↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Generating accurate 3D models of molecules is crucial for drug discovery. Current methods struggle with the multiscale nature of molecules, often failing to capture both coarse-grained structural features and fine-grained atomic details. Existing diffusion models are also not ideally suited to this task because random noise indiscriminately affects both scales. This paper addresses these challenges.
This work introduces Equivariant Blurring Diffusion (EBD), a novel method that uses a two-stage process. First, it generates a coarse-grained representation of the molecule using readily available tools. Second, a diffusion model is used to refine the atomic details, guided by the coarse-grained structure and ensuring that the final model is accurate and consistent. EBD significantly outperforms existing methods in terms of accuracy and diversity of generated conformers. The method is also computationally efficient, requiring far fewer steps to produce high-quality results.
Key Takeaways#
Why does it matter?#
This paper is highly important for researchers in cheminformatics and drug discovery due to its novel approach to generating 3D molecular conformers. It introduces a hierarchical, coarse-to-fine method that leverages the power of diffusion models and equivariant networks, achieving superior performance compared to existing methods. This opens new avenues for generating more accurate and diverse conformers which is crucial for drug design and development. The release of the code further enhances its impact.
Visual Insights#

This figure illustrates the concept of blurring diffusion generative processes in two different contexts: image generation and molecular conformer generation. For images, it shows how a blurry prior (average pixel intensities) is refined to a detailed image through a diffusion process. For molecules, it illustrates how a coarse-grained fragment-level 3D structure is refined into a fine-grained atomic-level structure.
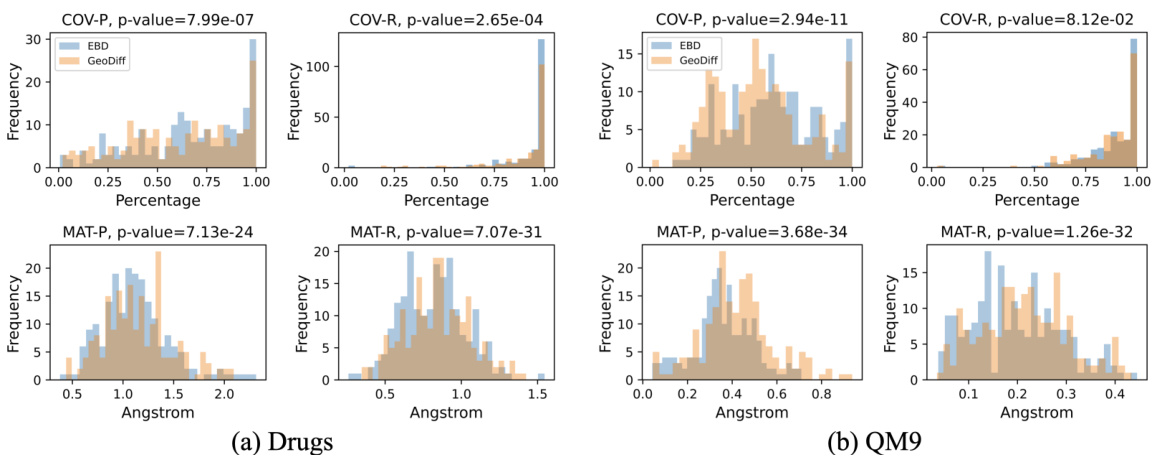
This table presents the quantitative comparison of the proposed EBD model to other state-of-the-art methods on the drug-like molecules dataset. The evaluation metrics include COV-R (coverage recall), MAT-R (matching recall), COV-P (coverage precision), and MAT-P (matching precision). Lower values for MAT-R and MAT-P, and higher values for COV-R and COV-P indicate better performance. The threshold δ for RMSD is set to 1.25Å for Drugs dataset.
In-depth insights#
Equivariant Diffusion#
Equivariant diffusion models represent a significant advancement in generative modeling, particularly for data with inherent symmetries like molecules or images. The core idea is to incorporate group equivariance directly into the diffusion process itself. This means the model’s output transforms in a predictable way under the action of a symmetry group (e.g., rotations for images, roto-translations for molecules). This property is crucial because it leads to more physically meaningful and realistic generations, avoiding artifacts that violate the underlying symmetries. Equivariant neural networks are essential building blocks, ensuring the model’s predictions respect these symmetries at each step of the diffusion process. A key challenge lies in designing the forward and reverse diffusion processes while preserving equivariance. This often involves carefully crafted data augmentation strategies and network architectures. The benefits extend to applications beyond molecular conformer generation including, image generation, 3D shape modeling, and various other domains where symmetry plays a significant role. However, creating truly efficient and effective equivariant diffusion models often requires dealing with the inherent complexity of both the diffusion process and the representation of group actions on high-dimensional data, posing a computational challenge.
Hierarchical Generation#
Hierarchical generative models offer a powerful approach to tackling complex data by breaking down the generation process into a hierarchy of scales. Starting with coarse-grained structures and progressively refining details, this method enhances both the efficiency and interpretability of the generative process. In the context of molecular conformer generation, hierarchical approaches leverage the multiscale nature of molecules. First, coarse-grained structural elements are generated, such as fragments, and then fine-grained atomic details are added while preserving the larger structural context. This strategy addresses the challenge of simultaneously managing both global and local properties of complex molecules. Equivariance, or the invariance of the generated structures under transformations like rotations and translations, becomes crucial in achieving chemically plausible conformers, ensuring that the generated structures are not artifacts of the coordinate systems used in the representation. While still a developing field, hierarchical generation promises significant advances in achieving both accurate and efficient molecular conformer generation.
