TL;DR#
Molecular property prediction (MPP) is vital but often hindered by limited data, leading to challenges in few-shot molecular property prediction (FSMPP). Existing methods often struggle with ineffective fine-tuning of pre-trained molecular encoders, partly due to the abundance of parameters relative to the scarce labeled data, as well as insufficient contextual understanding by the encoder. This limits the ability of these methods to effectively adapt to various FSMPP tasks.
The authors introduce Pin-Tuning, a parameter-efficient in-context tuning approach that addresses these issues. Pin-Tuning employs lightweight adapters for pre-trained message passing layers and Bayesian weight consolidation for embedding layers, effectively preventing overfitting and catastrophic forgetting. The method also incorporates contextual perceptiveness into the adapters, enhancing the encoder’s ability to leverage context-specific information for improved adaptability and predictive accuracy. Experimental results demonstrate that Pin-Tuning achieves superior performance with fewer tunable parameters compared to state-of-the-art methods across multiple benchmark datasets.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in drug discovery and material science due to its focus on few-shot molecular property prediction (FSMPP), a critical area hampered by data scarcity. The proposed Pin-Tuning method offers a parameter-efficient solution that enhances model adaptability and predictive performance, directly addressing a major limitation in current approaches. This work opens up avenues for improving few-shot learning in other domains with similar data constraints, and its emphasis on contextual perceptiveness provides valuable insights for improving model generalization. The methodology is rigorously evaluated, providing a strong foundation for future research building on this method.
Visual Insights#
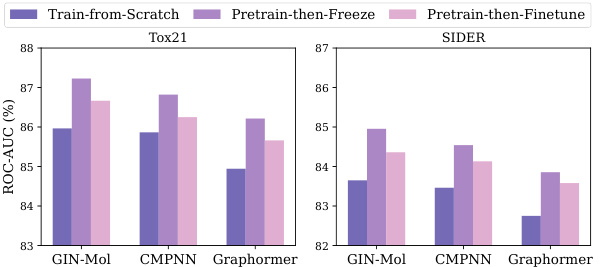
🔼 The figure compares the performance of three different molecular encoder training methods: training from scratch, pre-training then freezing the weights, and pre-training then fine-tuning the weights. It shows ROC-AUC scores across two datasets (Tox21 and SIDER) and three encoder architectures (GIN-Mol, CMPNN, and Graphormer). The results highlight the effectiveness of pre-training but reveal the limitations of current fine-tuning techniques, indicating that fine-tuning often results in performance inferior to simply freezing the pre-trained encoder weights.
read the caption
Figure 1: Comparison of molecular encoders trained via different paradigms: train-from-scratch, pretrain-then-freeze, and pretrain-then-finetune. The evaluation is conducted across two datasets and three encoder architectures [20, 47, 66]. The results consistently demonstrate that while pretraining outperforms training from scratch, the current methods do not yet effectively facilitate finetuning.
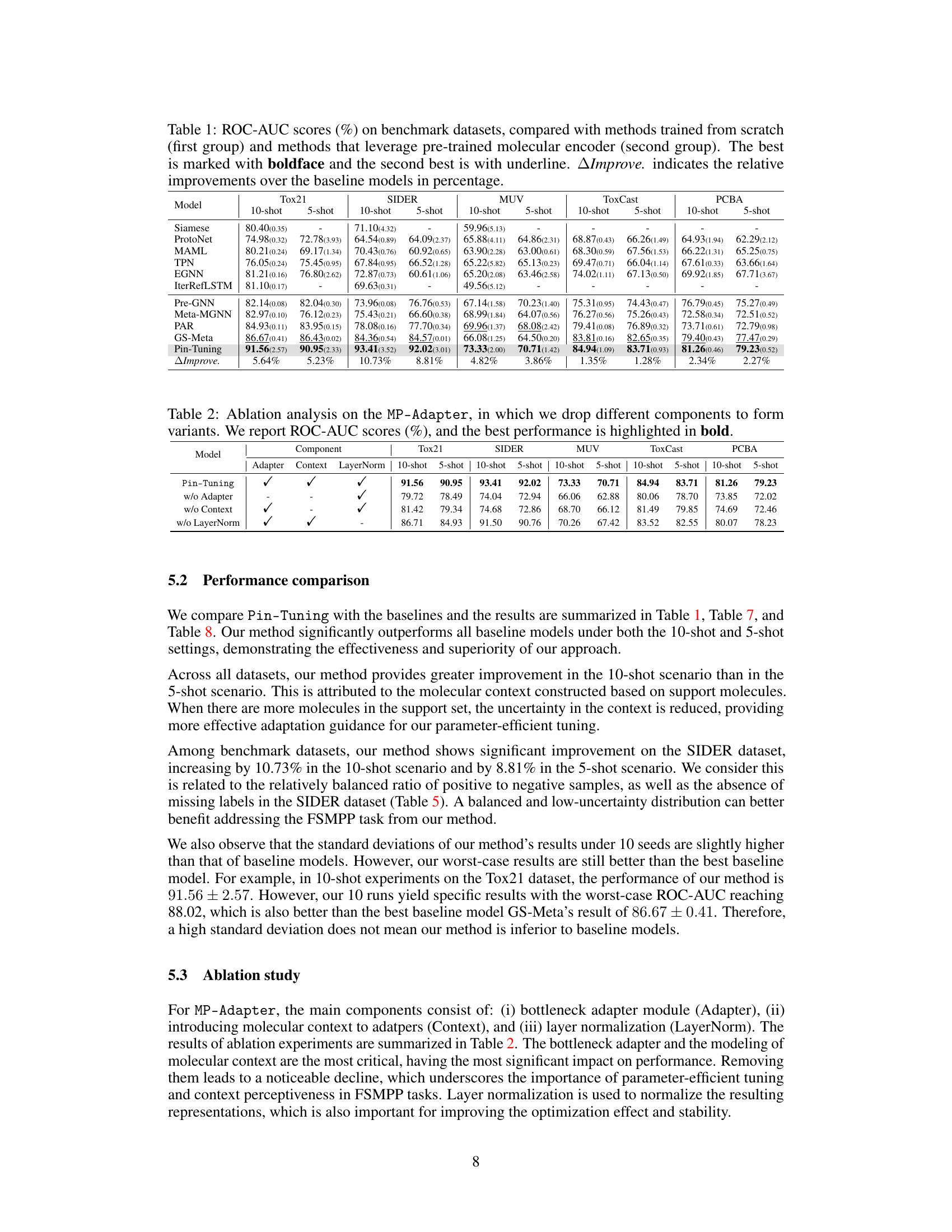
🔼 This table presents the ROC-AUC scores achieved by various methods on five benchmark datasets for few-shot molecular property prediction. It compares methods trained from scratch with those using pre-trained molecular encoders. The best performing method for each dataset and shot setting is highlighted, along with the percentage improvement relative to the best baseline method.
read the caption
Table 1: ROC-AUC scores (%) on benchmark datasets, compared with methods trained from scratch (first group) and methods that leverage pre-trained molecular encoder (second group). The best is marked with boldface and the second best is with underline. △Improve. indicates the relative improvements over the baseline models in percentage.
In-depth insights#
Pin-Tuning’s Promise#
