLP), and BUDDY models. The metrics reported are Hits@50 and MRR (Mean Reciprocal Rank). The * indicates results that were quoted from another paper.
In-depth insights#
Heterophily’s Impact#
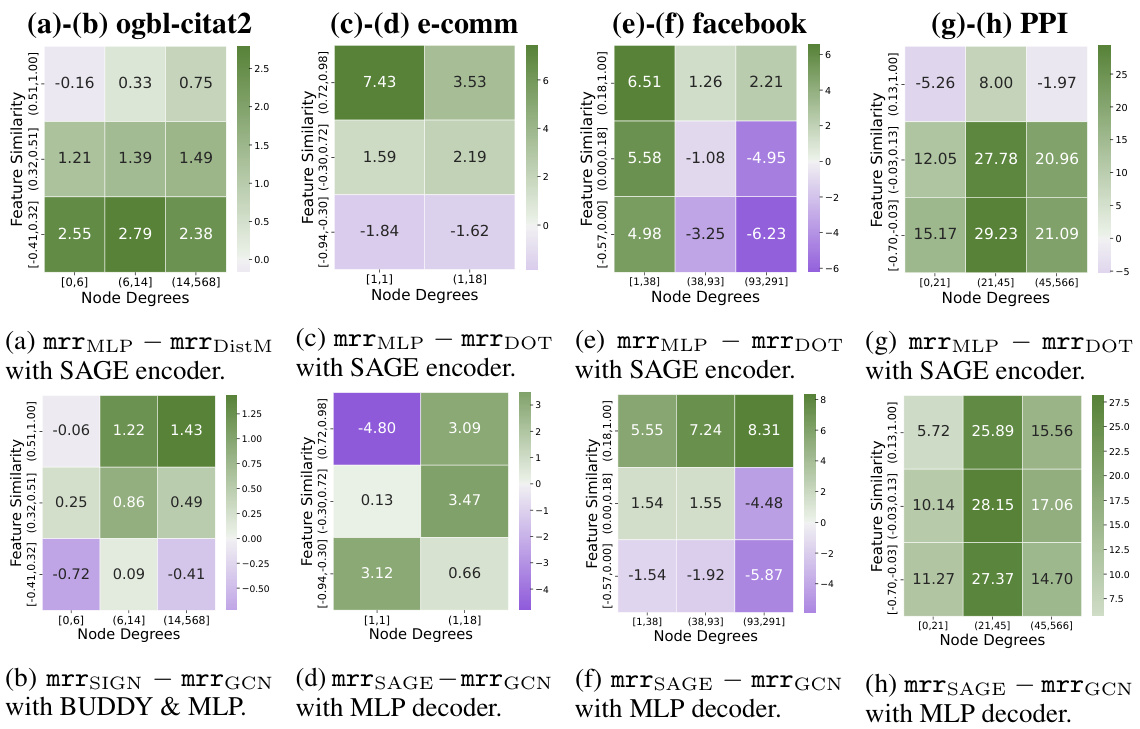
The concept of heterophily, where connected nodes in a network exhibit dissimilar features, significantly impacts link prediction using graph neural networks (GNNs). Homophilic GNNs, which assume similar nodes connect, struggle with heterophilic data. This is because they implicitly optimize for situations where similar features indicate a higher likelihood of a link. The paper systematically analyzes how different levels of feature heterophily affect GNN performance in link prediction. A key finding is the importance of using adaptable decoder mechanisms, such as MLPs (multi-layer perceptrons) instead of simpler dot products, to better capture complex feature relationships in heterophilic scenarios. Learnable decoders offer increased flexibility and the capacity to handle both homophilic and heterophilic link prediction tasks effectively. Furthermore, the study underscores the need for GNN encoders that separate ego and neighbor embeddings during message passing. This separation enhances the model’s capacity to learn robust representations, especially in situations with varying degrees of feature similarity and differing node degrees, thus improving performance across various homophily levels.
