↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Generating novel molecules with desired properties is crucial for drug discovery and materials science. However, existing methods struggle to efficiently explore the vast chemical space and often lack the ability to incorporate physical constraints. Current state-of-the-art equivariant diffusion models use pre-specified forward processes, limiting their generative capacity.
This paper introduces Equivariant Neural Diffusion (END), a novel diffusion model that addresses these limitations. END uses a learnable forward process, parameterized through a time- and data-dependent transformation equivariant to rigid transformations. Benchmark results demonstrate END’s superior performance in both unconditional and conditional molecule generation compared to existing methods. The results highlight the advantages of learnable forward processes in diffusion models, making it a promising approach for generating molecules with desired properties.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in molecule generation due to its introduction of Equivariant Neural Diffusion (END), a novel diffusion model that significantly improves molecule generation in 3D. END’s use of a learnable forward process enhances generative modeling and achieves state-of-the-art performance on benchmarks, opening new avenues in drug discovery and materials science. Its equivariance to Euclidean transformations ensures physically meaningful results, addressing a key challenge in the field. Furthermore, END’s conditional generation capabilities add a degree of controllability greatly needed for practical applications.
Visual Insights#
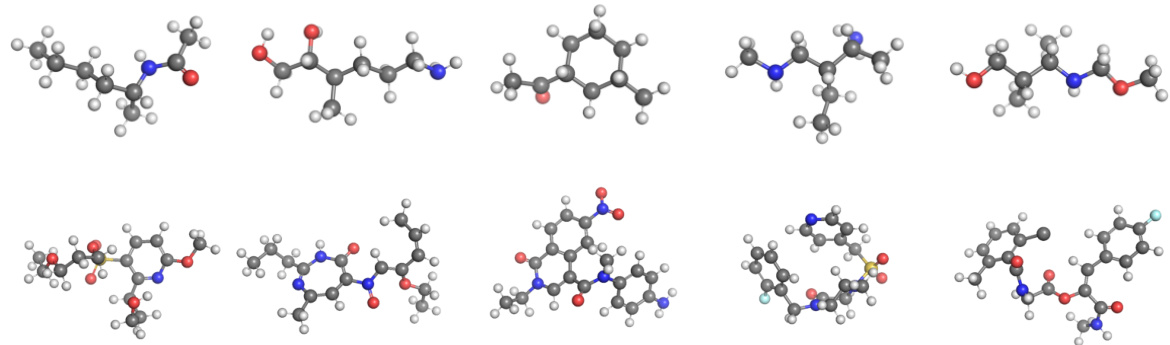
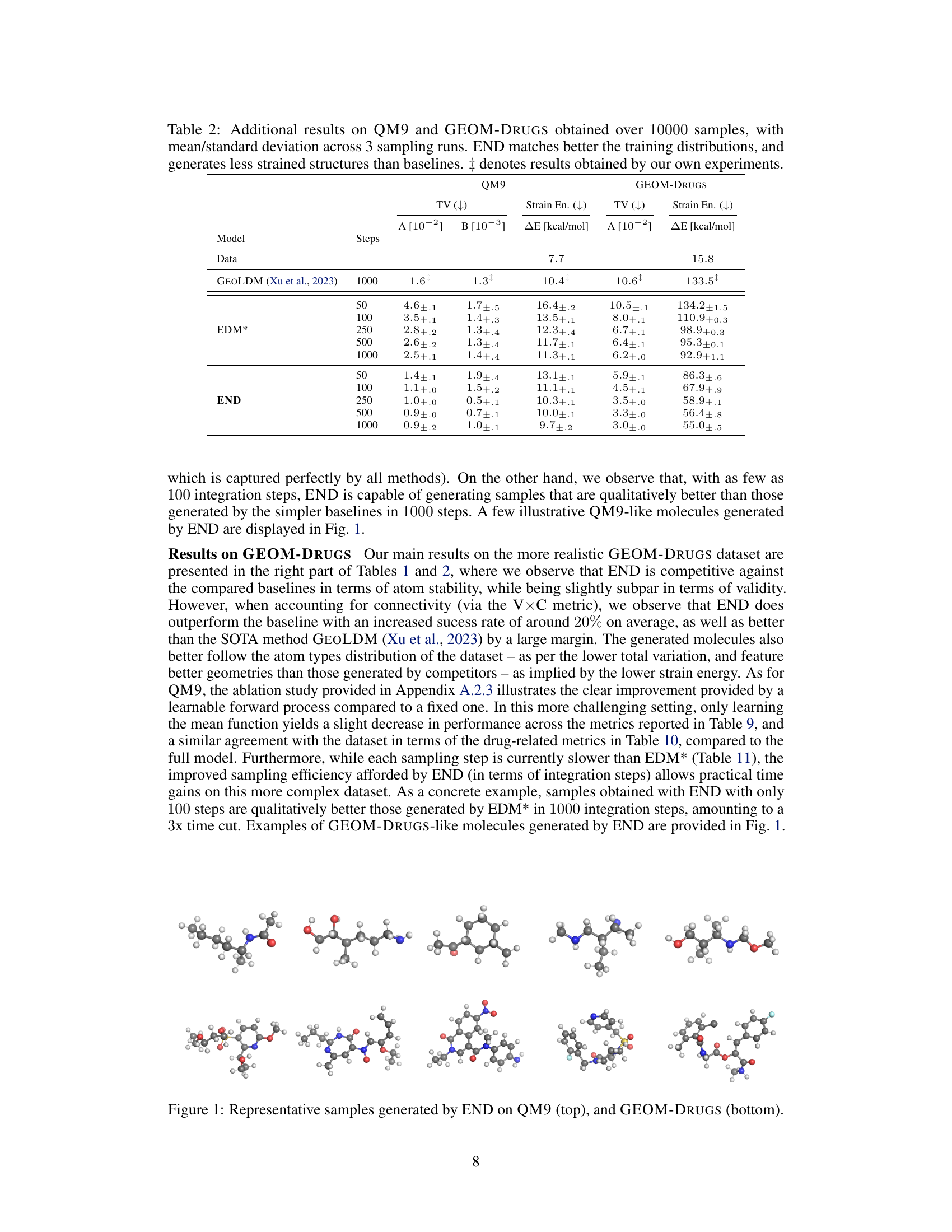
This figure shows example molecules generated by the Equivariant Neural Diffusion (END) model. The top row displays smaller molecules from the QM9 dataset, while the bottom row shows larger, more complex molecules from the GEOM-Drugs dataset. The figure visually demonstrates the model’s ability to generate diverse and chemically plausible molecules of varying sizes and complexities.
