TL;DR#
Incomplete clinical data significantly hinders accurate medical evaluations and reliable research. The challenge lies in integrating heterogeneous, noisy multi-modal biological data (proteomics, metabolomics) with clinical records for improved patient characterization. Existing imputation methods often struggle to capture the complex relationships between various data types and handle missing data effectively.
The proposed MPI framework innovatively addresses this by decoupling the modeling of biological and phenotypic data via graph neural networks. It leverages residual quantization to extract meaningful biological factors, encoding patient correlations from two separate graphs (biological and phenotype views). Cross-view contrastive knowledge distillation refines the imputation process, using the biological view to enhance the phenotype view. Results on real-world datasets demonstrate MPI’s superior performance over current state-of-the-art methods.
Key Takeaways#
Why does it matter?#
This paper is important because it addresses the critical issue of missing data in clinical research, a common problem that hinders accurate medical evaluation and limits the reliability of research findings. By proposing a novel framework that leverages multi-modal biological data and advanced machine learning techniques, this research opens new avenues for improving phenotype imputation and enhancing the precision of clinical studies. It also provides valuable insights into effective ways of integrating multi-modal data for better patient characterization. The proposed approach demonstrates superior performance compared to existing methods on real-world biomedical datasets, and its findings have significant implications for the future of biomedical data analysis.
Visual Insights#

🔼 This figure illustrates the concept of phenotype imputation using multi-modal biological data. It shows a matrix representing phenotypes (P1-P4) with some missing values represented by question marks. A second matrix shows corresponding multi-modal biological data (M1, M2) with missing data represented by dashes. The connection between the two is made via a central icon representing the integration of biological and clinical data to infer missing phenotypes. The resulting imputed phenotypes are shown in a third matrix.
read the caption
Figure 1: Phenotype imputation with multi-modal biological data. 'M1' denotes Modality 1, and 'M2' represents Modality 2. '-' refers to the missing modality and the red question mark refers to the phenotype that needs to be imputed.
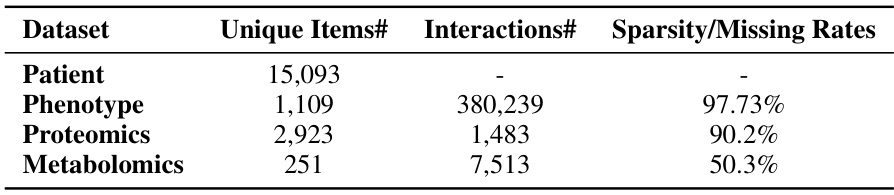
🔼 This table presents the statistics of the UK Biobank dataset used in the paper. It shows the number of unique patients, phenotypes, proteomics, and metabolomics, as well as the number of interactions between patients and phenotypes and the sparsity/missing rates for proteomics and metabolomics.
read the caption
Table 1: Dataset Statistics
In-depth insights#
Multimodal Phenotype Imputation#
Multimodal phenotype imputation tackles the challenge of predicting missing phenotypic data by leveraging information from multiple biological sources. This approach is crucial because clinical records are often incomplete, leading to biased research and suboptimal patient care. The integration of diverse data types, such as genomics, proteomics, and metabolomics, significantly enriches the information available for imputation. However, this also introduces complexities such as data heterogeneity and noise. Successful strategies involve techniques like view decoupling, separating the modeling of biological and phenotypic data to account for their differing characteristics, and knowledge distillation, transferring insights from one view to improve the other. Advanced methods such as graph neural networks are increasingly used to model the complex relationships between different data modalities and patients, leading to more accurate and robust phenotype predictions. A key focus of research is developing methods to manage the irregularity and sparsity of multi-modal data, and to mitigate the challenges of high dimensionality and noise. The ultimate goal is to improve the accuracy and reliability of medical evaluations and clinical research.