GNN Encoder Designs#
Effective GNN encoder design is crucial for handling feature heterophily in link prediction. The choice between architectures like GraphSAGE and GCN significantly impacts performance, with GraphSAGE demonstrating superiority due to its ego-neighbor embedding separation. This separation proves especially beneficial in real-world scenarios exhibiting variations in node degrees and feature similarities. The study highlights the importance of learnable decoders capable of capturing non-linear feature correlations, particularly in heterophilic or gated link prediction tasks. The theoretical analysis presented underscores the limitations of simple linear decoders like dot product in non-homophilic settings, advocating for more expressive alternatives such as MLP or DistMult which better address complex feature interactions. The findings strongly suggest that GNN encoders should be carefully selected and designed to improve model adaptability in situations beyond feature homophily.
Decoder Strategies#
Decoder strategies in link prediction using graph neural networks (GNNs) are crucial for translating learned node embeddings into accurate link probabilities. The choice of decoder significantly impacts performance, especially in the presence of feature heterophily where connected nodes exhibit dissimilar features. Simple decoders, like dot product, may be efficient but struggle to capture complex relationships between nodes, especially non-homophilic ones. More sophisticated decoders, such as multi-layer perceptrons (MLPs) or DistMult, offer greater expressiveness and the ability to learn non-linear relationships. MLP decoders, while powerful, can be computationally