Blurring Corruption#
The concept of “blurring corruption” in generative models, particularly within the context of diffusion models, presents a compelling alternative to traditional noise-based corruption methods. Instead of adding random noise, blurring strategically corrupts the data by progressively shifting it towards a lower resolution or coarser representation. This approach has several key advantages. First, it provides a more natural and interpretable corruption process, mirroring how we perceive the world at different scales. Second, blurring preserves crucial structural information at a coarser level, which can be leveraged during the reconstruction phase. This allows the model to focus on restoring fine details while retaining the overall structure, leading to improved generation quality. Third, the blurring process can be controlled and tailored, potentially offering more flexibility and granular control over the generative process compared to noise-based corruption. However, careful consideration needs to be given to the design of the blurring function and the blurring schedule, as poorly chosen settings could lead to artifacts or an inability to capture fine details. Future research should explore the effectiveness of different types of blurring functions, explore applications beyond images (e.g., 3D molecular structures or graphs), and investigate the interplay between blurring and other forms of data augmentation.
Ablation Studies#
Ablation studies systematically investigate the contribution of individual components within a machine learning model. In the context of this research paper, ablation studies likely probed the impact of different design choices on the performance of the proposed Equivariant Blurring Diffusion (EBD) model for molecular conformer generation. Key aspects potentially explored include the effect of varying fragment granularity (the size and type of molecular fragments used as building blocks), different data corruption methods (how the model introduces noise during training), and the design of the loss function (how model errors are measured). The results of these ablation studies would have provided critical insights into the effectiveness of each component, guiding design choices and ultimately demonstrating the robustness of the EBD model compared to existing alternatives. By systematically removing or modifying elements of the EBD model, the researchers could isolate the individual contributions of each part, leading to a more refined understanding of its underlying mechanisms. The findings may show, for example, whether a particular type of data corruption yields superior results, or highlight the importance of a specific loss function for achieving accurate conformer generation. Such insights are critical for advancing the model’s development and improving its generalization ability. Furthermore, the ablation study may have provided valuable information to future researchers on how to adapt and improve similar models. By examining how different components affect the model’s performance, future studies can learn from these experiments and build upon the existing research.