Pin-Tuning presents a promising approach to parameter-efficient fine-tuning of pre-trained molecular encoders for few-shot molecular property prediction (FSMPP). Its lightweight adapters and Bayesian weight consolidation strategies effectively address the challenge of data scarcity by mitigating overfitting and catastrophic forgetting. By enhancing these adapters with contextual perceptiveness, Pin-Tuning further improves the encoder’s ability to adapt to specific tasks. The method demonstrates superior performance compared to existing methods while using significantly fewer parameters, making it particularly suitable for resource-constrained settings and expanding the potential applications of FSMPP in drug discovery and materials science. However, further research is needed to address the relatively high standard deviations observed in some experiments, potentially through improved handling of uncertainty in context information. Despite this limitation, the potential of Pin-Tuning to improve efficiency and expand FSMPP’s applicability is significant.
Efficient Tuning#
Efficient tuning in the context of large language models and other deep learning architectures is crucial for balancing performance gains with computational costs and memory constraints. Parameter-efficient fine-tuning methods, such as adapters or low-rank updates, allow for adapting pre-trained models to downstream tasks using significantly fewer trainable parameters compared to full fine-tuning. This is particularly beneficial in scenarios with limited data or computational resources. However, challenges remain in ensuring these methods maintain the performance of fully-tuned models while avoiding issues like catastrophic forgetting. Furthermore, contextual awareness during tuning is increasingly important to effectively leverage the vast knowledge encapsulated within large pretrained models. Methods that incorporate context information during adaptation can significantly improve performance on specific tasks by guiding the tuning process towards relevant parts of the model’s knowledge base. Future research directions could focus on more sophisticated methods for integrating contextual information, addressing issues of catastrophic forgetting more robustly, and exploring new parameter-efficient techniques that may outperform existing methods. Developing a framework for understanding and comparing the trade-offs between various efficient tuning approaches across different architectures and datasets would be a significant advancement.
Contextualization#
Contextualization in few-shot learning, particularly within the domain of molecular property prediction, is crucial for effective model adaptation. Insufficient contextual information hinders the model’s ability to generalize to unseen tasks and molecules. Strategies for incorporating context include using contextual graphs, which represent relationships between molecules and properties. These graphs can be leveraged by a separate context encoder module, providing contextual awareness to the main molecular encoder. Parameter-efficient approaches are essential to avoid overfitting, as few labeled data samples are usually available. Incorporating context through methods like attention mechanisms could lead to further gains in accuracy. Careful selection of contextual features is important; incorporating irrelevant information can negatively impact model performance. Future research might explore more sophisticated methods for dynamically weighting the contribution of contextual information, as well as methods for handling uncertainty and noise within the contextual data itself.
FSMPP Challenges#
Few-shot molecular property prediction (FSMPP) faces significant challenges stemming from inherent data scarcity. Ineffective fine-tuning of pre-trained molecular encoders is a major hurdle, often leading to performance inferior to using a frozen encoder. This is attributed to the imbalance between abundant tunable parameters and limited labeled data, resulting in overfitting and catastrophic forgetting. Furthermore, existing methods often lack contextual perceptiveness within the encoder, hindering its ability to leverage the nuanced relationships between molecules and their properties. Addressing these challenges requires innovative approaches that prioritize parameter efficiency, prevent overfitting, and enhance contextual understanding within the model architecture for improved few-shot learning performance.