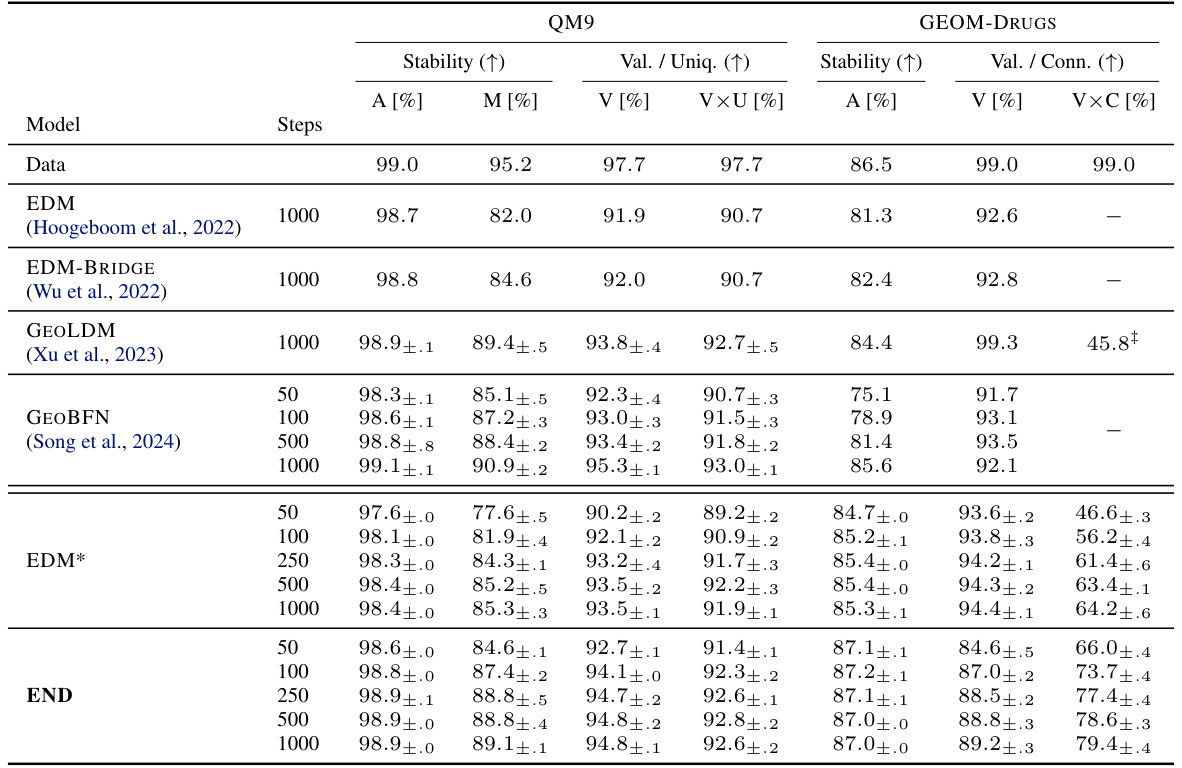
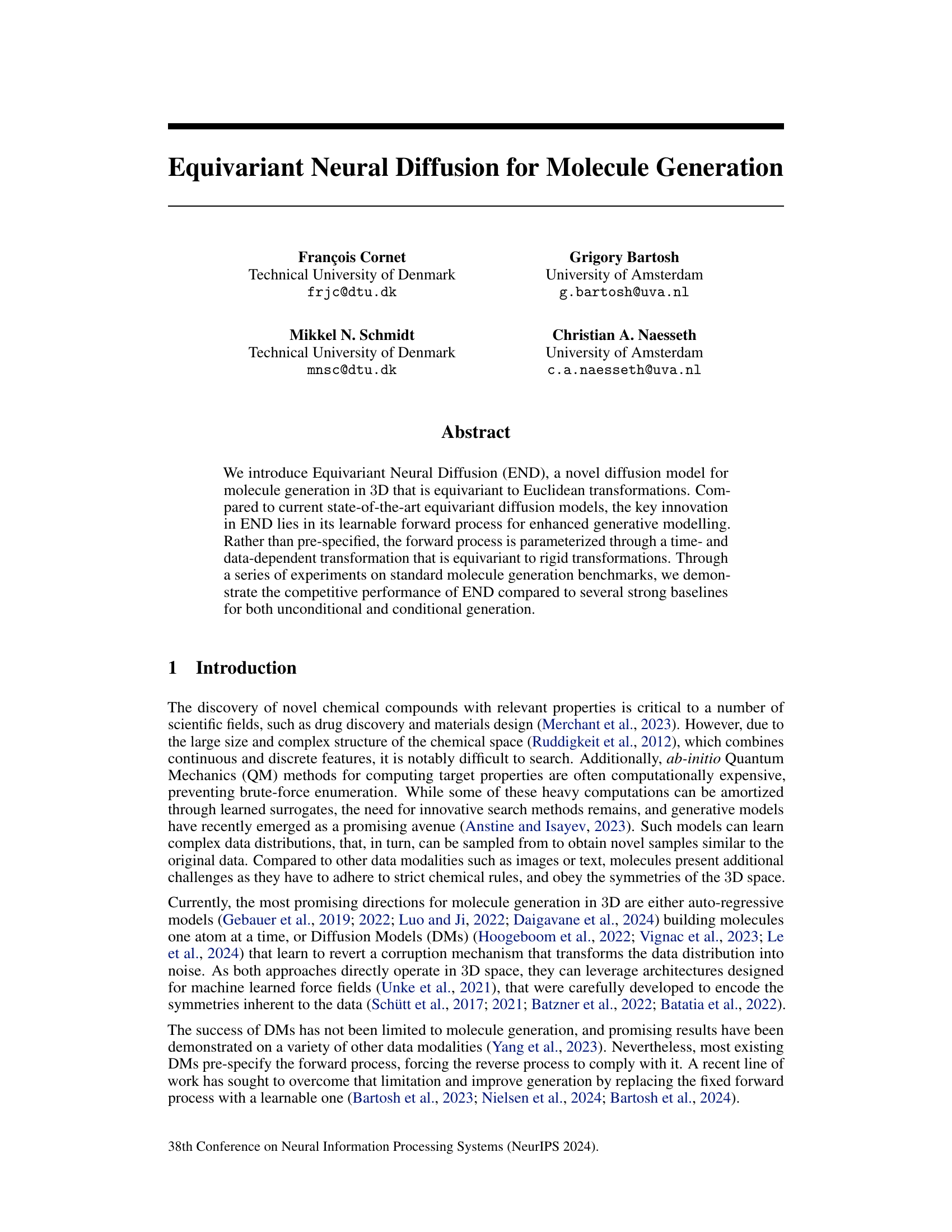
This table presents the stability and validity results of the END model and various baselines on QM9 and GEOM-DRUGS datasets. The metrics used are: stability (A[%], M[%], V[%]), validity/uniqueness (VxU[%]), and validity/connectivity (VxC[%]). The END model demonstrates competitive performance compared to the state-of-the-art models, particularly with a reduced number of sampling steps. The table highlights the consistent improvement of END across different evaluation metrics and datasets.
In-depth insights#
Equivariant Diffusion#
Equivariant diffusion models represent a significant advancement in generative modeling, particularly for data with inherent symmetries, such as molecules. Equivariance, meaning the model’s output transforms consistently with the input under symmetry transformations (e.g., rotations), is crucial for generating realistic and physically meaningful samples. Unlike standard diffusion models that often struggle with symmetries, equivariant versions directly incorporate them into the model architecture, leading to improved sample quality and reduced computational cost. Learnable forward processes are another key innovation; instead of relying on fixed noise schedules, these models learn the forward diffusion process, providing greater flexibility and control over the generation process. This adaptability allows for enhanced generative capabilities in conditional settings, enabling the generation of molecules with specific properties or structures. While showing promise, challenges remain in scaling to larger molecules and handling complex conditional constraints effectively. Future work could explore novel network architectures and training strategies to address these limitations further.
Learnable Forward Process#
The concept of a “Learnable Forward Process” in diffusion models represents a significant departure from traditional approaches. Instead of a pre-defined, fixed forward diffusion process (e.g., adding noise according to a predetermined schedule), this innovative approach allows the model to learn its own forward process. This offers several key advantages. Firstly, it enhances the flexibility and control over the generative process. A learned forward process can better adapt to the nuances and complexities of the