expensive, impacting scalability. DistMult, a linear model, balances representational capacity and efficiency, making it a suitable alternative for specific tasks. The choice of decoder should be informed by the nature of the data—homophilic datasets may favor simple, efficient decoders, while heterophilic datasets demand the greater expressivity of MLPs or DistMult. Furthermore, the interaction between decoder choice and encoder design should be considered. Optimizing decoder complexity in tandem with the encoder architecture is key for effectively leveraging both node features and structural graph information.
Benchmark Datasets#
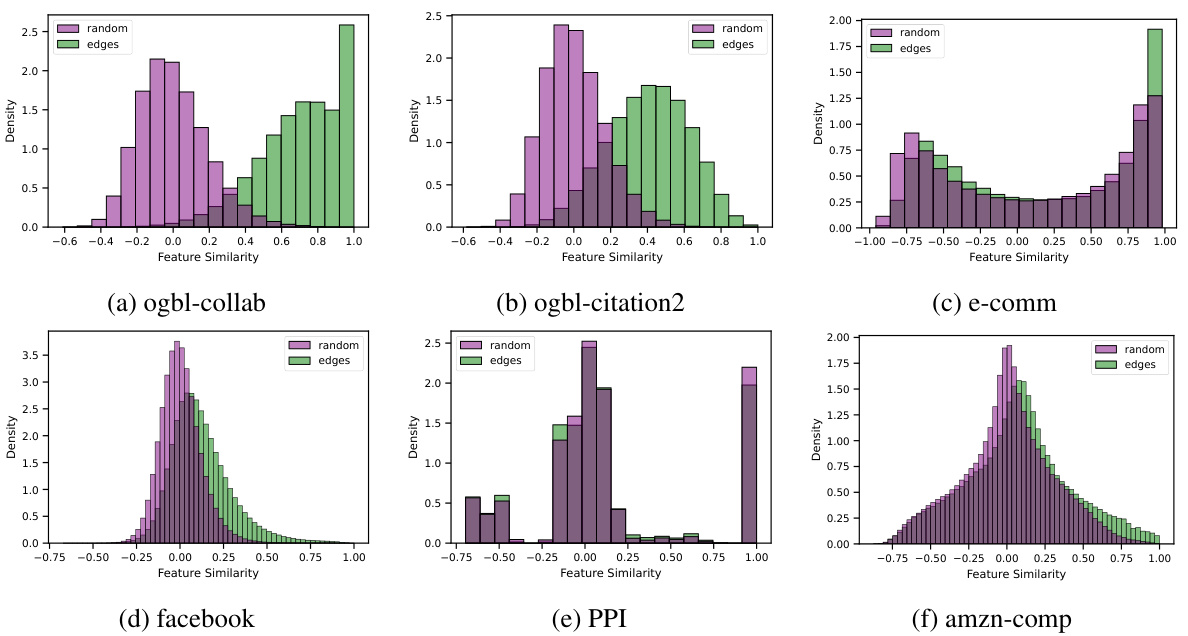
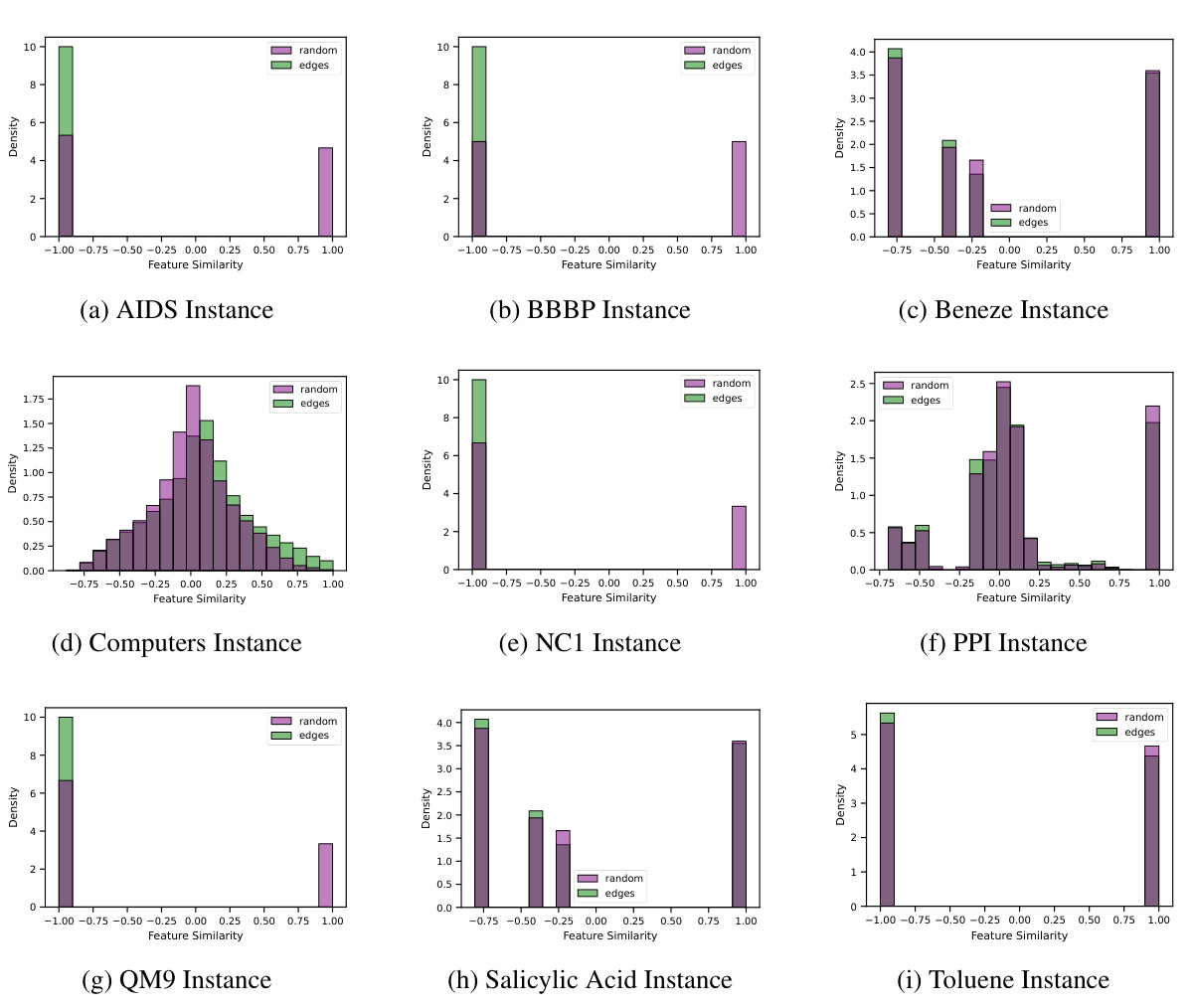
A robust benchmark dataset is crucial for evaluating the effectiveness of link prediction models, especially when dealing with varying levels of feature heterophily. Real-world datasets, such as those used in this paper (spanning from homophilic to heterophilic scenarios), offer valuable insights into model performance in diverse settings and uncover limitations. Synthetic datasets, while controlled, provide a crucial tool for systematic analysis of heterophily’s influence. The combination of both types is key: synthetic data allows for isolating the impact of heterophily, while real-world datasets validate findings and show model generalizability. Diversity in dataset characteristics (size, domain, edge count, feature dimensionality and distribution, and type of relationships) is essential for building a reliable benchmark. The choice of evaluation metrics (like Hits@K and MRR) is crucial, reflecting the performance aspects most relevant for the specific application.