Future Directions#
The ‘Future Directions’ section of a research paper on parameter-efficient in-context tuning for few-shot molecular property prediction could explore several promising avenues. Extending the approach to handle larger, more complex molecules is crucial, as current methods may struggle with the increased computational demands. Investigating the impact of different molecular representations and their suitability for various property prediction tasks is vital, as different encodings may highlight or obscure crucial structural features. Another key area is to improve the contextual understanding of the model. While the paper introduces context, refining how the model integrates and utilizes it for more accurate predictions would significantly enhance performance. Developing more sophisticated methods for handling uncertainty in few-shot settings is paramount. The paper mentions uncertainty in contextual information; addressing this through advanced statistical techniques or Bayesian frameworks could improve robustness. Finally, exploring transfer learning capabilities to adapt the model to new properties or datasets without extensive retraining would unlock its broader applicability and pave the way for more widespread use in various scientific disciplines.
More visual insights#
More on figures
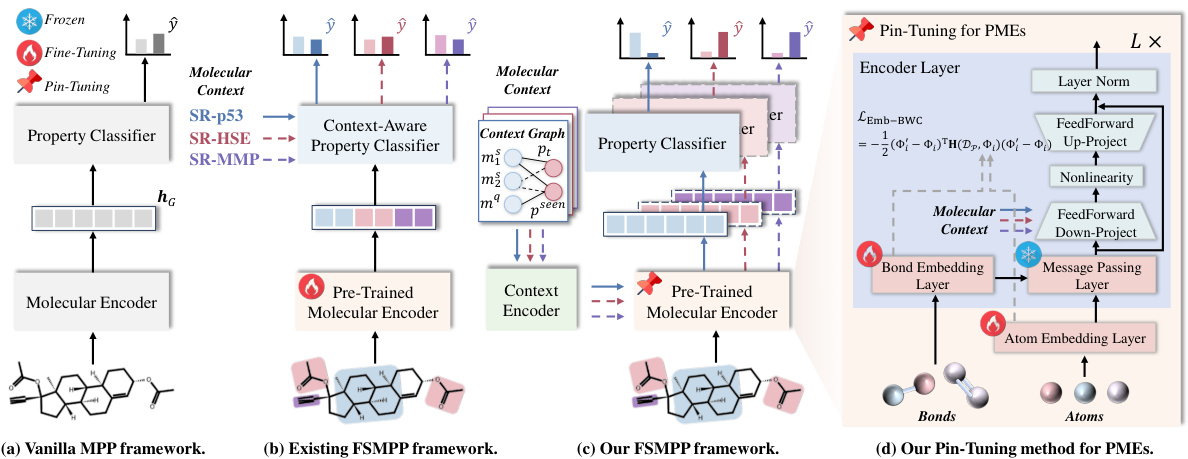
🔼 This figure illustrates the evolution of the molecular property prediction framework from vanilla MPP to the proposed FSMPP with Pin-Tuning. (a) shows the simple encoder-classifier framework. (b) highlights the existing FSMPP approach utilizing pre-trained molecular encoders and context-aware classifiers. (c) presents the proposed framework incorporating Pin-Tuning for parameter-efficient adaptation. (d) details the Pin-Tuning method applied to pre-trained molecular encoders, focusing on message passing layer and embedding layer adaptations.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
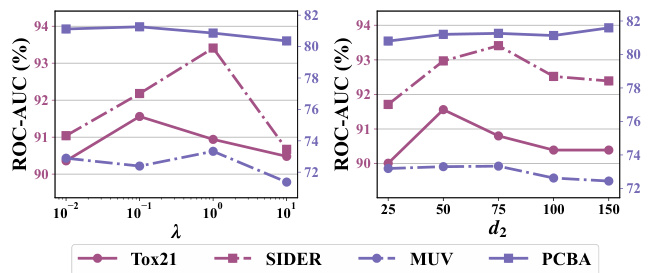
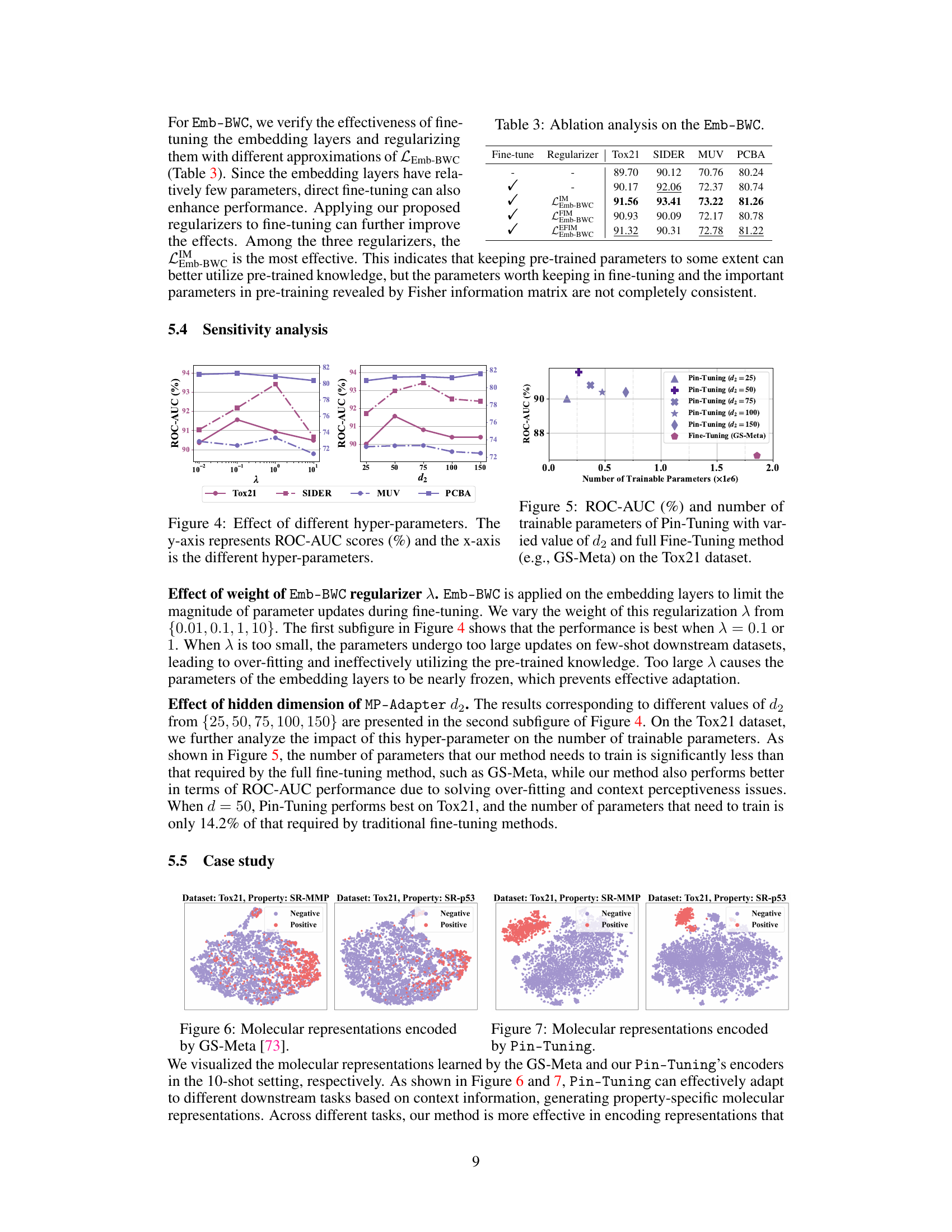