Geometric Evaluation#
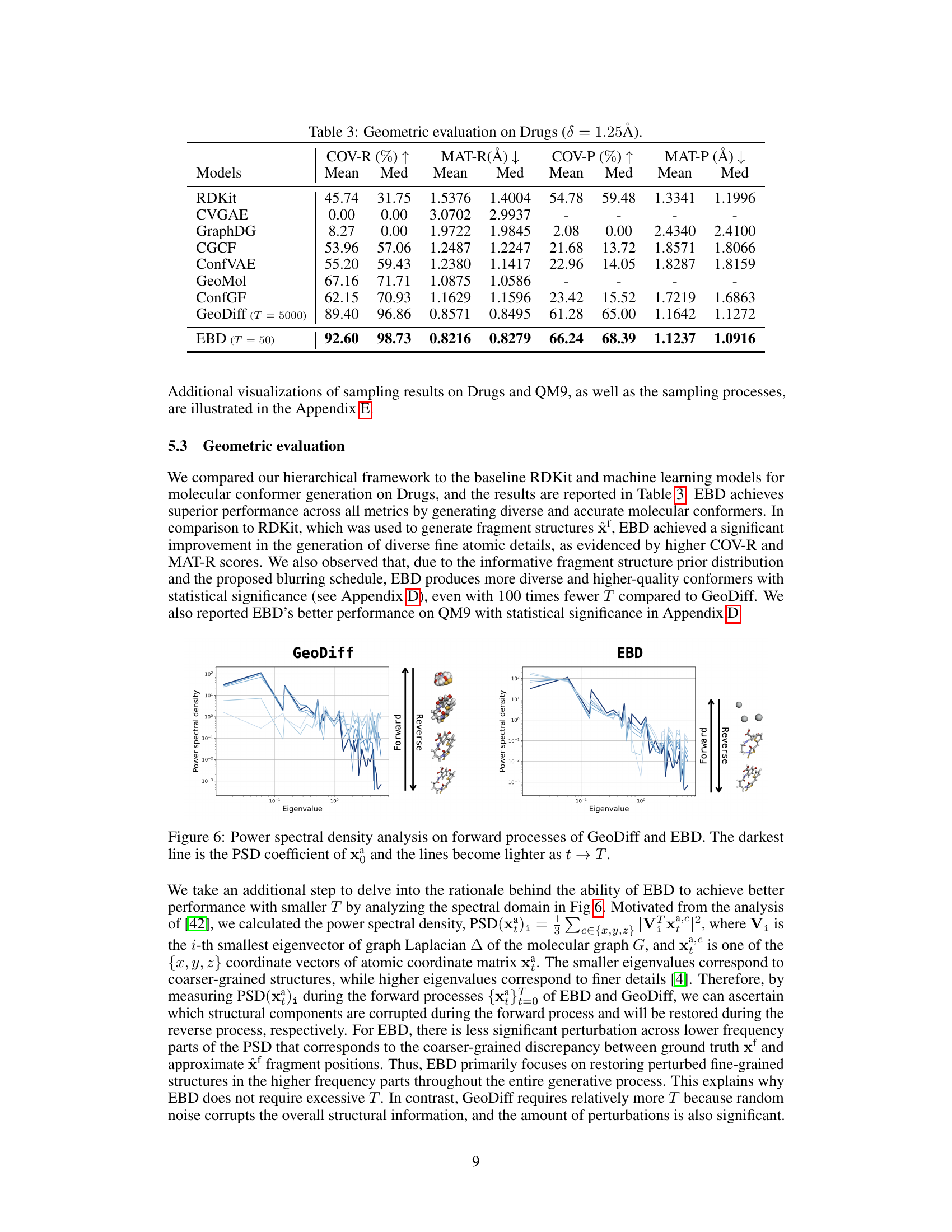
A geometric evaluation section in a research paper would systematically assess the accuracy and diversity of generated 3D molecular conformers. It would compare generated conformers against ground truth conformers using metrics such as RMSD (Root Mean Square Deviation) to quantify the geometric similarity. Coverage and matching metrics would further assess how well the generated conformers represent the structural diversity of the ground truth set. The evaluation would typically involve comparison against state-of-the-art methods to establish the proposed method’s effectiveness and potentially investigate the influence of hyperparameters on the quality of conformer generation. Statistical significance tests, like Wilcoxon signed-rank tests, would be vital in validating that the observed improvements were not merely due to chance. Visualization of both the generated conformers and the ground truth conformers is also essential to offer intuitive understanding and assessment of the geometric similarity and diversity. Analysis of the distribution of RMSD values can provide insights into the model’s tendency to produce conformers clustered around certain structures or spread across the conformational space. A comprehensive geometric evaluation is critical for validating the effectiveness of any molecular conformation generation method.
More visual insights#
More on figures
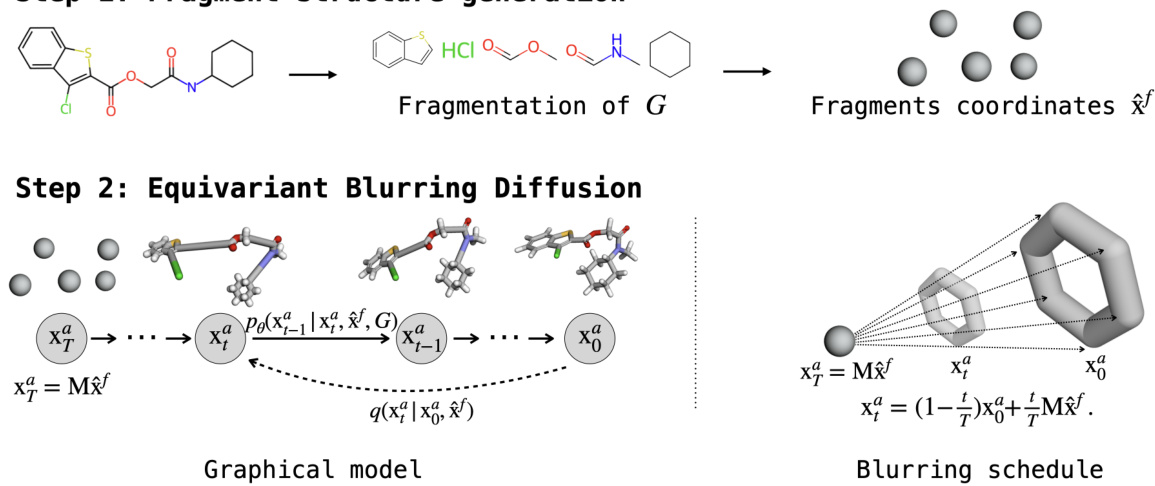
This figure illustrates the two-stage hierarchical model for molecular conformer generation. The first stage involves fragmenting the input molecular graph and generating coarse-grained 3D fragment coordinates. The second stage uses Equivariant Blurring Diffusion (EBD) to generate the fine-grained atomic details, conditioned on the fragment coordinates and the original molecular graph. The blurring schedule guides the diffusion process, moving from a coarse representation towards a refined atomic structure.

This figure presents the ablation study results for three key design choices of the proposed Equivariant Blurring Diffusion (EBD) model. It shows how model performance (measured by MAT-R and MAT-P metrics) changes based on: (a) the granularity of the fragment vocabulary used (coarser vs. finer fragments), (b) what state is used as the target for the state estimator (previous state vs. ground truth state), and (c) the type of data corruption process used (EBD’s blurring corruption vs. DecompDiff’s corruption). The results highlight the impact of each design decision on the model’s ability to generate accurate and diverse molecular conformers.