data distribution, leading to more effective reverse diffusion (generation) and potentially higher-quality samples. Secondly, it provides opportunities for improved model efficiency. By learning a data-dependent transformation, the model might discover more efficient ways to inject noise compared to a universal method. However, it is crucial to address potential drawbacks. A learnable forward process increases model complexity, thus potentially increasing computational costs and making training more challenging. Moreover, it requires careful design and training to prevent the model from learning a trivial or ineffective forward process that does not facilitate effective reverse sampling. This is addressed by enforcing constraints that preserve the essential equivariance properties of the model, ensuring that the learned forward process aligns with the symmetries and structure of the data.
3D Molecule Generation#
3D molecule generation is a significant challenge in cheminformatics due to the complexity and high dimensionality of the chemical space. Equivariant neural networks are particularly well-suited for this task because they inherently capture the rotational and translational symmetries of molecules, leading to more robust and physically meaningful representations. Diffusion models have emerged as a powerful approach for generative modeling, and combining them with equivariant architectures offers significant advantages. A learnable forward process, rather than a pre-defined one, allows for more flexibility and potentially better generative capabilities, as demonstrated by the introduction of END (Equivariant Neural Diffusion). The effectiveness of END is highlighted by its competitive performance on benchmark datasets, showing improved stability and validity compared to existing methods. Conditional generation, driven by composition or substructure constraints, is another crucial aspect that benefits from the symmetries captured by equivariant diffusion. The ability to generate molecules with specific properties or substructures is key to applications such as drug discovery and materials science. Future work should explore further enhancements to diffusion models for 3D molecule generation, including efficient sampling strategies and more sophisticated ways to incorporate additional chemical information or constraints.