View Decoupling Framework#
A View Decoupling Framework, in the context of multi-modal data for phenotype imputation, is a powerful technique to address the inherent heterogeneity of data sources. It acknowledges that biological data and phenotype data provide distinct, yet complementary, perspectives on a patient’s health. By decoupling the modeling of these views, the framework avoids the limitations of forcing a joint representation that struggles to capture the nuances of each data type. Instead, it leverages separate graph neural networks, one for each view. The biological view graph focuses on uncovering latent factors within biological data that reveal patient correlations, while the phenotype view graph focuses on phenotype co-occurrence patterns. A crucial element is the cross-view contrastive knowledge distillation, which leverages insights from the richer biological data to improve the accuracy and robustness of phenotype imputation in the phenotype view. This approach mitigates the effects of noise and irrelevant information in the biological data, enhancing the overall performance and interpretability of the imputation model. The resulting framework is better equipped to handle missing data and data variability, thereby leading to more reliable and comprehensive medical evaluations.
Residual Quantization#
Residual quantization, as described in the context of the research paper, is a technique used for disentangling complex, heterogeneous factors within multi-modal biological data. It leverages a multi-level vector quantizer to convert residuals into a series of codes, offering a hierarchical approach that approximates the data from coarse to fine granularity. This is particularly useful for handling the high-dimensionality and continuous characteristics of data such as proteomics and metabolomics. By using separate codebooks for different levels, the method effectively captures different granularity levels. The resulting disentangled biological factors provide a more robust and interpretable representation of patient biological states, paving the way for more effective downstream tasks like phenotype imputation. This approach contrasts with traditional methods that might struggle to disentangle complex relationships within the data, leading to less reliable and less informative representations. The use of residual quantization represents a significant improvement in pre-processing biological data for applications in biomedical graph neural network modeling.
Cross-view Distillation#
Cross-view distillation, in the context of multi-modal data integration for phenotype imputation, is a powerful technique to address the challenges posed by noisy and heterogeneous biological data. It leverages the strengths of both biological and phenotypic views, mitigating the limitations of each. The core idea is to distill knowledge from a teacher model (trained on a more reliable biological view) to enhance a student model (focused on the often incomplete phenotype view). This distillation process helps refine the student model’s ability to predict missing phenotypes by leveraging the latent patterns discovered in the comprehensive biological data, while simultaneously mitigating the influence of noise and irrelevant information within the biological dataset. The method’s effectiveness lies in its ability to transfer relevant biological insights to improve the accuracy and reliability of phenotype predictions, bridging the gap between the two views and ensuring a more holistic and robust imputation model. By carefully selecting positive and negative sample pairs during training, this technique enables a fine-grained knowledge transfer and avoids over-reliance on potentially inaccurate biological information.
Future Research#
Future research directions stemming from this phenotype imputation work could profitably explore several avenues. Expanding the modality scope to encompass additional biological data types, such as genomics and imaging, could significantly enhance predictive power. Investigating the robustness of the model across diverse patient populations and disease contexts is crucial to establish generalizability. Further work could focus on developing more efficient algorithms to improve computational speed and scalability, particularly vital for very large datasets. A key area for future work lies in exploring how the framework could be adapted to address different types of missing data patterns beyond the irregular missingness addressed in this study. Finally, research could delve into the interpretability of the learned latent factors, potentially facilitating deeper biological insights and a better understanding of disease mechanisms. Such research would significantly increase the clinical utility of the approach.