🔼 This figure shows the sensitivity analysis of two hyperparameters: λ (lambda), the weight of the Emb-BWC regularizer, and d2, the hidden dimension of the MP-Adapter. The plots show ROC-AUC scores on four datasets (Tox21, SIDER, MUV, PCBA) for different values of λ and d2. The results demonstrate the impact of these hyperparameters on the model’s performance. Optimal values are found for both λ and d2, showing the importance of tuning these parameters for optimal results.
read the caption
Figure 4: Effect of different hyper-parameters. The y-axis represents ROC-AUC scores (%) and the x-axis is the different hyper-parameters.

🔼 This figure shows the trade-off between the number of trainable parameters and the ROC-AUC score achieved by the proposed Pin-Tuning method compared to the full fine-tuning method (GS-Meta) on the Tox21 dataset. Different values of the hyperparameter d2 (hidden dimension of the MP-Adapter) are tested, demonstrating that Pin-Tuning achieves comparable or better performance with significantly fewer trainable parameters.
read the caption
Figure 5: ROC-AUC (%) and number of trainable parameters of Pin-Tuning with varied value of d2 and full Fine-Tuning method (e.g., GS-Meta) on the Tox21 dataset.

🔼 This figure shows four different frameworks for molecular property prediction (MPP) tasks. (a) depicts the basic encoder-classifier framework. (b) shows the standard few-shot MPP framework using a pre-trained encoder and context-aware classifier. (c) illustrates the proposed framework incorporating the Pin-Tuning method. Finally, (d) provides a detailed overview of the Pin-Tuning method for adapting pre-trained molecular encoders, highlighting the modifications made to the message passing layers and embedding layers.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure illustrates the evolution of molecular property prediction (MPP) frameworks from a basic encoder-classifier model to the few-shot learning (FSL) adaptation with context information and finally to the proposed Pin-Tuning method. (a) shows the standard MPP setup. (b) introduces the use of pre-trained molecular encoders and context-aware classifiers, typical in FSMPP. (c) presents the authors’ proposed approach, incorporating their Pin-Tuning method to improve upon existing FSMPP frameworks. Lastly, (d) provides a detailed diagram of their proposed Pin-Tuning technique for adapting pre-trained molecular encoders.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.