Conditional Generation#
The section on ‘Conditional Generation’ in this research paper explores the ability of the model to generate molecules subject to specific constraints or conditions. This is a crucial step beyond unconditional generation, moving toward more practical applications in drug discovery or materials science. The experiments focus on two key conditioning methods: composition-conditioned generation (specifying the desired elemental composition) and substructure-conditioned generation (specifying the presence of particular molecular substructures). The results demonstrate that the model exhibits a high degree of controllability in both scenarios, significantly outperforming baselines on metrics such as Tanimoto similarity (for substructure matching) and composition accuracy. This suggests a superior ability to leverage conditional information during the generative process. The success of conditional generation highlights the model’s potential for targeted molecule design and offers valuable insights into the capabilities and limitations of using diffusion models for such a complex task. The incorporation of conditional information into the learnable forward process is particularly noteworthy, suggesting it is a critical aspect for successful conditional generation within the diffusion framework.
Future Directions#
Future research directions for equivariant diffusion models in molecule generation could explore several promising avenues. Improving sampling efficiency remains crucial; current methods are slower than conventional approaches. Investigating alternative parameterizations of the forward and reverse processes, perhaps leveraging normalizing flows, could significantly enhance speed and scalability. Expanding the types of conditioning information beyond composition and substructure, to incorporate other properties or 3D spatial relationships, would further broaden applicability. The