More visual insights#
More on figures

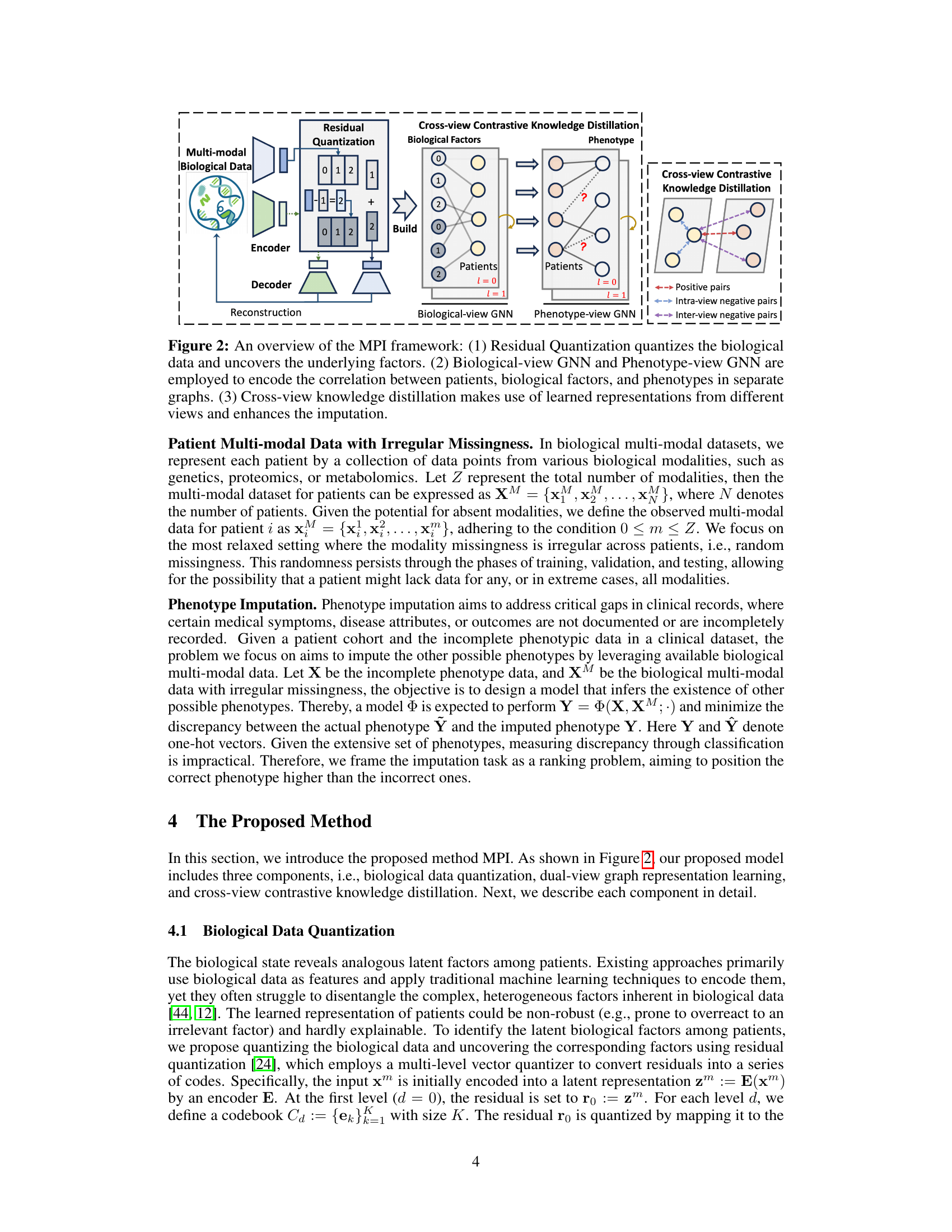
🔼 This figure illustrates the MPI (Multimodal Phenotype Imputation) framework, which consists of three main components. First, Residual Quantization processes the multi-modal biological data to identify latent biological factors. Second, two separate graph neural networks (GNNs) model the relationships between patients and their biological factors (biological-view GNN), and the relationships between patients and their phenotypes (phenotype-view GNN). Finally, cross-view contrastive knowledge distillation leverages information from both views to improve phenotype imputation, enhancing the model’s accuracy and robustness.
read the caption
Figure 2: An overview of the MPI framework: (1) Residual Quantization quantizes the biological data and uncovers the underlying factors. (2) Biological-view GNN and Phenotype-view GNN are employed to encode the correlation between patients, biological factors, and phenotypes in separate graphs. (3) Cross-view knowledge distillation makes use of learned representations from different views and enhances the imputation.