🔼 This figure shows four different frameworks for molecular property prediction. (a) shows a basic encoder-classifier model. (b) shows a few-shot learning model that uses a pre-trained encoder. (c) shows the proposed Pin-Tuning model, which adds an adapter to the pre-trained encoder. (d) shows a detailed diagram of the Pin-Tuning adapter.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure shows four different frameworks for molecular property prediction. (a) shows a basic encoder-classifier framework. (b) shows a framework using a pre-trained molecular encoder and a context-aware classifier, common in few-shot molecular property prediction. (c) presents the authors’ proposed framework which uses Pin-Tuning to update the pre-trained encoder. (d) provides detailed information of the proposed Pin-Tuning method for pre-trained molecular encoders.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure shows four different frameworks for molecular property prediction (MPP). (a) is a basic encoder-classifier framework. (b) shows an existing framework for few-shot MPP (FSMPP) that utilizes a pre-trained molecular encoder. (c) shows the proposed FSMPP framework that incorporates a Pin-Tuning method to improve the adaptation of the pre-trained encoder. (d) provides a detailed illustration of the Pin-Tuning method applied to pre-trained molecular encoders.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure shows four different frameworks for molecular property prediction. (a) shows the basic encoder-classifier framework for general molecular property prediction. (b) illustrates an existing framework for few-shot molecular property prediction (FSMPP) that incorporates a pre-trained molecular encoder to leverage pre-existing knowledge and a context-aware classifier to utilize the many-to-many relationships between molecules and properties. (c) presents the proposed FSMPP framework, named Pin-Tuning, which enhances the pre-trained encoder with a parameter-efficient in-context tuning method to prevent over-fitting and catastrophic forgetting. Finally, (d) details the proposed Pin-Tuning method for adapting the pre-trained molecular encoder, specifically targeting the message passing and embedding layers with lightweight adapters and Bayesian weight consolidation.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.

🔼 This figure shows four different frameworks for molecular property prediction (MPP). (a) shows a basic encoder-classifier framework. (b) shows a framework for few-shot molecular property prediction (FSMPP) that uses a pre-trained molecular encoder and a context-aware classifier. (c) shows the authors’ proposed framework which uses Pin-Tuning to update the pre-trained molecular encoder. (d) shows a detailed diagram of the Pin-Tuning method.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.

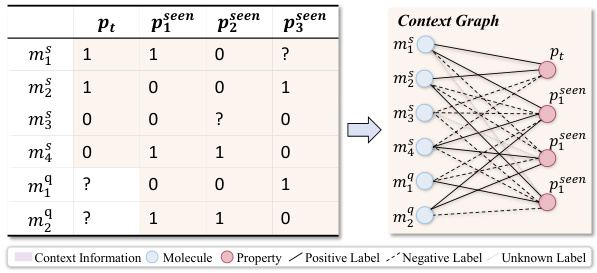
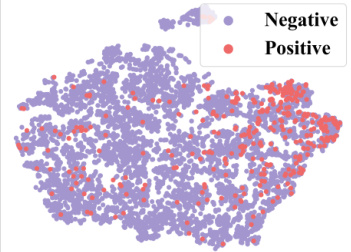
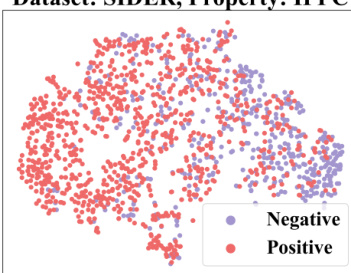
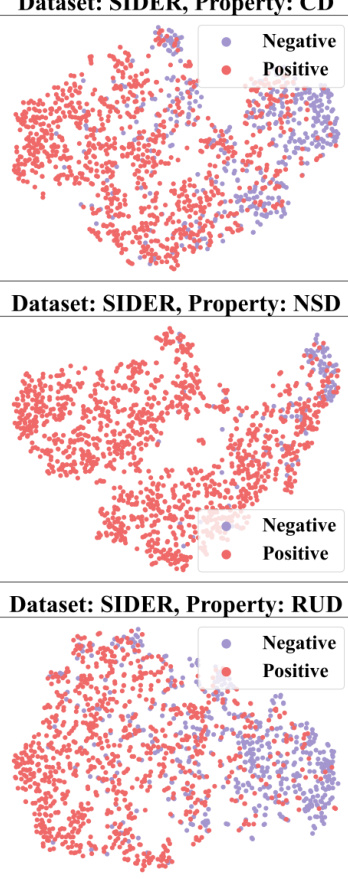
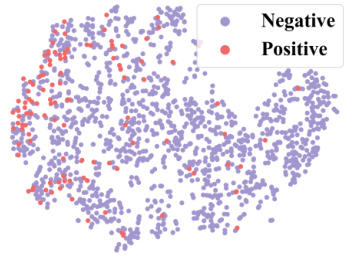
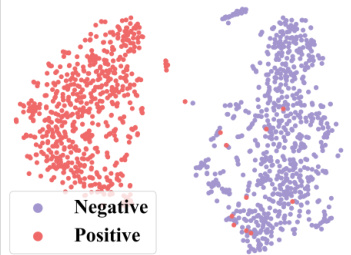
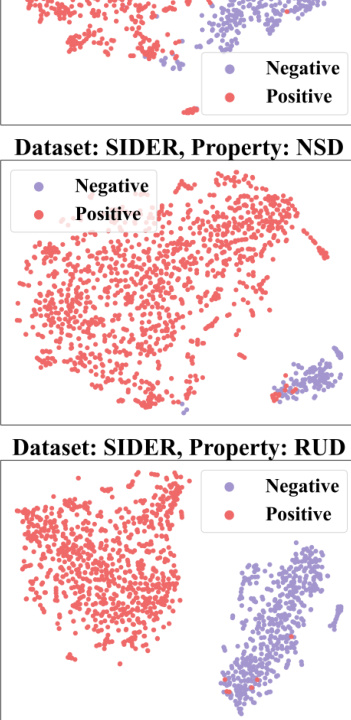
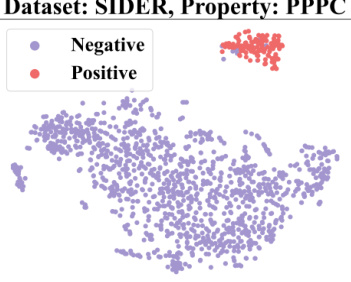
🔼 This figure demonstrates how context information from a 2-shot episode is converted into a context graph. The left table shows the labels of molecules for the target property (pt) and seen properties (pseen). The question marks (?) represent labels that are unknown and need to be predicted. This information is then transformed into a context graph (shown on the right) with molecule nodes (m) and property nodes (p), and different types of edges indicating relationships between molecules and properties. This graph is then used as input to a GNN-based context encoder, enabling the model to perceive and utilize the context when performing few-shot predictions.