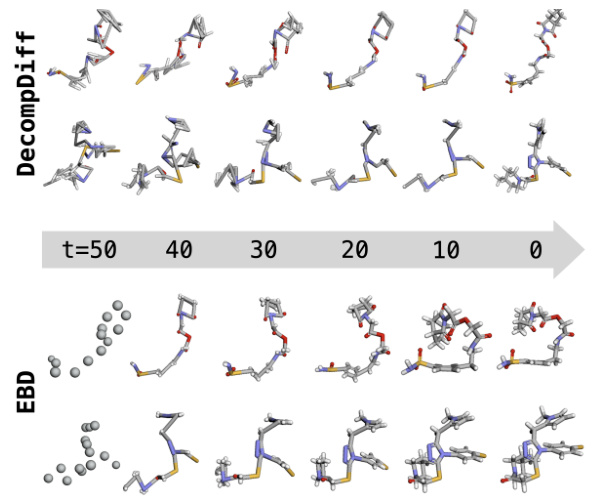
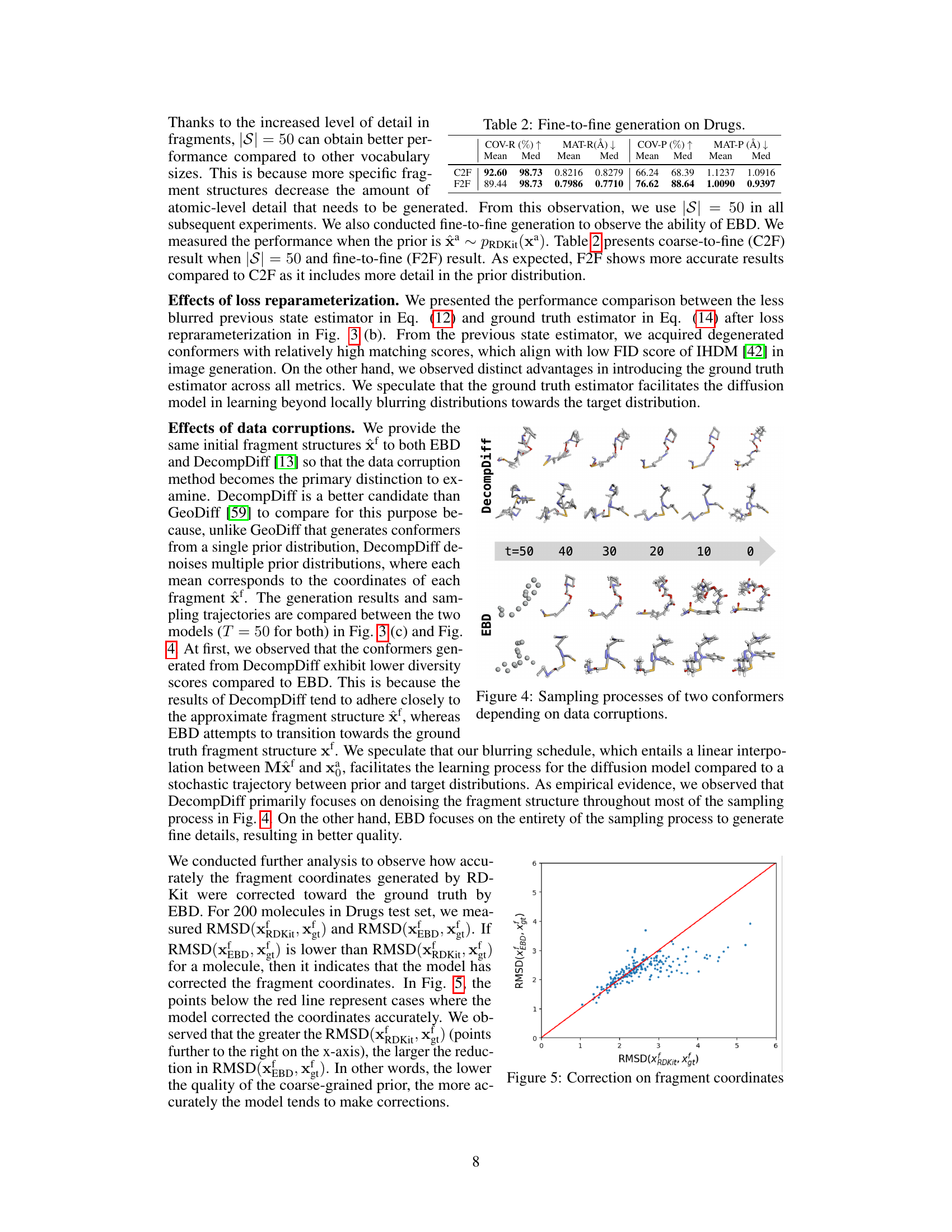
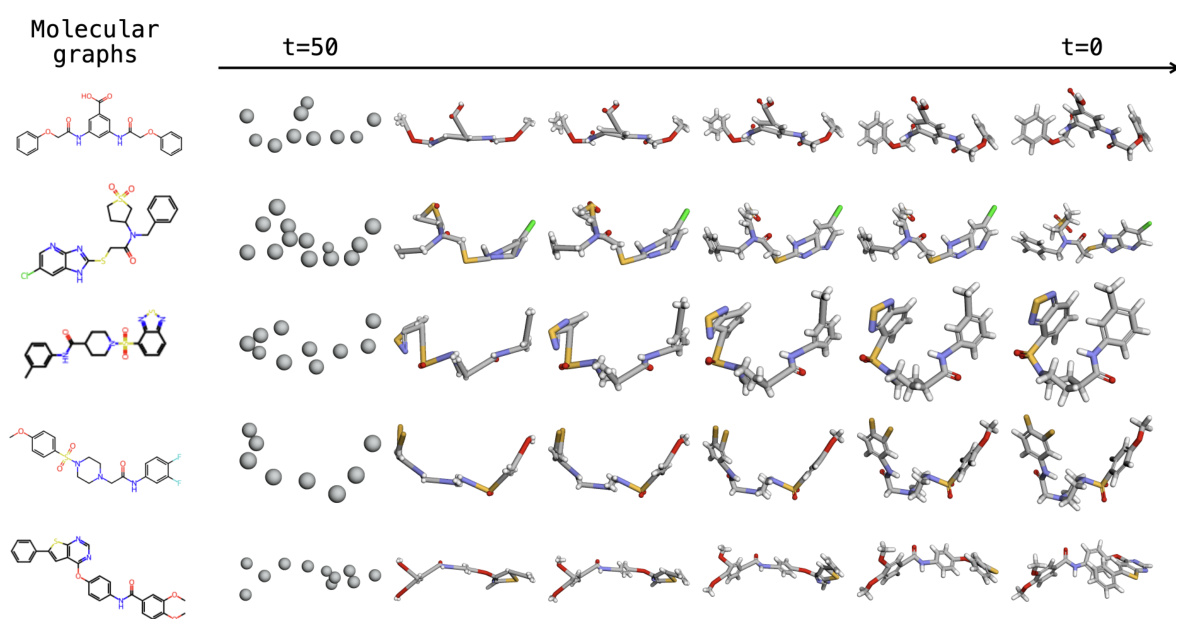
This figure compares the sampling processes of two conformers generated by DecompDiff and EBD. It visually demonstrates how the data corruption methods (blurring vs. decomposition) affect the generation process. The top row shows DecompDiff’s sampling process, where the conformer starts from a relatively complete structure and gradually refines toward the final structure. The bottom row shows EBD’s sampling process, where the conformer starts from a very coarse-grained representation and gradually incorporates fine details, highlighting its coarse-to-fine approach. The figure shows different stages in the sampling process, as indicated by the time (t) values ranging from 50 to 0.