development of more robust and generalizable architectures is essential to handle larger, more complex molecules and diverse chemical spaces. Addressing the challenges of efficiently handling discrete features (like atom types) is needed. Exploring the potential of hybrid approaches, combining diffusion models with autoregressive methods, might leverage the strengths of both techniques. Finally, rigorous benchmarking and comparison against a wider range of baselines, using both standard and newly developed metrics, is vital to objectively assess progress in the field.
More visual insights#
More on tables
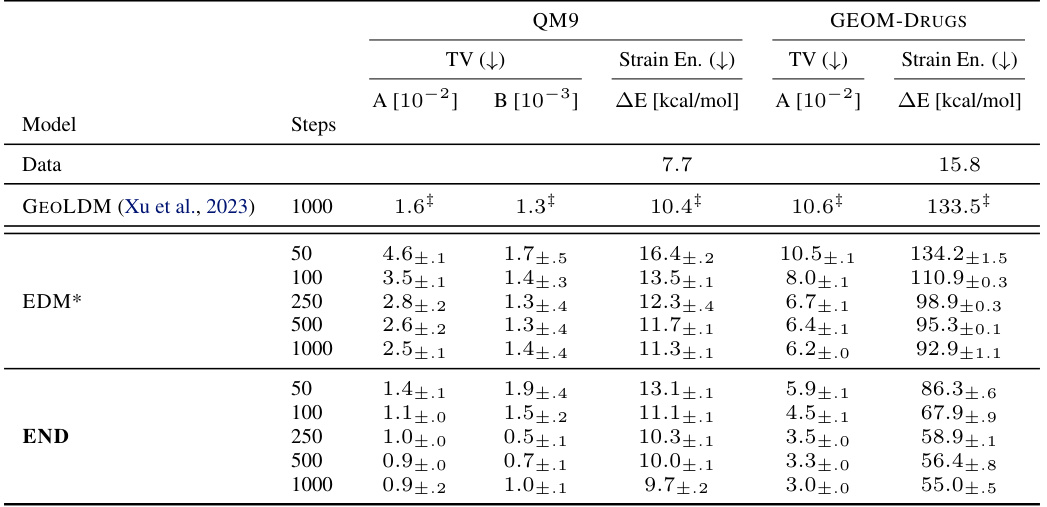
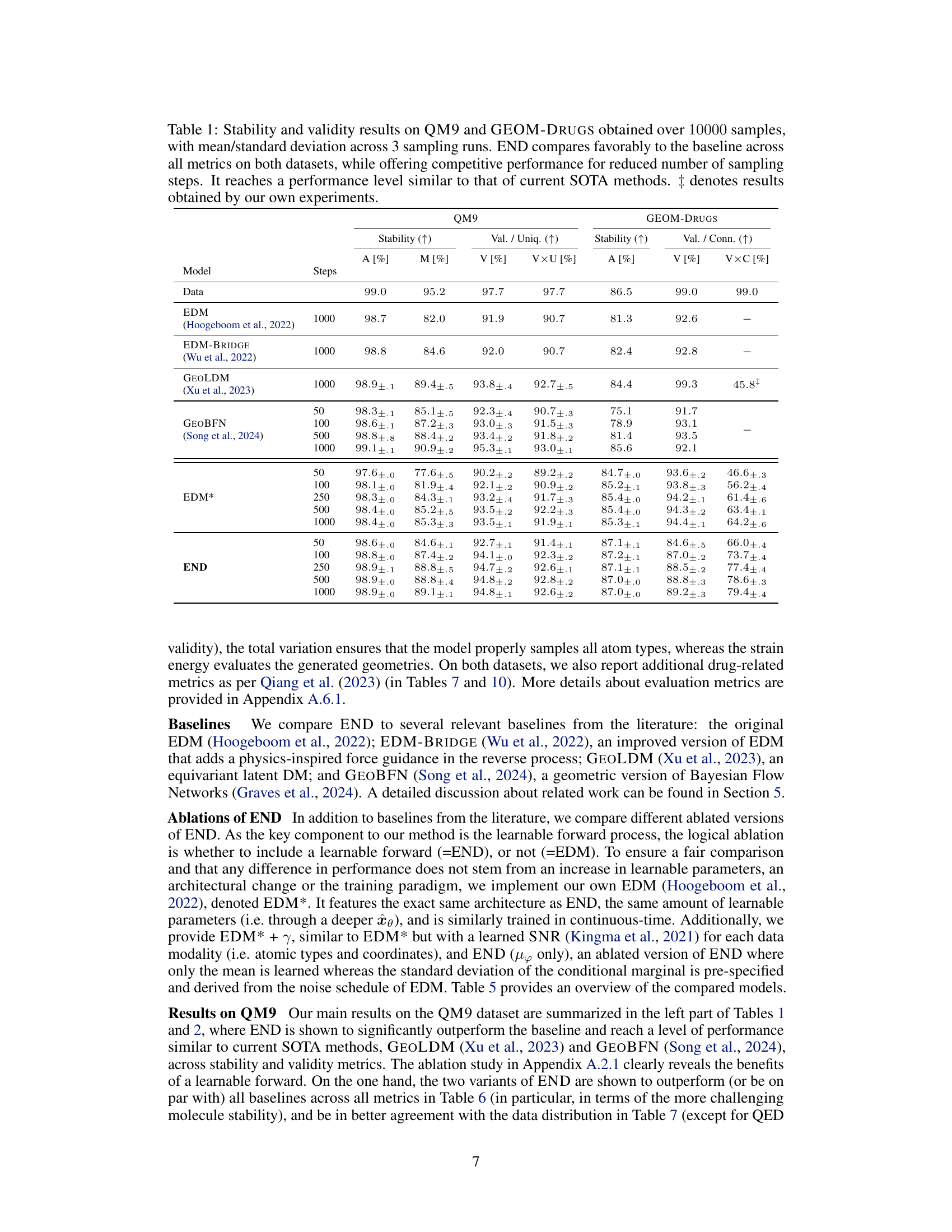
This table presents additional quantitative results on the QM9 and GEOM-Drugs datasets to complement Table 1. It shows the total variation (TV), atom types distribution (A and B), and strain energy (ΔE) for different models and numbers of integration steps. Lower TV values indicate better agreement with the training data distribution, and lower strain energy (ΔE) signifies that the model generates less strained structures. The results demonstrate that END consistently outperforms baselines in terms of matching the training distribution and producing less strained molecules.
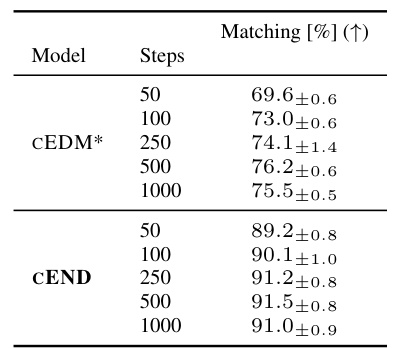
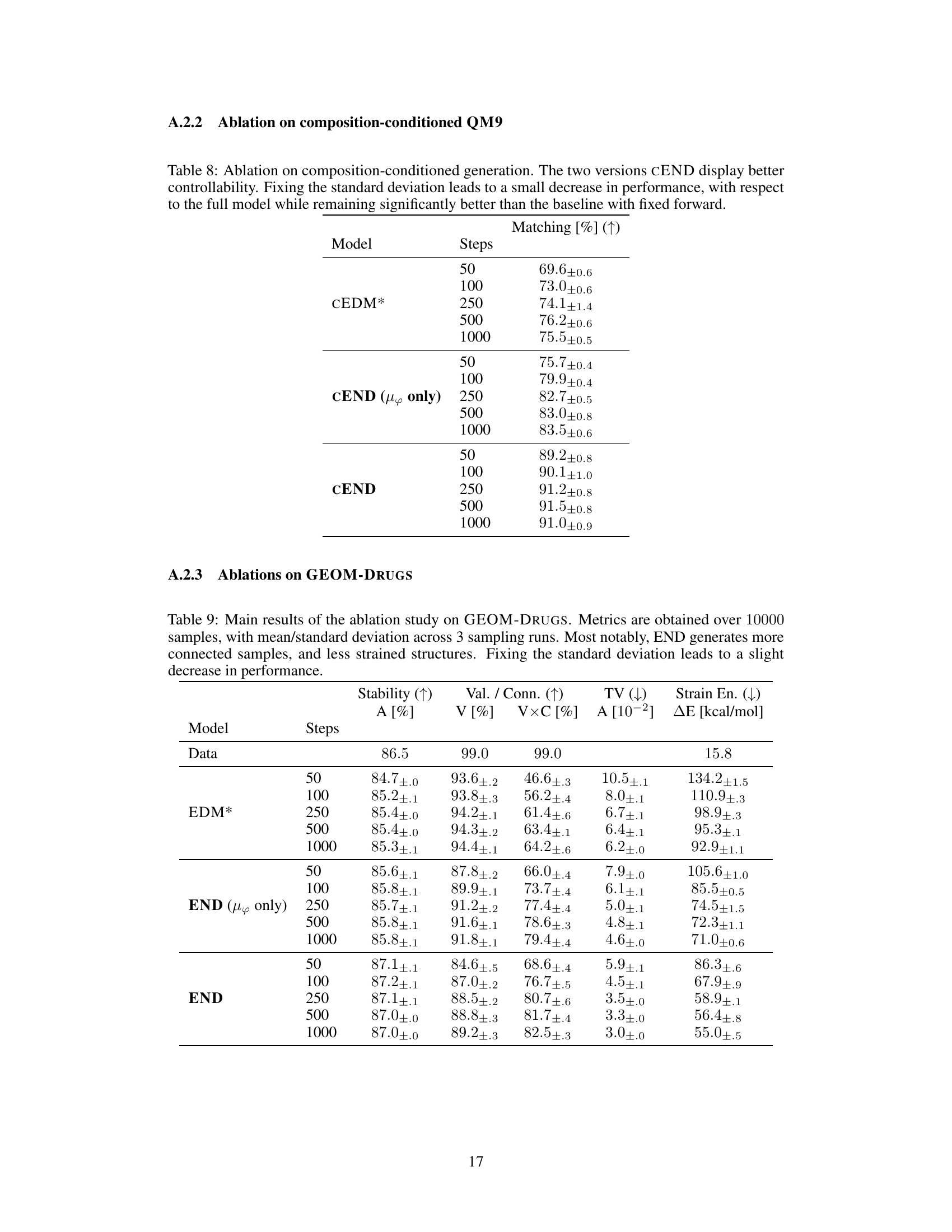
This table presents the results of composition-conditioned molecule generation experiments. The experiment aimed to generate molecules with a specific composition (defined by the number of each atom type). The table compares the performance of CEND (the proposed model) against CEDM* (a baseline model) across different numbers of sampling steps. The ‘Matching [%]’ column shows the percentage of generated molecules that successfully matched the target composition. The results demonstrate CEND’s superior ability to control the generated molecule’s composition.