🔼 This figure presents an overview of the proposed Multimodal Phenotype Imputation (MPI) framework. It shows three main stages: (1) Residual Quantization processes the multi-modal biological data to extract latent biological factors. (2) Two separate graph neural networks (GNNs) model patient relationships—one based on the biological factors (biological view) and one based on phenotype co-occurrences (phenotype view). (3) Cross-view contrastive knowledge distillation is employed to refine phenotype imputation by transferring knowledge from the biological-view GNN to the phenotype-view GNN.
read the caption
Figure 2: An overview of the MPI framework: (1) Residual Quantization quantizes the biological data and uncovers the underlying factors. (2) Biological-view GNN and Phenotype-view GNN are employed to encode the correlation between patients, biological factors, and phenotypes in separate graphs. (3) Cross-view knowledge distillation makes use of learned representations from different views and enhances the imputation.
🔼 This figure provides a visual overview of the proposed Multimodal Phenotype Imputation (MPI) framework. It details three key components: 1. Residual Quantization: This step processes the multi-modal biological data, reducing noise and identifying underlying biological factors using a residual quantization technique. 2. Dual-view Graph Neural Networks (GNNs): The processed data is fed into two separate GNNs: one focusing on patient-phenotype relationships, and the other focusing on relationships between patients based on their biological factors. These GNNs generate embeddings representing each patient from two different perspectives. 3. Cross-view Contrastive Knowledge Distillation: This component integrates knowledge from the two separate GNNs. It leverages information from the biological view to improve the accuracy of the phenotype imputation using a contrastive knowledge distillation strategy.