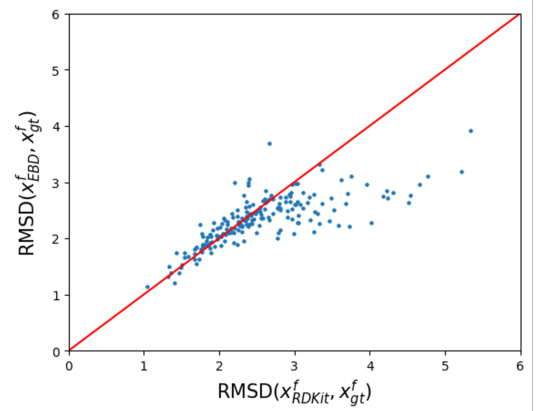
This figure shows a scatter plot that compares the root mean square deviation (RMSD) between the fragment coordinates generated by RDKit and the ground truth conformers (x-axis) against the RMSD between the fragment coordinates generated by EBD and the ground truth conformers (y-axis). Each point represents a molecule. The red line represents the ideal scenario where the RMSD values are equal. The plot shows that as the RDKit generated coordinates deviate more from the ground truth, EBD corrects them more accurately. This indicates the effectiveness of EBD in refining the initial fragment coordinates to produce more accurate molecular conformers.

This figure shows the power spectral density (PSD) analysis of the forward diffusion processes in GeoDiff and EBD. The PSD is computed for different time steps (t), with darker lines representing earlier time steps and lighter lines representing later steps. The PSD measures the distribution of power across different frequencies (eigenvalues). The figure aims to illustrate how the forward diffusion process changes the distribution of information across frequency components. The visualizations alongside the plots show example molecules during the forward and reverse processes.
This figure illustrates the two-stage hierarchical approach for generating molecular conformers. The first stage involves fragmenting the input molecular graph and generating coarse-grained fragment coordinates. The second stage utilizes Equivariant Blurring Diffusion to refine these coordinates and generate the atom-level details of the conformer, guided by a blurring schedule that progressively refines the structure.
This figure illustrates the two-step hierarchical model for generating molecular conformers. First, the input molecular graph G is fragmented, and the coordinates of these fragments (xf) are generated. Second, the Equivariant Blurring Diffusion (EBD) model takes these fragment coordinates and the original molecular graph as input to generate the final, detailed atomic coordinates (xa). The process is guided by a blurring schedule that gradually refines the atomic details from the coarse-grained fragment structure.