This table presents the results of substructure-conditioned molecule generation experiments. It compares the performance of the proposed CEND model against the EEGSDE baseline model in terms of Tanimoto similarity scores. Higher Tanimoto similarity indicates a better match between the generated molecules’ substructures and the target substructures specified as conditions. The results show that CEND significantly outperforms EEGSDE, demonstrating its superior ability to control the generation process based on provided substructure information.

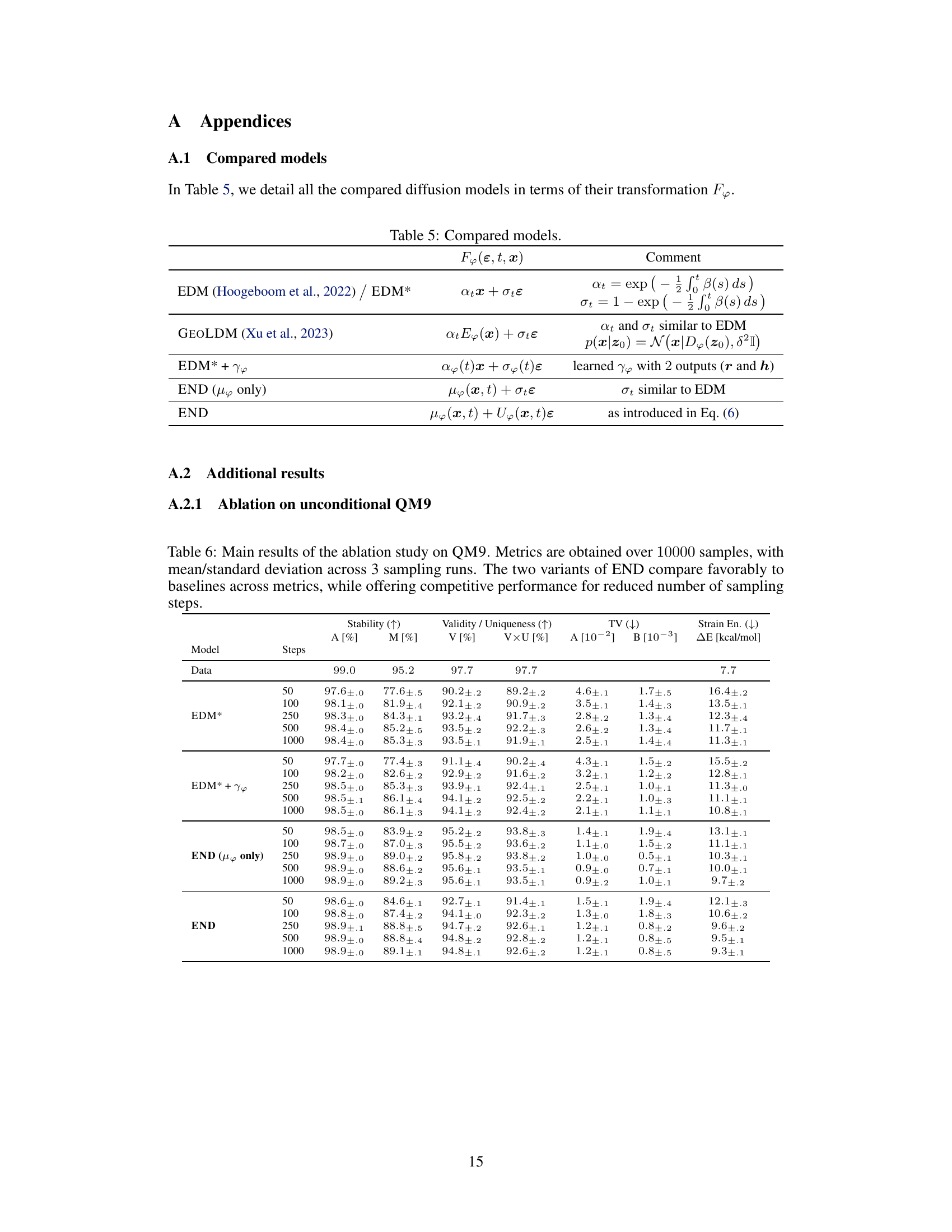
This table compares different diffusion models used in the paper, focusing on their forward transformation function, F. It shows how each model defines F, highlighting the differences in their approaches. In particular, it shows the equations used to calculate the transformation and the parameters involved for each model. The models include EDM, GEOLDM, EDM* + Υφ, END (μφ only), and END. The ‘Comment’ column provides additional context and explanations for each model’s approach.