read the caption
Figure 2: An overview of the MPI framework: (1) Residual Quantization quantizes the biological data and uncovers the underlying factors. (2) Biological-view GNN and Phenotype-view GNN are employed to encode the correlation between patients, biological factors, and phenotypes in separate graphs. (3) Cross-view knowledge distillation makes use of learned representations from different views and enhances the imputation.
More on tables

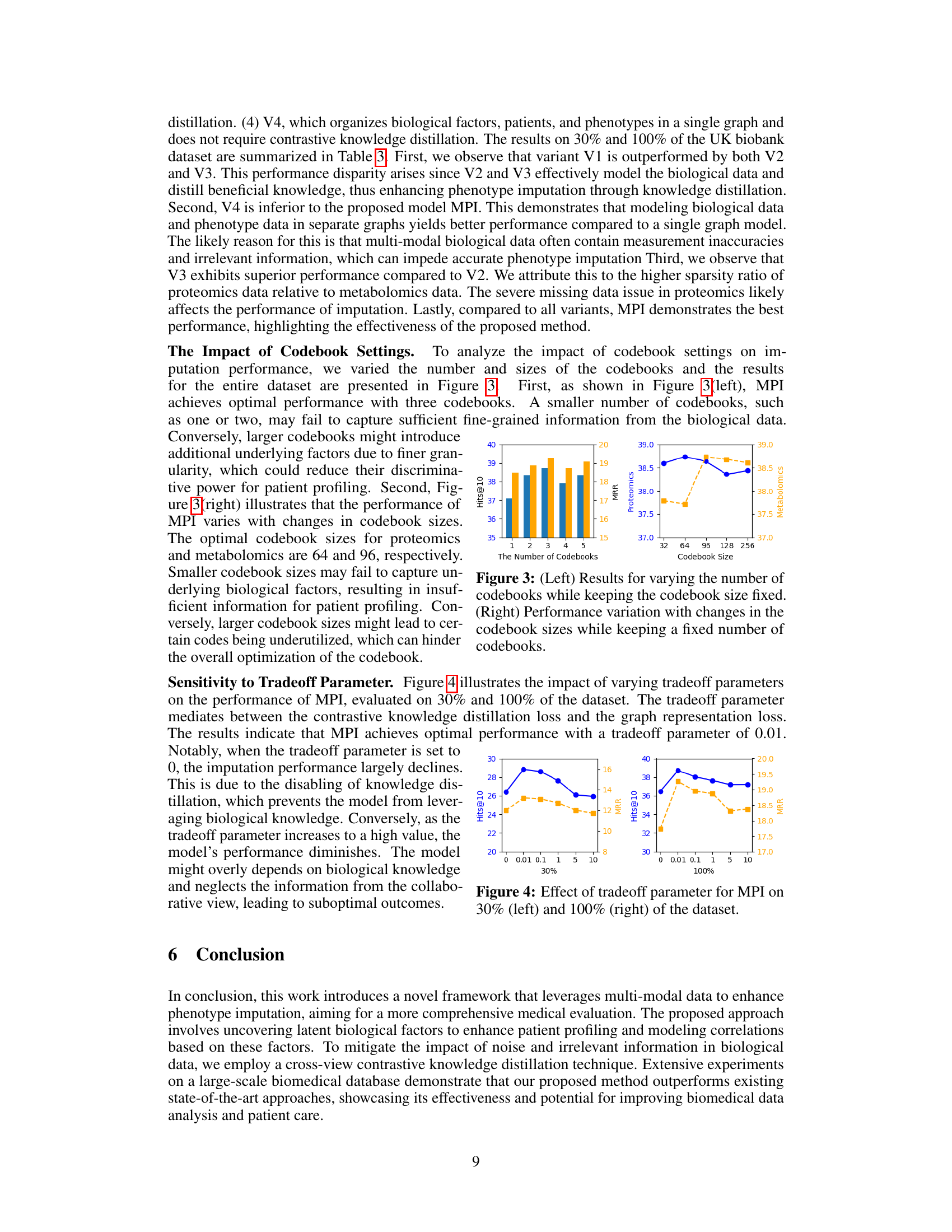
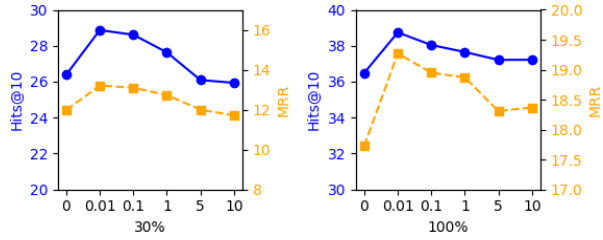
🔼 This table presents the performance of the proposed MPI model and several baseline models on different proportions of the UK Biobank dataset (30%, 50%, 70%, 90%, and 100%). The metrics used to evaluate performance are Hit@10, Hit@20, Hit@50, and MRR (Mean Reciprocal Rank). The table allows for a comparison of the models’ ability to handle varying amounts of missing data and highlights the relative strengths and weaknesses of each model in different data scarcity scenarios.
read the caption
Table 2: Performance comparison for different models on varying dataset proportions.
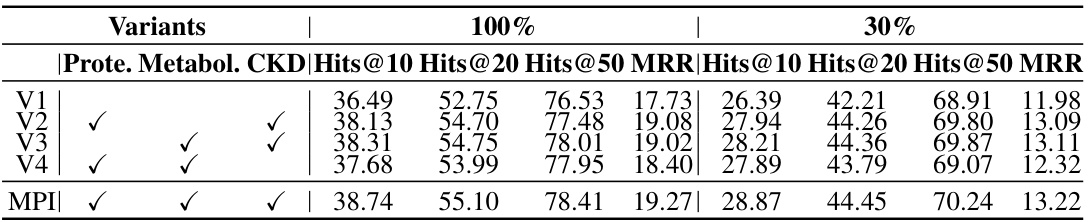
🔼 This table presents the results of an ablation study comparing the proposed method (MPI) with several variants. The variants systematically remove components of the MPI framework to evaluate their individual contributions. Results are presented for both 30% and 100% of the dataset, showing the impact of each component on performance metrics including Hits@10, Hits@20, Hits@50, and MRR.
read the caption
Table 3: Ablation study of variants comparison on 30% and 100% of the dataset.
Full paper#