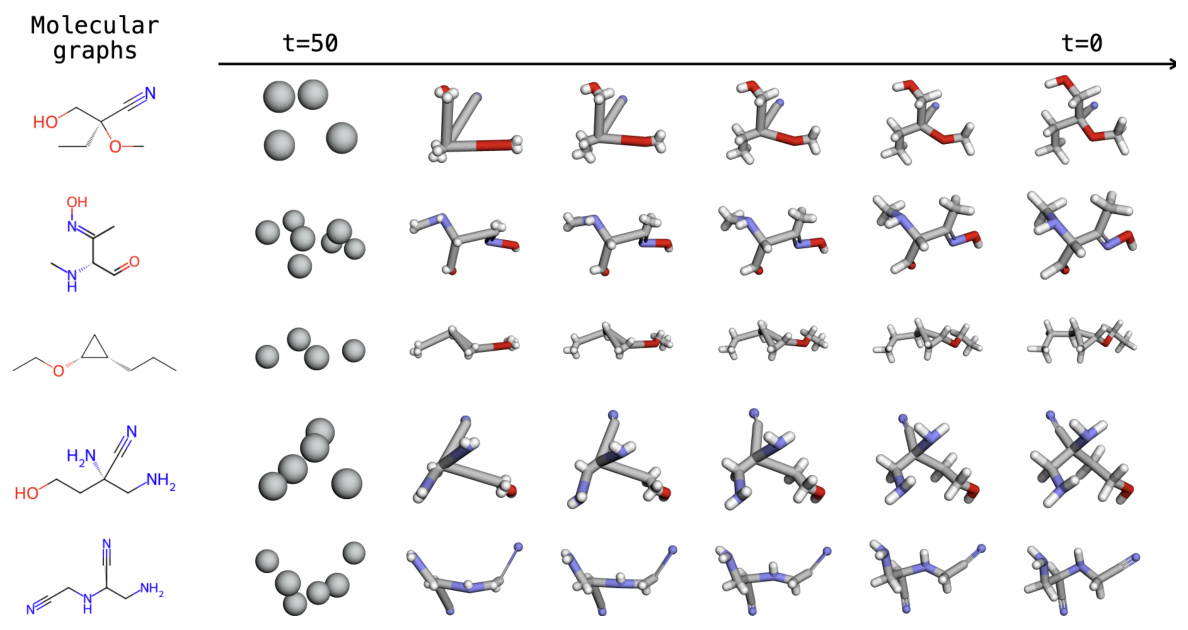
This figure shows five examples of molecular graphs from the Drugs dataset and their corresponding sampling processes using the EBD model. Each row represents a different molecule, and each column shows the intermediate states generated during the diffusion process at different timesteps. The leftmost column shows the initial state of the sampling process, where only the fragment-level structure is available. As the timesteps increases (moving from left to right), the model progressively refines the atomic details of the conformers, starting with a blurred representation of the atoms and gradually sharpening the details until the final conformer is obtained (rightmost column). The figure demonstrates that EBD can accurately generate the conformers starting from coarse-grained information by progressively adding details at finer scales.
This figure illustrates the two-step hierarchical process for generating molecular conformers. The first step involves fragmenting the input molecular graph and generating coarse-grained fragment coordinates. The second step utilizes Equivariant Blurring Diffusion to refine these coordinates by generating fine-grained atomic details, guided by a blurring schedule that gradually refines the structure from coarse to fine.

This figure shows a comparison of molecular graphs, their corresponding ground truth conformers, and conformers generated by the Equivariant Blurring Diffusion (EBD) model. It visually demonstrates the model’s ability to generate diverse and plausible conformers based on the input molecular graph. The comparison highlights the model’s capacity to capture the structural features from the molecular graph and translate them into realistic 3D conformations.