read the caption
Figure 3: Convert the context information of a 2-shot episode into a context graph.

🔼 The figure illustrates how context information from a 2-shot episode is converted into a context graph. The left side shows a table with the labels of molecules for the target property (pt) and other seen properties (pseen). The shaded values represent available context. The right side displays the resulting context graph, which contains molecule nodes and property nodes connected by various edges representing different relationships.
read the caption
Figure 3: Convert the context information of a 2-shot episode into a context graph.
🔼 This figure illustrates the different frameworks used for molecular property prediction (MPP) and few-shot molecular property prediction (FSMPP). (a) shows the basic encoder-classifier framework. (b) shows the framework commonly used in FSMPP which uses a pre-trained molecular encoder and a context-aware classifier. (c) presents the proposed framework, which incorporates the Pin-Tuning method to improve the pre-trained encoder. (d) details the Pin-Tuning method’s application to pre-trained molecular encoders. The figure highlights the evolution of the framework from vanilla MPP to the proposed FSMPP method.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure shows four different frameworks for molecular property prediction (MPP) tasks. (a) shows a simple encoder-classifier framework. (b) shows a common framework for few-shot MPP (FSMPP) which uses a pre-trained molecular encoder and a context-aware classifier. (c) shows the proposed framework which uses a Pin-Tuning method for updating pre-trained encoders. (d) illustrates the details of the Pin-Tuning method. This method is designed to address the limitations of existing FSMPP methods by improving the parameter efficiency and incorporating contextual information.
read the caption
Figure 2: (a) The vanilla encoder-classifier framework for MPP. (b) The framework widely adopted by existing FSMPP methods, which contains a pre-trained molecular encoder and a context-aware property classifier. (c) Our proposed framework for FSMPP, in which we introduce a Pin-Tuning method to update the pre-trained molecular encoder followed by the property classifier. (d) The details of our proposed Pin-Tuning method for pre-trained molecular encoders. In (b) and (c), we use the property names like SR-HSE to denote the molecular context in episodes.
🔼 This figure demonstrates how context information is converted into a context graph. The left table shows the labels for two molecules (m1, m2) for a target property (pt). Other shaded values represent the labels of query molecules for other properties. This information is converted into a context graph (Gt = (Vt, At, Xt)) with M molecule nodes and P property nodes. Three types of edges represent the relationship between the nodes.
read the caption
Figure 3: Convert the context information of a 2-shot episode into a context graph.
More on tables

🔼 This table presents the ROC-AUC scores achieved by various models on five benchmark datasets for both 5-shot and 10-shot settings. It compares models trained from scratch with those using pre-trained molecular encoders. The best performing model for each dataset and shot scenario is highlighted, along with the relative improvement over the baseline model.
read the caption
Table 1: ROC-AUC scores (%) on benchmark datasets, compared with methods trained from scratch (first group) and methods that leverage pre-trained molecular encoder (second group). The best is marked with boldface and the second best is with underline. △Improve. indicates the relative improvements over the baseline models in percentage.

🔼 This table presents the results of ablation experiments conducted on the MP-Adapter component of the Pin-Tuning method. The researchers systematically removed different parts of the MP-Adapter (adapter module, context integration, and layer normalization) to assess their individual contribution to the model’s overall performance. The ROC-AUC scores are reported for each ablation variant across five different datasets (Tox21, SIDER, MUV, ToxCast, and PCBA) under both 10-shot and 5-shot settings, allowing for a comprehensive evaluation of the impact of each component.
read the caption
Table 2: Ablation analysis on the MP-Adapter, in which we drop different components to form variants. We report ROC-AUC scores (%), and the best performance is highlighted in bold.
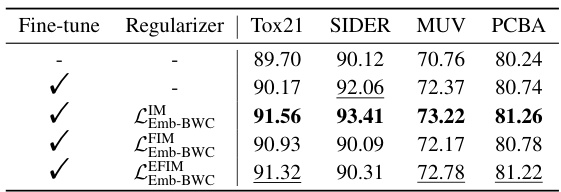
🔼 This ablation study analyzes the impact of different Emb-BWC regularizers on the model’s performance. It compares the results of fine-tuning the embedding layers with and without different regularizers (Identity, Fisher Information Matrix, and Embedding-wise Fisher Information Matrix). The table shows the ROC-AUC scores achieved for each dataset (Tox21, SIDER, MUV, PCBA) with different configurations.
read the caption
Table 3: Ablation analysis on the Emb-BWC.

