TL;DR#
Traditional methods for kidney tumor segmentation in multi-sequence MRI require extensive annotations for each sequence, a time-consuming and labor-intensive process. Unsupervised domain adaptation (UDA) offers a solution by aligning cross-modal features but most UDA methods are limited to one-to-one adaptation, proving inefficient for multi-sequence data. This creates a significant bottleneck in clinical applications.
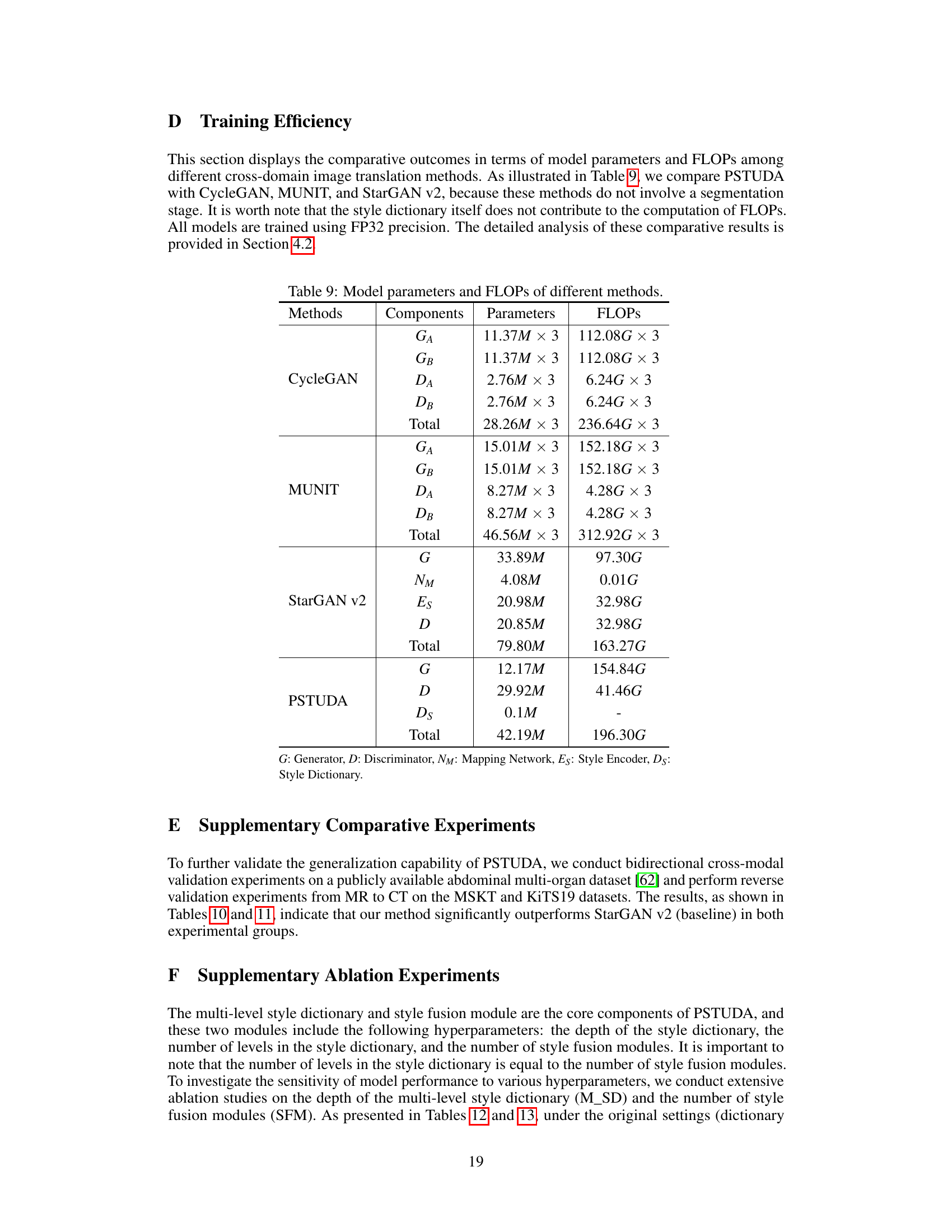
The paper introduces PSTUDA, a novel one-to-multiple progressive style transfer UDA framework. PSTUDA uses a multi-level style dictionary to manage style information across multiple target domains and employs multiple cascading style fusion modules with point-wise instance normalization. This significantly improves multi-sequence kidney tumor segmentation accuracy (Dice Similarity Coefficients increased by at least 1.8% and 3.9%) and efficiency (reduces computation by 72% and model parameters by 50%). PSTUDA demonstrates superior performance and efficiency compared to existing methods.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image analysis and unsupervised domain adaptation. It addresses the critical need for efficient multi-target domain adaptation, particularly relevant in medical imaging where multiple image sequences are common. The proposed method, PSTUDA, offers significant improvements in efficiency and accuracy, opening new avenues for research in multi-modal medical image analysis and advancing clinical applications.