This figure visualizes the results of the Equivariant Blurring Diffusion (EBD) model on a subset of the Drugs dataset. It shows the molecular graphs, ground truth conformers (from the dataset), and samples generated by EBD. Each row represents a different molecule, demonstrating the model’s ability to generate diverse and plausible conformers based on the given molecular graph. The visualization highlights the model’s capacity to capture both the coarse-grained structural information from the input graph and generate the fine-grained atomic details of the conformers.
More on tables

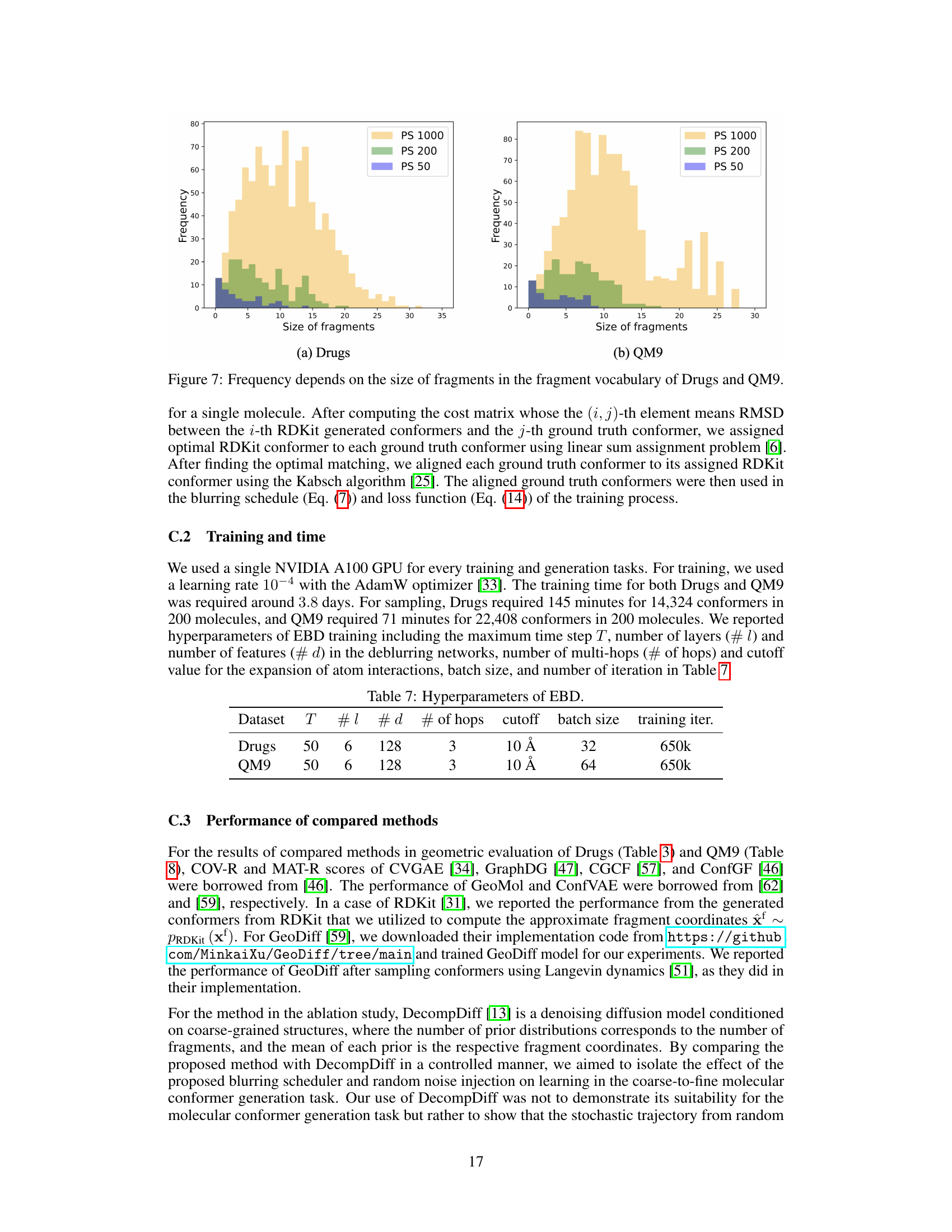
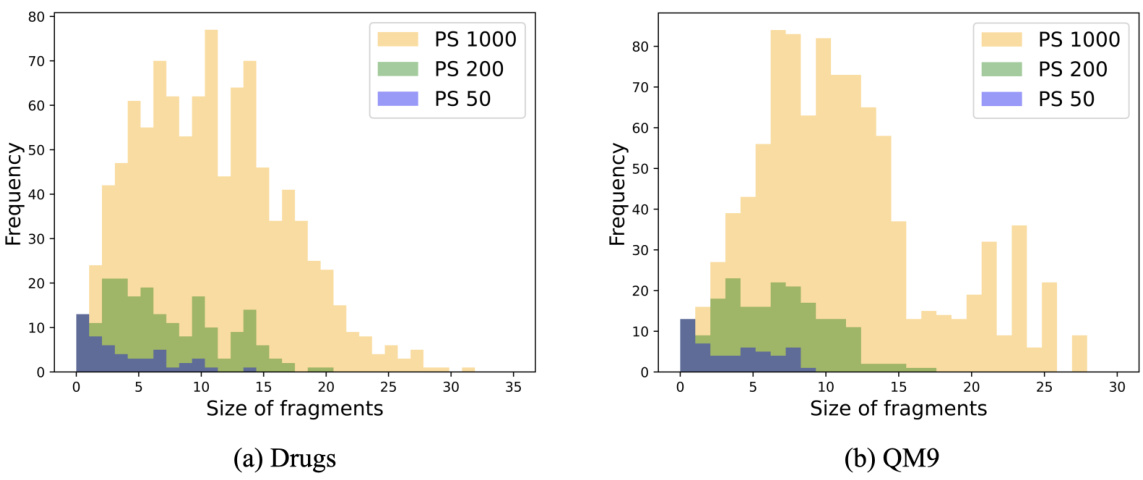
This table shows the statistics of fragment vocabulary size, average number of fragments per graph, and average number of atoms per fragment for three different fragment vocabulary sizes (50, 200, and 1000) in the Drugs dataset. The size of the vocabulary affects the level of detail and granularity of fragment structures during the conformer generation process.
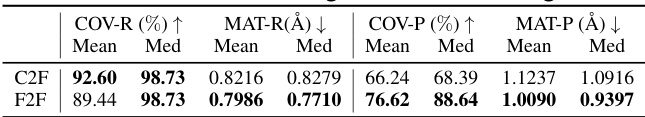
This table compares the performance of coarse-to-fine (C2F) and fine-to-fine (F2F) generation methods on the Drugs dataset. The metrics used are COV-R (coverage recall), MAT-R (matching recall), COV-P (coverage precision), and MAT-P (matching precision). Lower MAT-R and MAT-P values indicate better performance, while higher COV-R and COV-P values are better. The results show that the fine-to-fine approach, which uses a more detailed prior distribution, achieves better accuracy (lower MAT-R and MAT-P) compared to the coarse-to-fine approach.
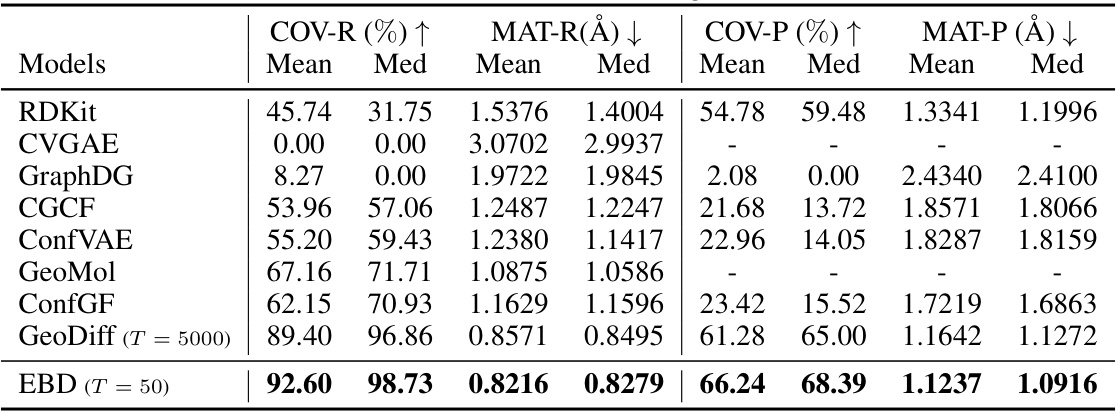
This table presents a comparison of different molecular conformer generation models on the Drugs dataset. The metrics used are COV-R (coverage recall), MAT-R (matching recall), COV-P (coverage precision), and MAT-P (matching precision), all calculated using root-mean-square deviation (RMSD) with a threshold of 1.25Å. Lower values of MAT-R and MAT-P indicate higher accuracy, while higher values of COV-R and COV-P indicate higher diversity and coverage, respectively. The table shows that EBD outperforms other methods across all metrics.