🔼 This table presents the ROC-AUC scores achieved by various methods on five benchmark datasets for few-shot molecular property prediction. The methods are categorized into two groups: those trained from scratch and those leveraging pre-trained molecular encoders. The table highlights the best and second-best performing methods for each dataset and each shot setting (5-shot and 10-shot). It also shows the percentage improvement of the best-performing method compared to the baseline methods.
read the caption
Table 1: ROC-AUC scores (%) on benchmark datasets, compared with methods trained from scratch (first group) and methods that leverage pre-trained molecular encoder (second group). The best is marked with boldface and the second best is with underline. △Improve. indicates the relative improvements over the baseline models in percentage.
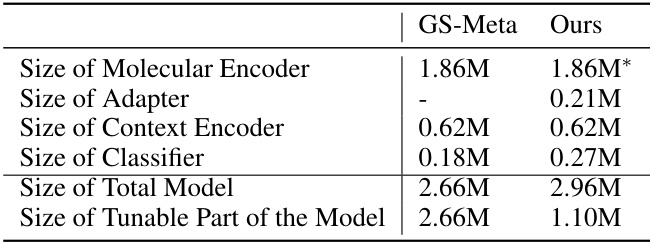
🔼 This table compares the total size of the models used in the experiments, differentiating between the GS-Meta baseline and the proposed Pin-Tuning method. It shows a breakdown of the model size into its components: molecular encoder, adapter, context encoder, and classifier. Importantly, it highlights that while the overall model size is slightly larger for Pin-Tuning, the number of tunable parameters is significantly reduced compared to GS-Meta, emphasizing the parameter efficiency of the proposed approach.
read the caption
Table 4: Comparison of total model size. * indicates that the parameters are frozen.

🔼 This table presents the statistics of five datasets used in the paper’s experiments. For each dataset, it shows the number of compounds, the number of properties, the number of properties used for training and testing, and the percentage of positive, negative, and unknown labels.