Visual Insights#
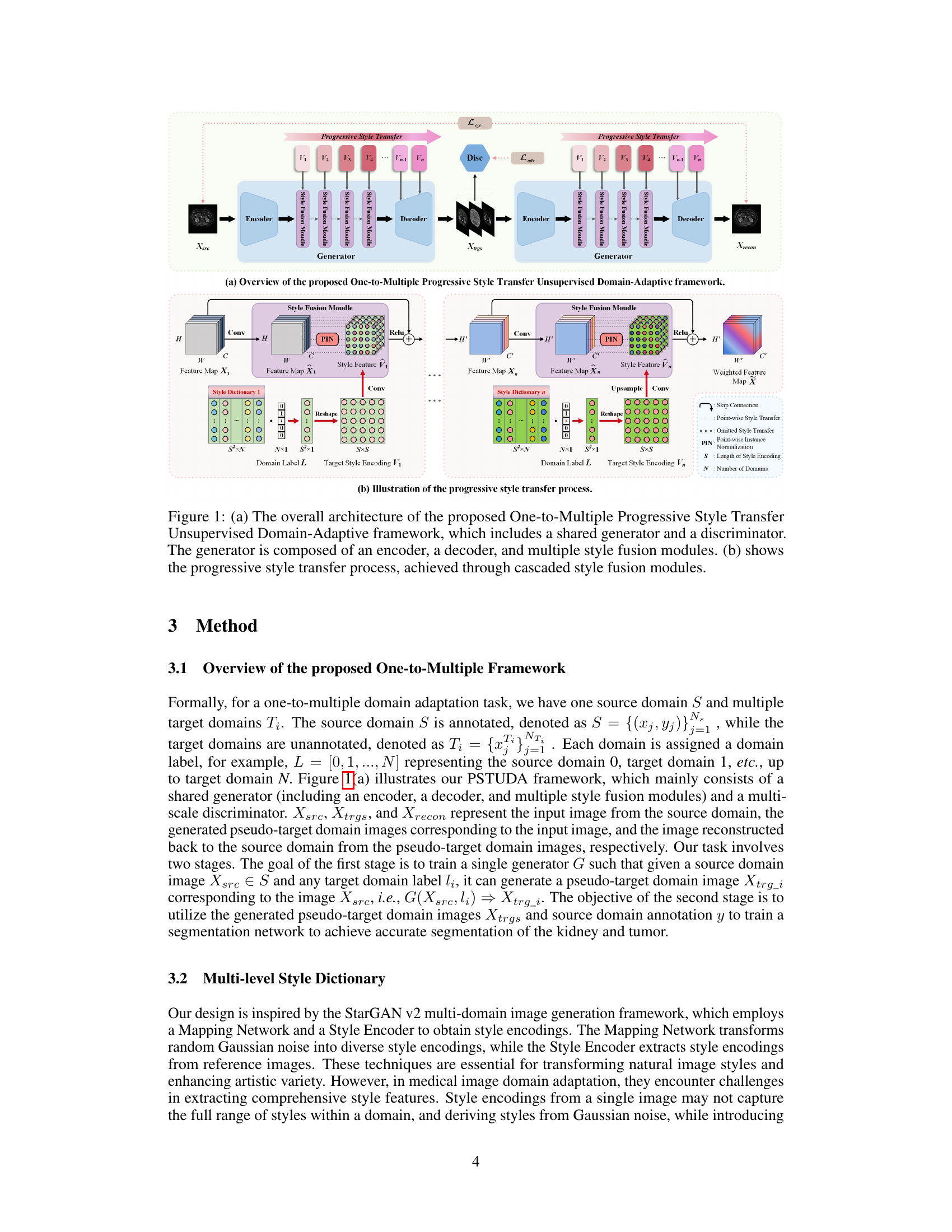
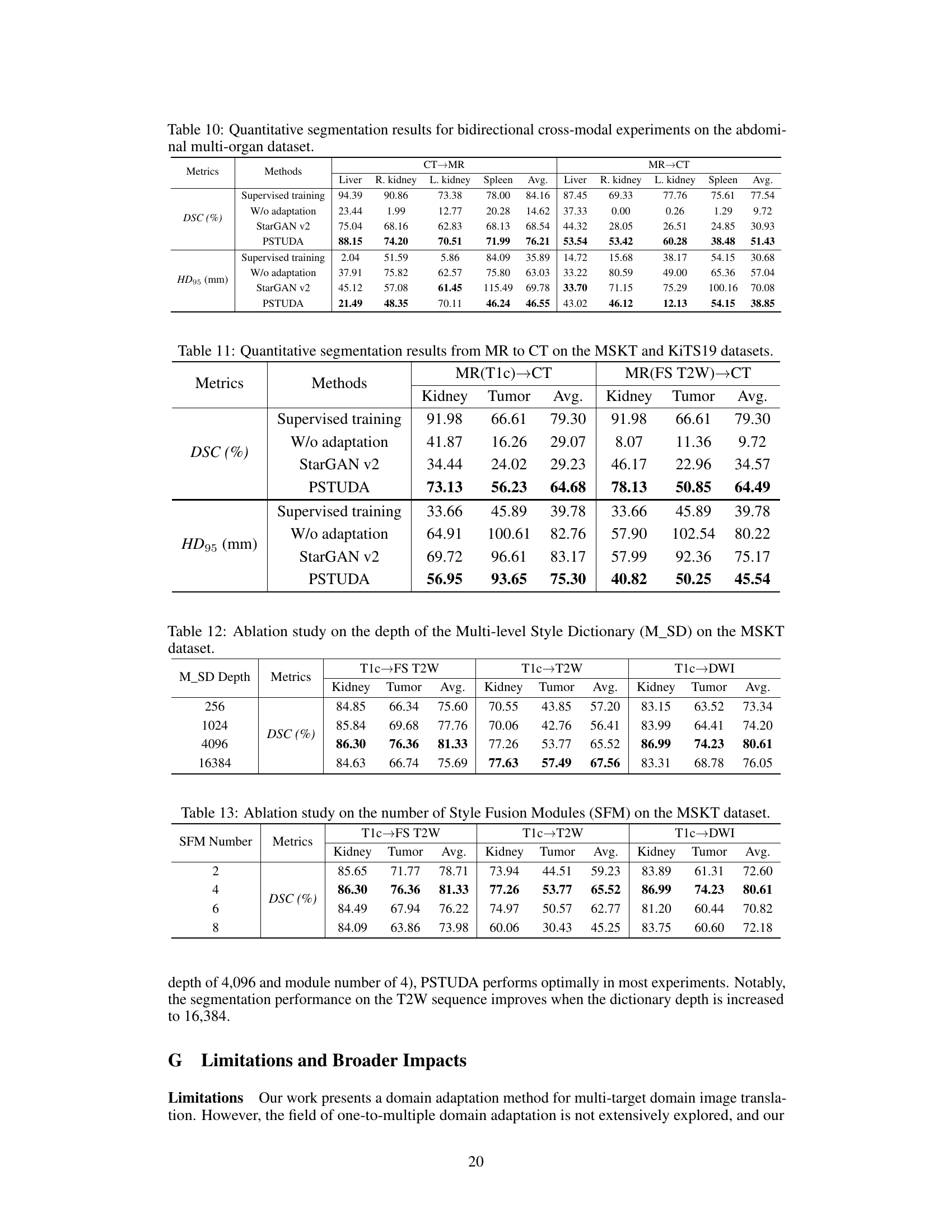
🔼 This figure shows the architecture of the proposed One-to-Multiple Progressive Style Transfer Unsupervised Domain-Adaptive (PSTUDA) framework. Panel (a) provides a high-level overview of the framework, illustrating the shared generator and discriminator, as well as the encoder, decoder, and multiple style fusion modules within the generator. Panel (b) delves into the detail of the progressive style transfer process, showing how content and style features are combined at multiple levels using cascaded style fusion modules, point-wise instance normalization, and style dictionaries.
read the caption
Figure 1: (a) The overall architecture of the proposed One-to-Multiple Progressive Style Transfer Unsupervised Domain-Adaptive framework, which includes a shared generator and a discriminator. The generator is composed of an encoder, a decoder, and multiple style fusion modules. (b) shows the progressive style transfer process, achieved through cascaded style fusion modules.