Future Directions#
Future research could explore more sophisticated methods for handling feature heterogeneity in link prediction, going beyond simple feature transformations. Developing novel GNN architectures specifically designed for heterophilic graphs is crucial. Investigating the interplay between structural information and feature similarity in link prediction is also important. More advanced decoders, capable of capturing complex interactions beyond dot products or MLPs, could significantly improve performance. The theoretical analysis of different link prediction methodologies under various heterophily levels needs further investigation. Creating more comprehensive benchmark datasets with controlled feature distributions and varied homophily levels would enhance the robustness and reliability of future research, and encourage the development of more generalized and adaptable GNN models. Finally, exploring applications to real-world problems where heterophily is prevalent, such as biological networks or recommendation systems, is needed to fully assess the practical implications of this work.
More visual insights#
More on figures

This figure visualizes the results of Theorem 1, showing the relationship between the predicted link score (ŷu’v’) and feature similarity (k(u’,v’)) for both homophilic and heterophilic link prediction tasks. In homophilic settings (yellow line), the predicted score increases linearly with feature similarity. Conversely, in heterophilic settings (blue line), the predicted score decreases linearly with increasing feature similarity. The threshold ‘M’ (0.5 in this example) separates the positive and negative samples in the feature similarity space, influencing the slope of these lines. This illustrates how the optimization of link prediction decoders differs fundamentally for homophilic versus heterophilic tasks.