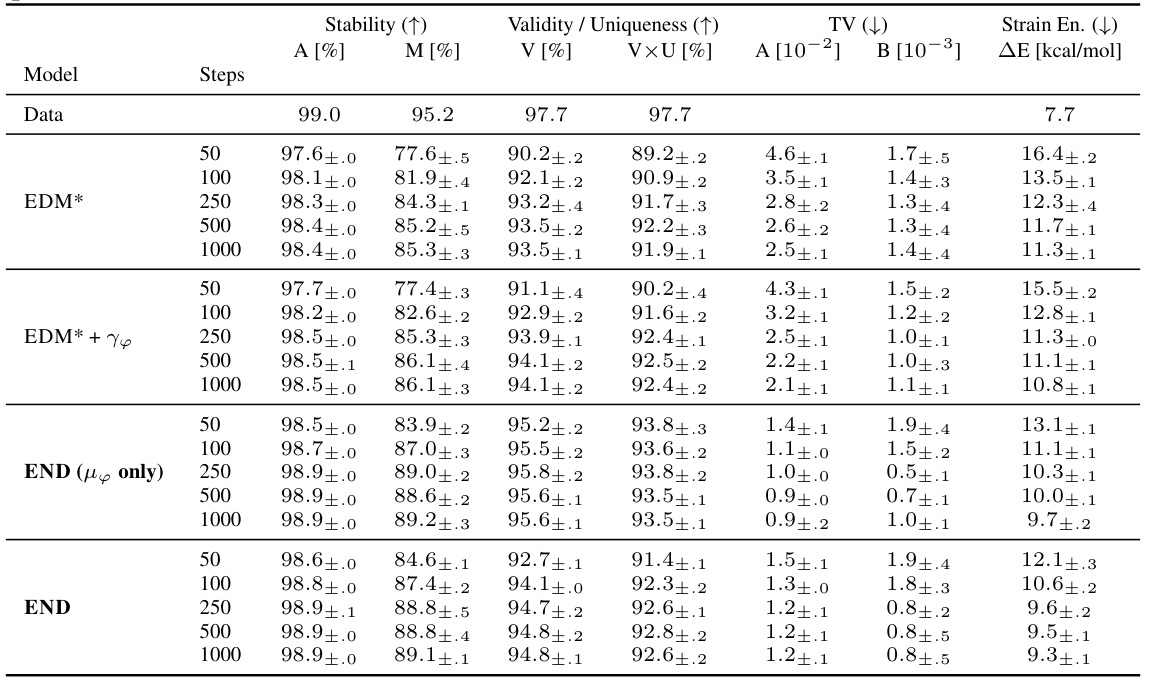
This table presents the results of stability and validity experiments on two datasets (QM9 and GEOM-Drugs) for molecule generation. It compares the performance of the proposed END model against several baselines, using different numbers of sampling steps. The metrics evaluated include stability (A[%], M[%]), validity (V[%]), and uniqueness (V×U[%]). The table highlights END’s competitive performance and its ability to achieve comparable results with fewer steps.
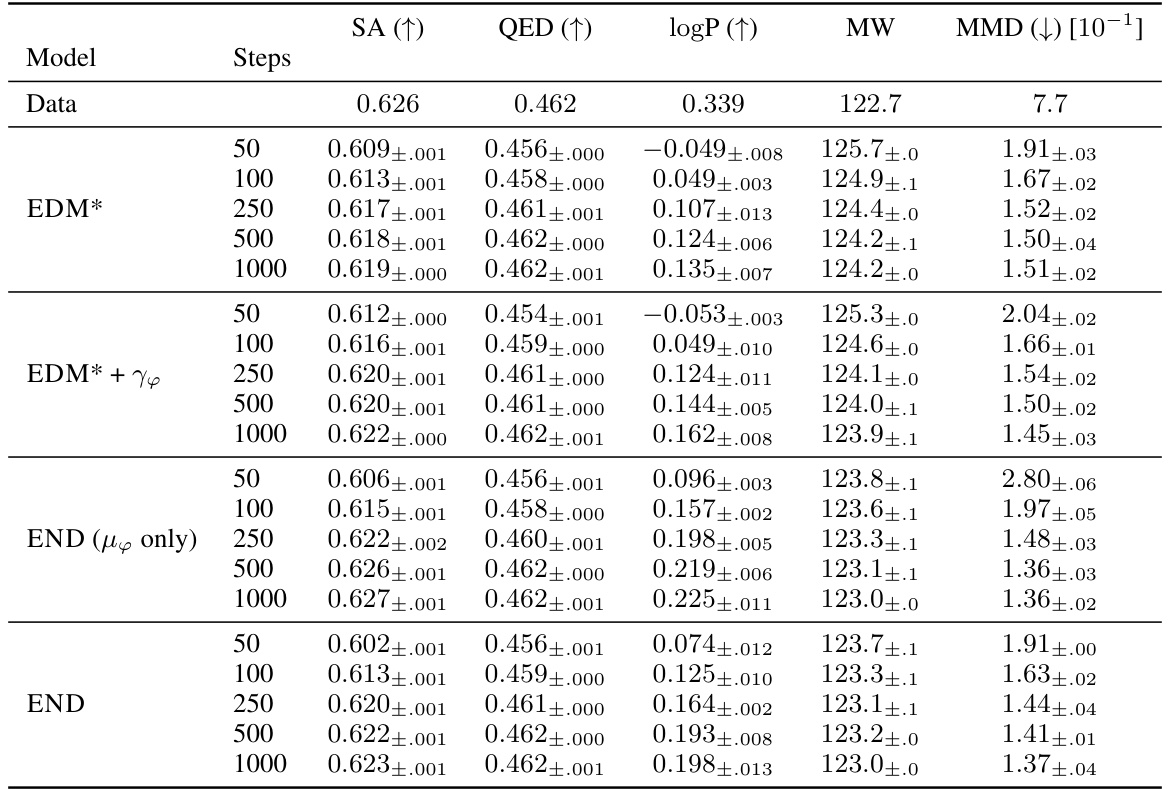
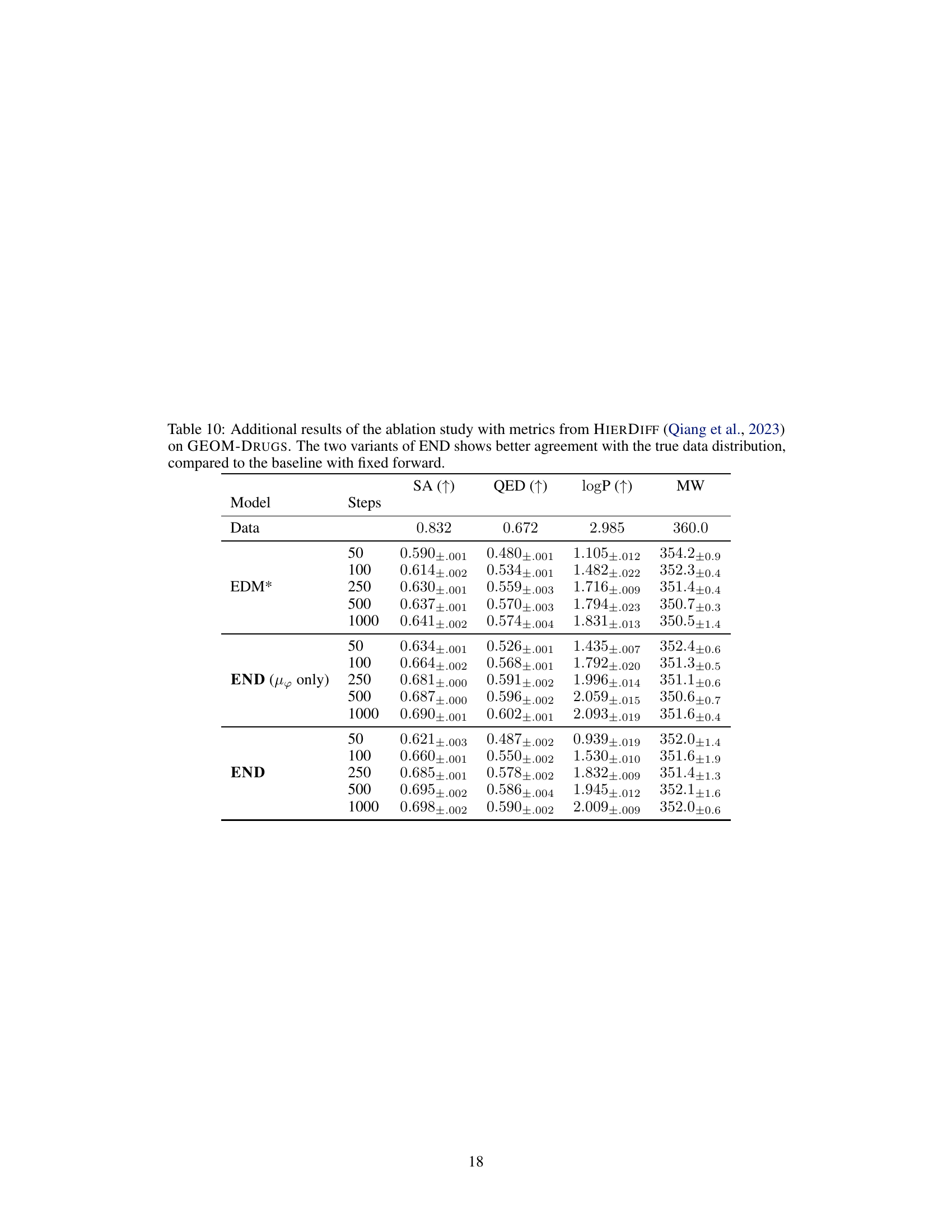
This table presents additional evaluation metrics for the ablation study on the QM9 dataset. It compares the performance of different model variations (END, EDM*, EDM*+Υφ, END (µφ only)) using metrics from HIERDIFF and MMD. The focus is on how well the models capture the true data distribution. The results show that END variants generally perform better or on par with the baselines.

This table presents the results of composition-conditioned generation experiments. It compares the performance of three models (CEDM*, CEND (µφ only), and CEND) across different numbers of sampling steps (50, 100, 250, 500, 1000). The key metric is ‘Matching [%]’, which represents the percentage of generated samples that perfectly match the target composition. Higher percentages indicate better controllability of the generation process.

This table presents the stability and validity results of the END model and several baselines on the QM9 and GEOM-DRUGS datasets. The metrics used to evaluate the models include stability, validity, uniqueness, and various other metrics for drug-related properties. The table shows that END outperforms or is comparable to existing state-of-the-art methods across different evaluation metrics and sampling steps, highlighting its robustness and efficiency.

This table compares the performance of the END model against several baselines on two datasets, QM9 and GEOM-DRUGS, using various metrics related to stability, validity, and uniqueness of generated molecules. The results show that END performs competitively with or surpasses the state-of-the-art methods, especially with fewer sampling steps.

This table presents the stability and validity results of the END model and several baselines on the QM9 and GEOM-DRUGS datasets. The metrics used include stability, validity, uniqueness (QM9), connectivity (GEOM-DRUGS), and combinations thereof. The results show END performing comparably to or better than state-of-the-art methods, especially with fewer sampling steps, indicating efficiency gains.
Full paper#