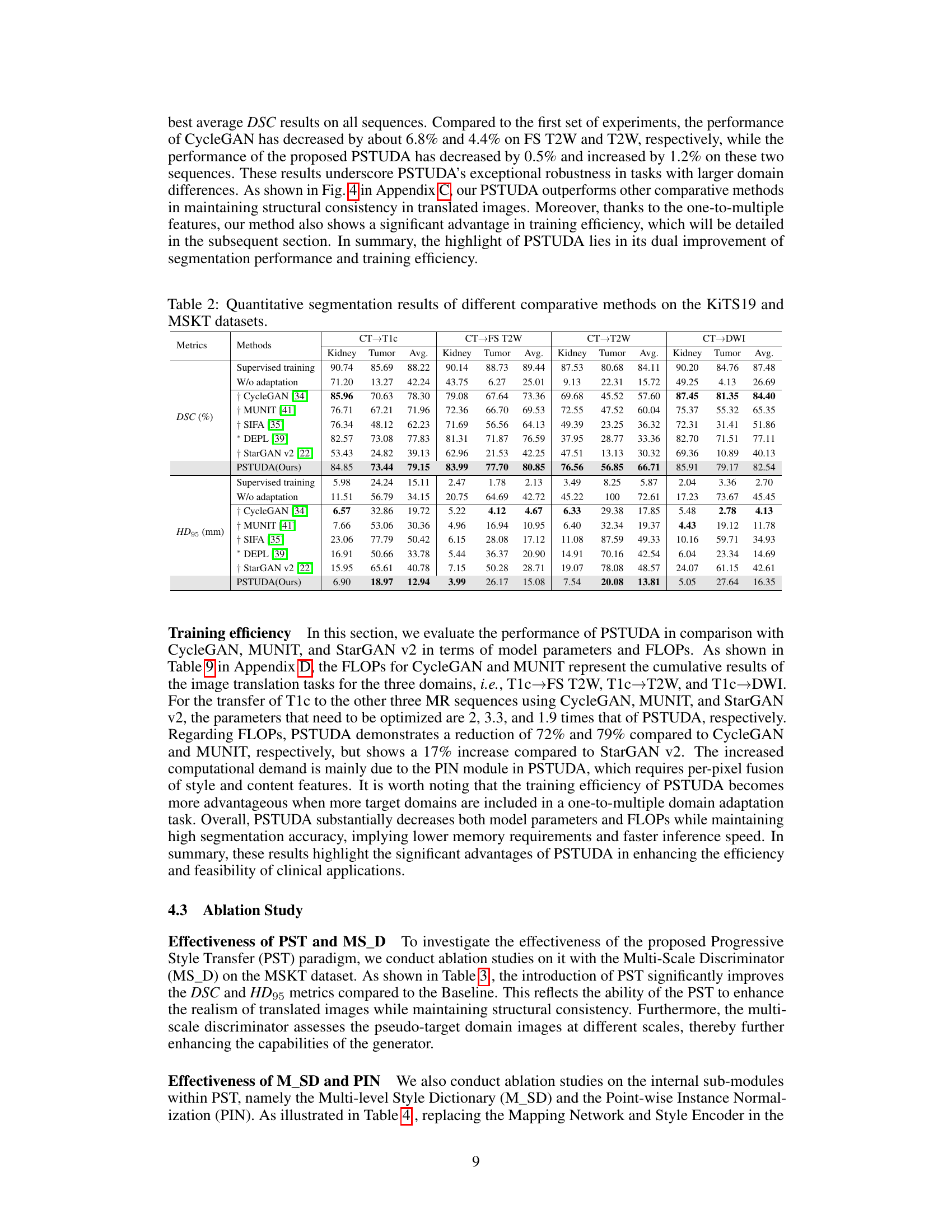
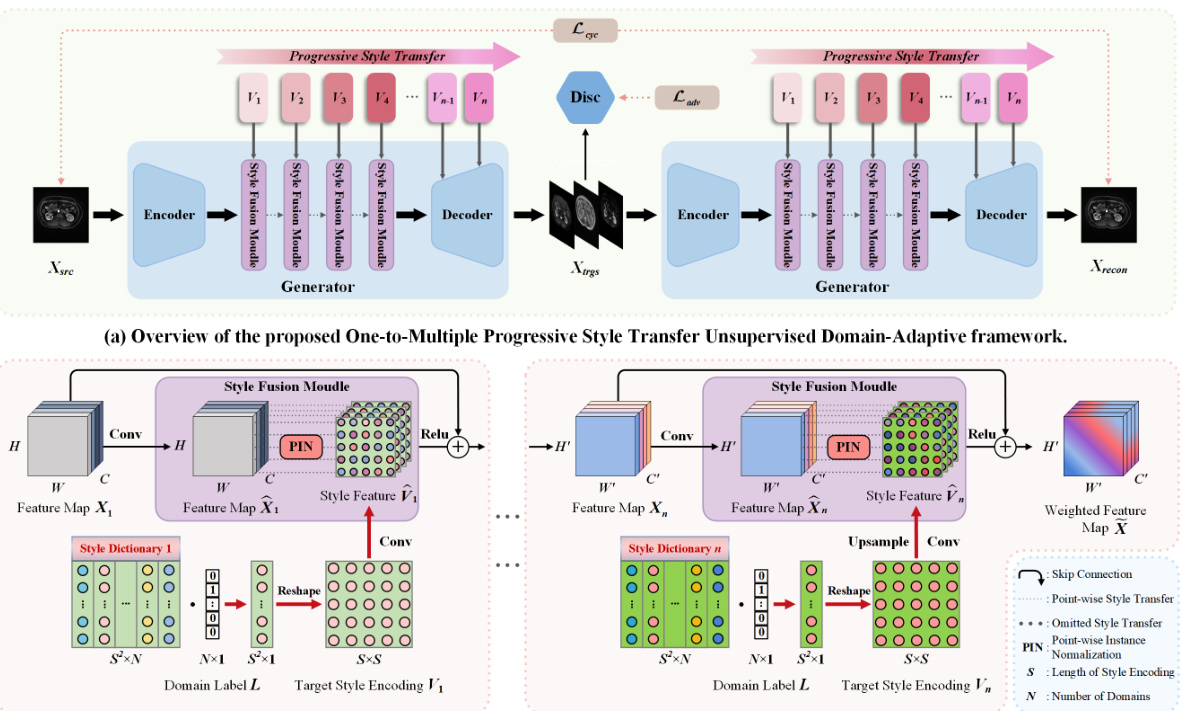
🔼 This table presents a quantitative comparison of different methods’ performance on the MSKT dataset for kidney and tumor segmentation across various MRI sequences (T1c, FS T2W, T2W, and DWI). Metrics include Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95). The table allows for a direct comparison of the proposed PSTUDA method against several state-of-the-art unsupervised domain adaptation techniques. Results are presented separately for kidney and tumor segmentation, providing a detailed view of each method’s strengths and weaknesses for each MRI sequence. The ‘Supervised training’ and ‘W/o adaptation’ rows provide baseline performance benchmarks.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
In-depth insights#
Progressive Style Transfer#
Progressive Style Transfer, as a concept, involves the gradual refinement of style features throughout an image translation process. Instead of a single, abrupt transformation, this method applies style changes incrementally across multiple stages. This approach is particularly useful when dealing with complex domains or tasks where a direct mapping might lead to artifacts or inconsistencies. By applying style adjustments progressively, this method aims to preserve structural integrity while achieving more natural and detailed stylistic changes. The iterative nature allows for effective decoupling of content and style, facilitating more precise control over the overall transformation. The advantage lies in the potential for improved quality of generated images, with greater fidelity to both the original content and the desired style. This method is particularly promising for multi-domain adaptation tasks, where the gradual style adjustment can handle the complexities of transferring across multiple target domains more effectively than one-to-one mapping.
Multi-Domain Adaptation#
Multi-domain adaptation tackles the challenge of training models that generalize across multiple, distinct domains. Unlike single-source adaptation, it aims for simultaneous transfer to several target domains, thus increasing efficiency. This is particularly crucial in scenarios like medical image analysis where data annotation is expensive and time-consuming. The core challenge lies in learning domain-invariant features while effectively capturing the unique characteristics of each target domain. Effective strategies often involve disentangling content and style representations, using techniques like style transfer or adversarial training to bridge domain gaps. Multi-level style dictionaries and progressive style fusion modules are innovative approaches designed to manage the complexity of simultaneous transfer to multiple domains. This approach allows for a more gradual and controlled style integration, potentially leading to better image quality and improved model performance on downstream tasks. The success of multi-domain adaptation hinges on robust normalization methods, ensuring structural consistency across domains and efficient alignment of features. The approach shows promise for handling multi-modal data, paving the way for more efficient and versatile AI-driven applications in various fields.