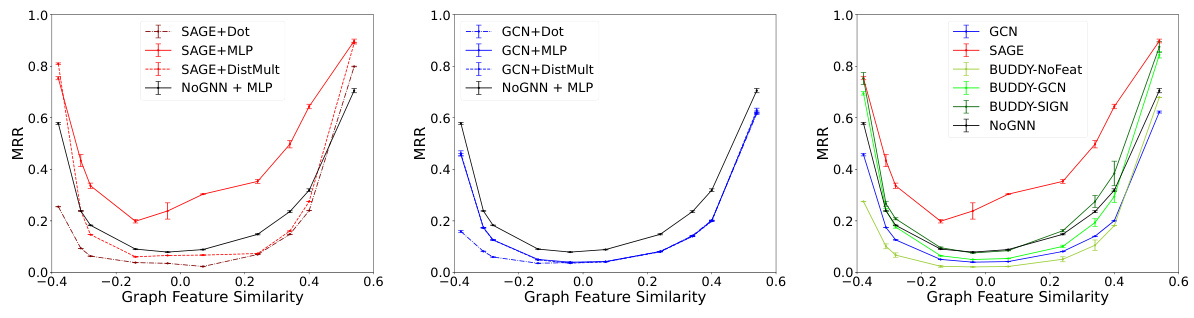
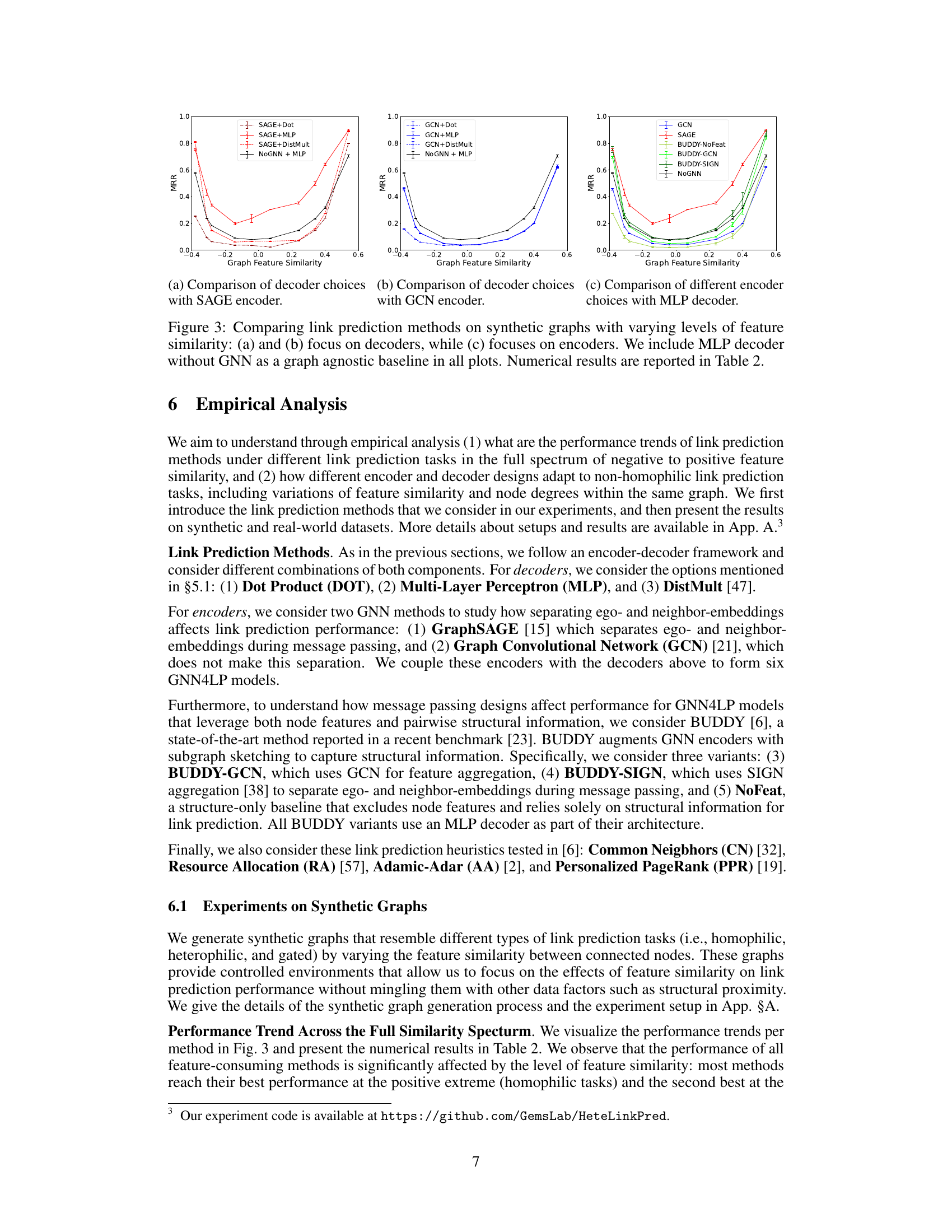
This figure compares different link prediction methods on synthetic graphs with varying levels of feature similarity. Panels (a) and (b) show the performance of different decoders (dot product, MLP, DistMult) while keeping the encoder (GraphSAGE) constant, highlighting the impact of decoder choice on heterophilic graphs. Panel (c) shows performance of different encoders (GCN, GraphSAGE, BUDDY variants) with a consistent MLP decoder, illustrating the impact of encoder design. A graph-agnostic MLP decoder is included as a baseline for comparison in all three plots. Table 2 provides the corresponding numerical results.
The figure compares different link prediction methods on synthetic graphs with varying levels of feature similarity. Panels (a) and (b) show the performance of different decoders (Dot Product, Multi-Layer Perceptron, and DistMult) using the same encoder (SAGE in (a), GCN in (b)). Panel (c) compares the performance of different encoders (GCN, SAGE, and BUDDY variants), using the same decoder (MLP). A graph agnostic baseline (NoGNN + MLP) is included for comparison in all plots. Table 2 provides the numerical results. The overall performance trend indicates that the link prediction methods perform best at the positive extreme (homophilic tasks) and second best at the negative extreme (heterophilic tasks).
This figure categorizes link prediction tasks into three types based on how the distributions of feature similarity scores for positive (edges) and negative (non-edges) node pairs are separated. Homophilic tasks show positive similarity scores clearly above a threshold, heterophilic show them below, and gated show a more complex, overlapping distribution.