read the caption
Table 5: Dataset statistics.

🔼 This table shows the statistics of sub-datasets from ToxCast dataset used in the paper. It includes the number of compounds, the number of properties, the number of training and test properties, and the percentage of positive, negative, and missing labels for each sub-dataset. This information is essential to understanding the characteristics of the data used for evaluation and comparing different models.
read the caption
Table 6: Statistics of sub-datasets of ToxCast.
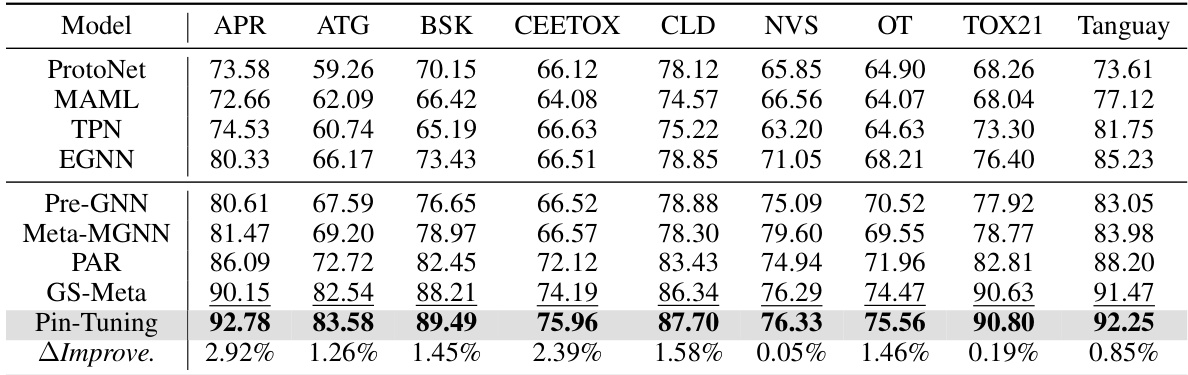
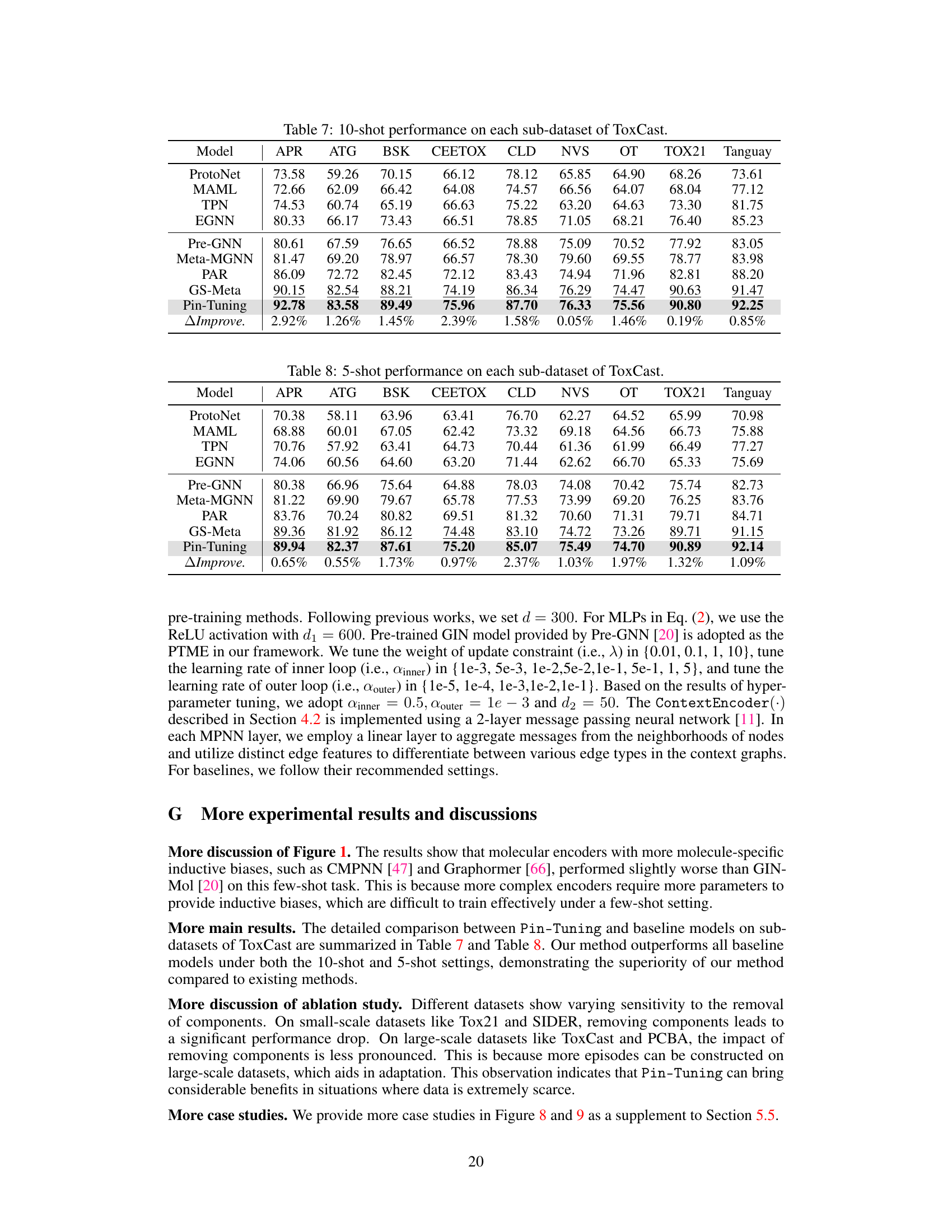
🔼 This table presents the 10-shot performance results on each sub-dataset of ToxCast for various models, including baselines and the proposed Pin-Tuning method. It shows the ROC-AUC scores for each model on various sub-datasets of ToxCast, allowing for a comparison of model performance across different tasks and datasets. The final row displays the relative improvement (%) of Pin-Tuning over the best baseline model.
read the caption
Table 7: 10-shot performance on each sub-dataset of ToxCast.
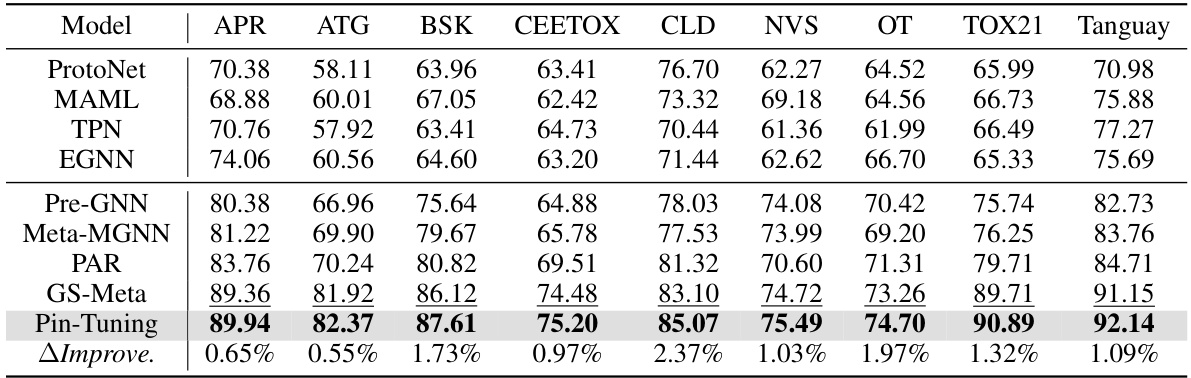
🔼 This table presents the 5-shot performance results for various models on the sub-datasets of ToxCast. It shows the ROC-AUC scores achieved by different methods, including baselines (ProtoNet, MAML, TPN, EGNN, Pre-GNN, Meta-MGNN, PAR, GS-Meta) and the proposed Pin-Tuning method. The last row indicates the relative improvement of Pin-Tuning over the best performing baseline for each dataset. This allows for a comparison of the effectiveness of each approach in few-shot scenarios.
read the caption
Table 8: 5-shot performance on each sub-dataset of ToxCast.
Full paper#