Multi-Level Style Dict#
A multi-level style dictionary is a novel approach for efficiently managing style information in a progressive style transfer framework. Unlike traditional methods that rely on a single generator to handle multiple target domains, the proposed framework uses a multi-level dictionary to explicitly store style information for each target domain at various stages of the style transfer process. This approach offers several advantages: First, it reduces the computational burden on the generator by decoupling content and style representation. Second, it enables effective style transfer across multiple domains by allowing the generator to focus on a specific level of style detail at each stage. Third, the multi-level structure facilitates progressive style fusion, enabling gradual and more accurate integration of style features with content features. This results in high-quality style transfer while preserving structural consistency. Overall, the multi-level style dictionary is a key innovation enabling the efficiency and effectiveness of the proposed framework.
Kidney Tumor Seg.#
Kidney tumor segmentation is a crucial task in medical image analysis with significant implications for diagnosis, treatment planning, and prognosis. Accurate segmentation is challenging due to the variability in tumor size, shape, location, and the presence of surrounding tissues. Traditional methods often rely on manual delineation, which is time-consuming and subjective. Unsupervised domain adaptation (UDA) techniques offer a promising alternative by leveraging data from multiple sources to improve segmentation accuracy, even with limited labeled data. Multi-sequence magnetic resonance imaging (MRI) presents both opportunities and challenges, as different sequences offer varied contrast but also introduce inter-domain differences. Successful methods must address the issue of aligning cross-modal features and maintaining structural consistency while minimizing the need for extensive manual annotation. This is an active area of research, with ongoing developments focusing on novel network architectures, loss functions, and data augmentation strategies to achieve robust and accurate kidney tumor segmentation.
UDA Framework#
This research paper explores Unsupervised Domain Adaptation (UDA) for kidney tumor segmentation in multi-sequence MRI. A core challenge addressed is the inefficiency of traditional one-to-one UDA methods when dealing with multiple MRI sequences. The proposed framework innovatively employs a one-to-multiple progressive style transfer approach, effectively decoupling content and style information for efficient cross-modal alignment. Multi-level style dictionaries store style information at various stages, reducing the burden on the generator. The method uses cascading style fusion modules with point-wise instance normalization to progressively recombine content and style features, improving structural consistency. Experiments demonstrate improved segmentation accuracy and efficiency compared to existing UDA methods. The framework’s novelty lies in its scalable design for handling multiple target domains simultaneously, resulting in a more efficient and practical solution for clinical applications.
More visual insights#
More on figures
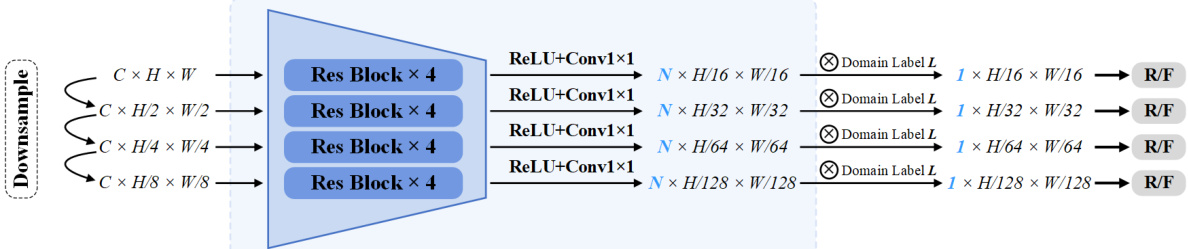
🔼 The figure shows the architecture of a Multi-Scale Discriminator used in the PSTUDA framework. It’s composed of multiple residual blocks. The input image is downsampled through several stages. At each stage, the feature map is processed by four residual blocks and then a 1x1 convolutional layer followed by ReLU activation. The output from each stage is concatenated with a domain label and fed into a final ReLU and convolutional layer. The output of the final layer is then fed into a classification layer that produces the final classification output. The discriminator is designed to be effective in detecting whether an image is real or fake, and it helps improve the quality of generated images by encouraging the generator to produce more realistic images.
read the caption
Figure 2: Architecture of the Multi-Scale Discriminator, composed of multiple residual blocks.