This figure categorizes link prediction tasks into homophilic, heterophilic, and gated types based on the distributions of feature similarity scores for positive (edges) and negative (non-edges) node pairs. Homophilic tasks show a clear separation between positive and negative similarity score distributions, with positive scores significantly higher than negative scores. Heterophilic tasks display the opposite, with negative scores being substantially greater than positive ones. Gated tasks represent a more complex scenario where positive and negative scores are not clearly separated by a single threshold.

This figure compares different link prediction methods on synthetic graphs with varying levels of feature similarity. The three subfigures (a), (b), and (c) show the Mean Reciprocal Rank (MRR) results across different feature similarity levels. Subfigures (a) and (b) focus on the decoder’s influence (using DOT, MLP, and DistMult decoders), while subfigure (c) focuses on the encoder’s influence (using GCN and GraphSAGE). A graph-agnostic MLP decoder is also included as a baseline for comparison.
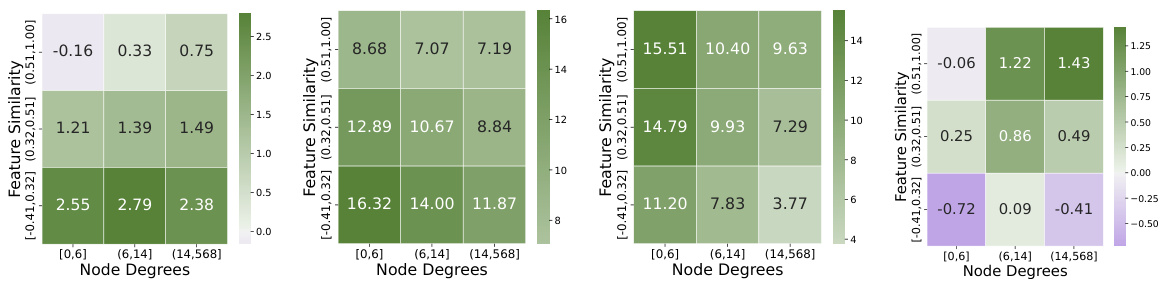
This figure compares the performance of various link prediction methods on synthetic graphs with different levels of feature similarity. Panels (a) and (b) show the impact of different decoder choices (dot product, multi-layer perceptron, and DistMult) while keeping the encoder (GraphSAGE or GCN) constant. Panel (c) focuses on the effect of different encoder choices (GraphSAGE, GCN, and BUDDY variants) with a fixed MLP decoder. A graph-agnostic MLP decoder (NoGNN) is used as a baseline for comparison. The x-axis in all plots represents the graph feature similarity, and the y-axis represents the mean reciprocal rank (MRR).

This figure compares the performance of different link prediction methods on synthetic graphs with varying levels of feature similarity. The plots show how different decoders (dot product, MLP, DistMult) and encoders (GraphSAGE, GCN) perform under different feature similarity conditions. A graph agnostic MLP decoder (without GNN) is included as a baseline. The results highlight that the choice of decoder and encoder significantly impacts performance, especially under non-homophilic settings (heterophily).
The figure categorizes link prediction tasks into homophilic, heterophilic, and gated types based on the distribution of feature similarity scores for edges (positive samples) and non-edges (negative samples). Homophilic tasks show positive similarity scores clearly separated from negative scores by a threshold (M). Heterophilic tasks display the opposite, with negative scores separated from positive. Gated tasks are more complex, lacking a single threshold to clearly separate positive and negative distributions.