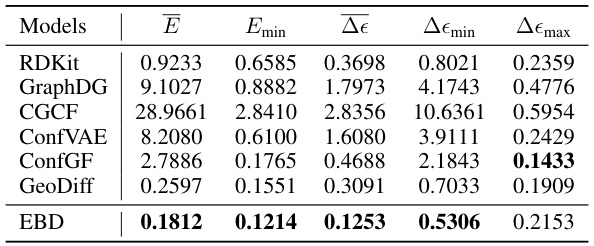
This table presents the mean absolute errors between the generated and ground truth ensemble properties for various models, including RDKit, GraphDG, CGCF, ConfVAE, ConfGF, GeoDiff, and the proposed EBD model. The properties considered are energy (E), minimum energy (Emin), average HOMO-LUMO gap (Δe), minimum HOMO-LUMO gap (ΔEmin), and maximum HOMO-LUMO gap (Δemax). Lower values indicate better performance in generating molecules with more accurate chemical properties.
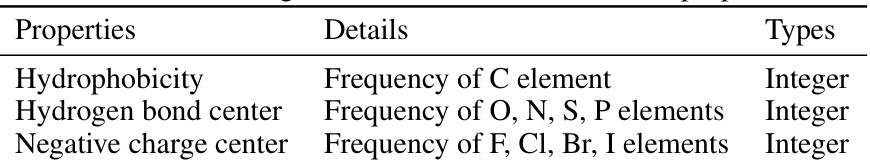
This table shows the features used to represent fragments in the model. Each fragment is characterized by three features: hydrophobicity, hydrogen bond center, and negative charge center. The hydrophobicity feature is represented by the frequency of carbon (C) atoms; the hydrogen bond center by the frequency of oxygen (O), nitrogen (N), sulfur (S), and phosphorus (P) atoms; and the negative charge center by the frequency of fluorine (F), chlorine (Cl), bromine (Br), and iodine (I) atoms. Each of these features is an integer value.

This table shows the average number of fragments per graph and the average number of atoms per fragment for different fragment vocabulary sizes (|S|) in the Drugs and QM9 datasets. The results are based on the Principal Subgraph (PS) fragmentation method used in the paper. A smaller vocabulary size leads to a larger average number of fragments per graph and smaller average number of atoms per fragment.

This table shows the hyperparameters used in the training of the Equivariant Blurring Diffusion (EBD) model for the Drugs and QM9 datasets. The hyperparameters include the maximum number of diffusion steps (T), the number of layers (#l) and number of features (#d) in the deblurring networks, the number of hops considered in the message passing, the cutoff distance used in defining neighborhood relations, the batch size used during training, and the total number of training iterations.
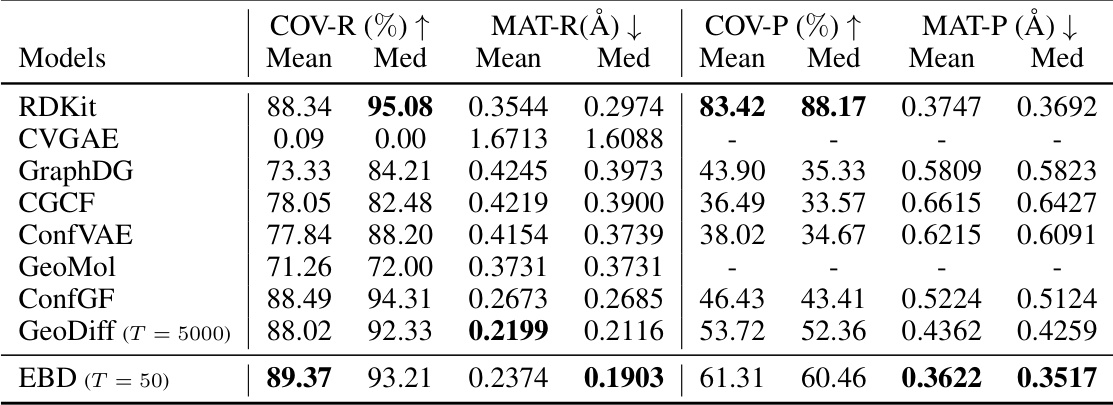
This table presents the results of a geometric evaluation comparing the performance of several models on the Drugs dataset. The metrics used are COV-R (coverage recall), MAT-R (matching recall), COV-P (coverage precision), and MAT-P (matching precision). Lower values for MAT-R and MAT-P indicate better accuracy, while higher values for COV-R and COV-P suggest better diversity. The threshold (δ) used for RMSD comparison is 1.25Å. The table allows for a direct comparison of the accuracy and diversity of conformer generation achieved by different models.
Full paper#