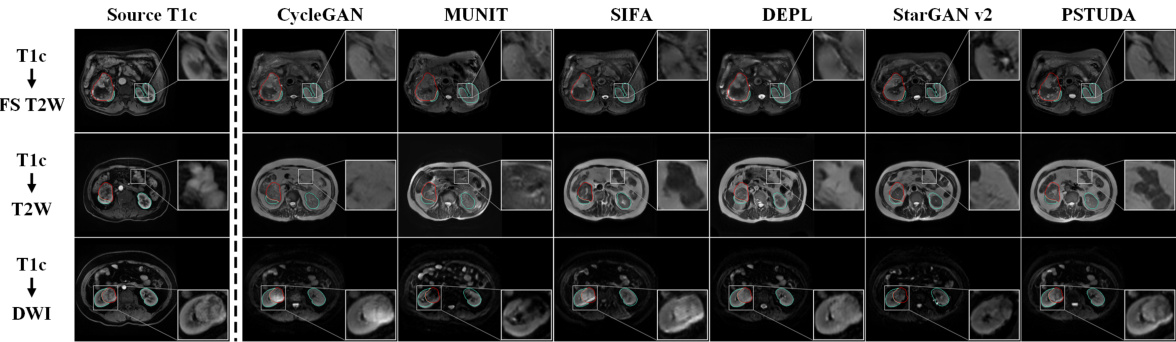
🔼 This figure shows the qualitative results of the image translation task from T1c MRI sequence to FS T2W, T2W, and DWI MRI sequences, using different methods: CycleGAN, MUNIT, SIFA, DEPL, StarGAN v2 and PSTUDA. The results demonstrate the superiority of PSTUDA in maintaining the structural consistency while translating images across different domains. Other methods show distorted structures or missing details.
read the caption
Figure 3: Qualitative results for T1c → FS T2W, T2W, and DWI on the MSKT dataset. Blue and red bounding boxes indicate the annotated boundaries of the kidney and tumor, respectively (Same below).
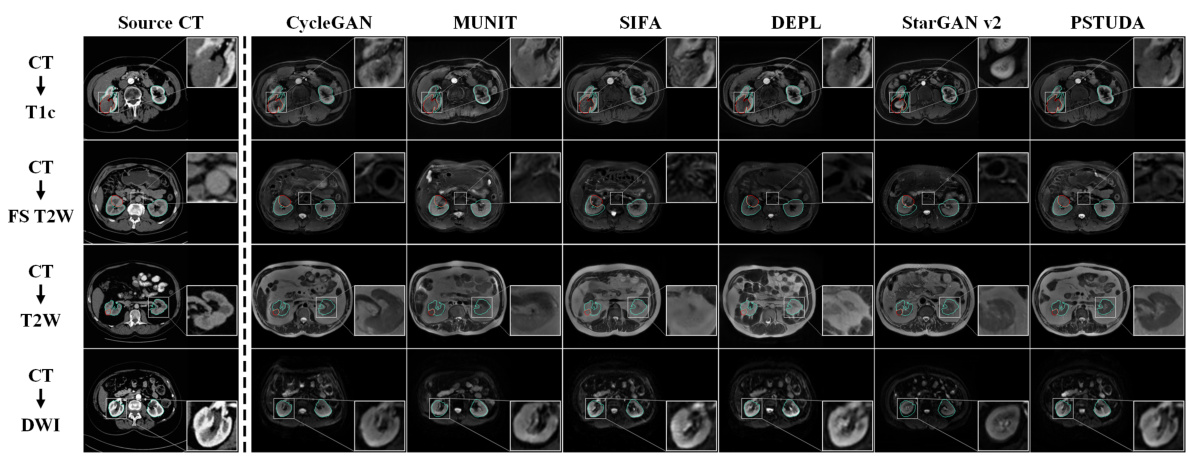
🔼 This figure shows a qualitative comparison of the results obtained by different methods (CycleGAN, MUNIT, SIFA, DEPL, StarGAN v2, and PSTUDA) for the task of translating CT images to four different MRI sequences (T1c, FS T2W, T2W, and DWI). The source CT images are shown in the first column, and the results of each method are displayed in the subsequent columns. The blue and red bounding boxes indicate the annotated boundaries of the kidney and tumor, respectively. This figure demonstrates the ability of PSTUDA to generate images that are visually similar to the target domain images while maintaining structural consistency and detailed information.
read the caption
Figure 4: Qualitative results for CT → T1c, FS T2W, T2W, and DWI on the KiTS19 and MSKT datasets.