More on tables

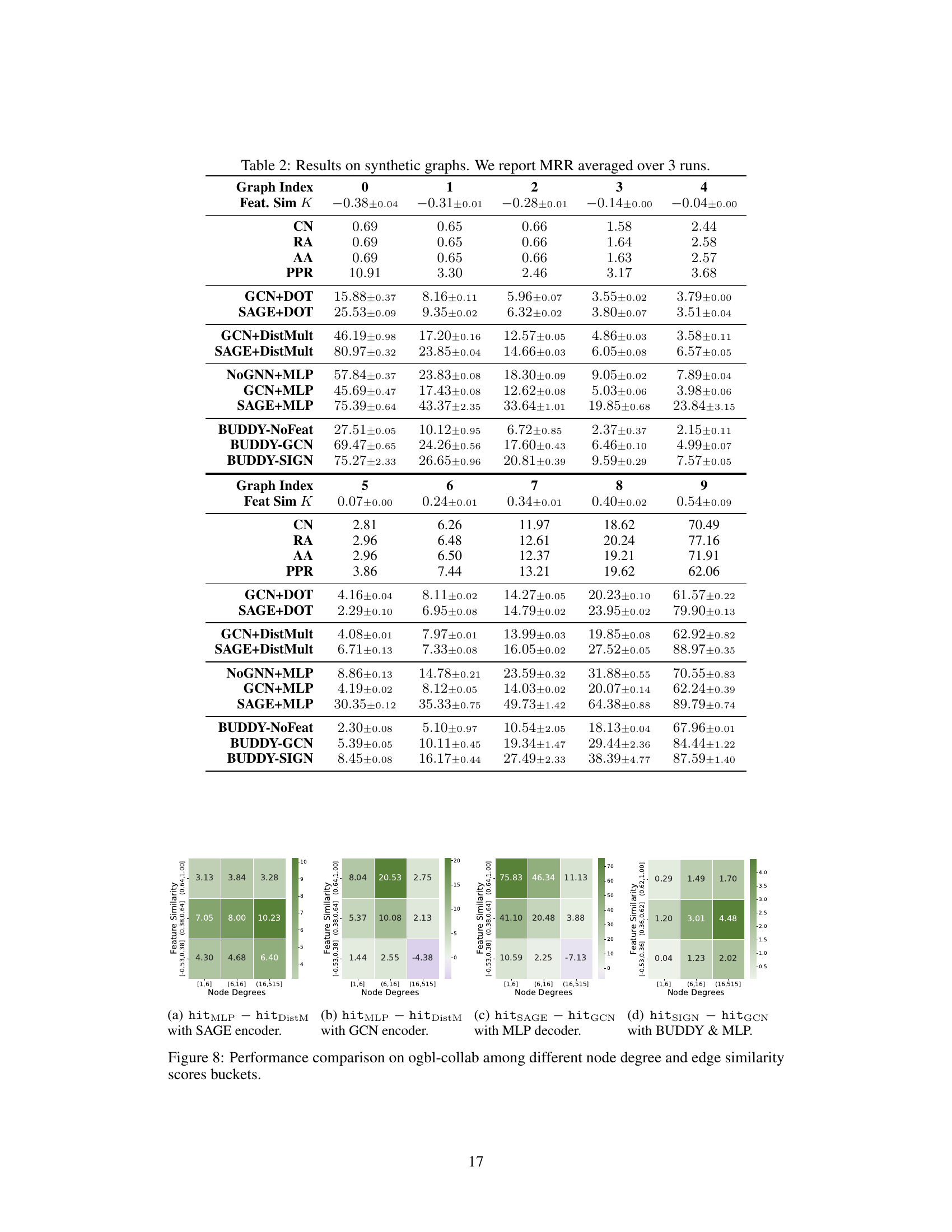
This table presents the mean reciprocal rank (MRR) results for link prediction on synthetic graphs with varying levels of feature similarity. The results are averaged over three runs and show the performance of various link prediction methods under different levels of homophily/heterophily. The table helps to evaluate the impact of different decoders and encoders in handling feature similarity variations across a spectrum from negative (heterophilic) to positive (homophilic) similarity.
This table presents the results of link prediction experiments on six real-world datasets using various methods. The datasets vary in size and feature similarity, ranging from homophilic to heterophilic. The results are presented for multiple GNN models with different encoders and decoders, as well as traditional link prediction heuristics. The metrics used are Hits@50 and MRR (Mean Reciprocal Rank). Results marked with ‘*’ are taken from a previous study ([6]) for comparison.

This table presents the results of the link prediction task on six real-world datasets using various methods. The datasets vary in size and feature similarity, ranging from homophilic to heterophilic. Metrics include Hits@50 and MRR. Different encoder (GCN, GraphSAGE, NoGNN, BUDDY variants) and decoder (DOT, DistMult, MLP) combinations are evaluated.
Full paper#