More on tables
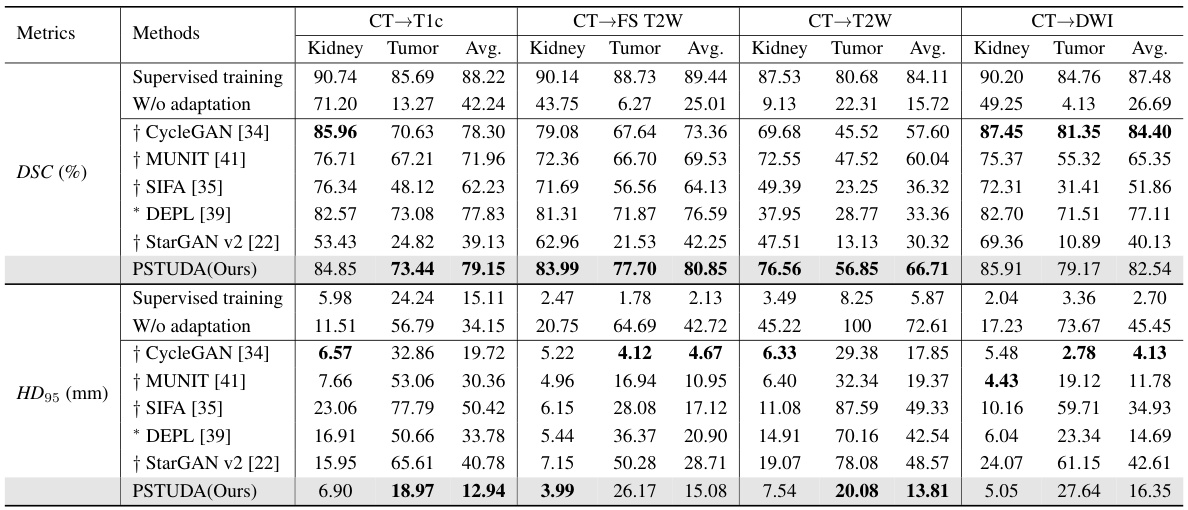
🔼 This table presents a quantitative comparison of different methods for kidney and tumor segmentation on the MSKT dataset. It shows the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) for each method across different MRI sequences (T1c, FS T2W, T2W, and DWI). The results are presented separately for kidney and tumor segmentation, and an average is also provided. The table allows for a direct comparison of the performance of the proposed PSTUDA method against other state-of-the-art unsupervised domain adaptation techniques.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
🔼 This table presents a quantitative comparison of different methods for kidney and tumor segmentation on the MSKT dataset. It shows the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) achieved by various methods across different MRI sequences (T1c, FS T2W, T2W, and DWI). The results highlight the superior performance of the proposed PSTUDA method compared to other state-of-the-art unsupervised domain adaptation (UDA) techniques.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
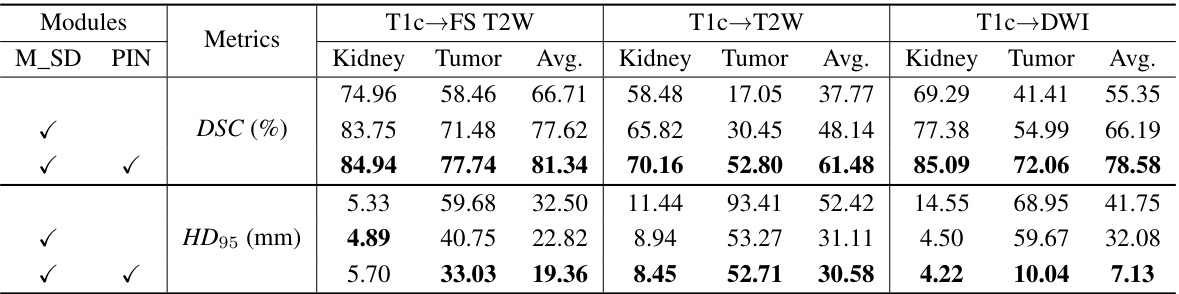
🔼 This table presents a quantitative comparison of different methods for kidney and tumor segmentation on the MSKT dataset. The metrics used are Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95), calculated for both kidney and tumor segmentation across three different MRI sequences (FS T2W, T2W, and DWI). The table allows for a direct comparison of the performance of the proposed PSTUDA method against several other state-of-the-art unsupervised domain adaptation (UDA) methods.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
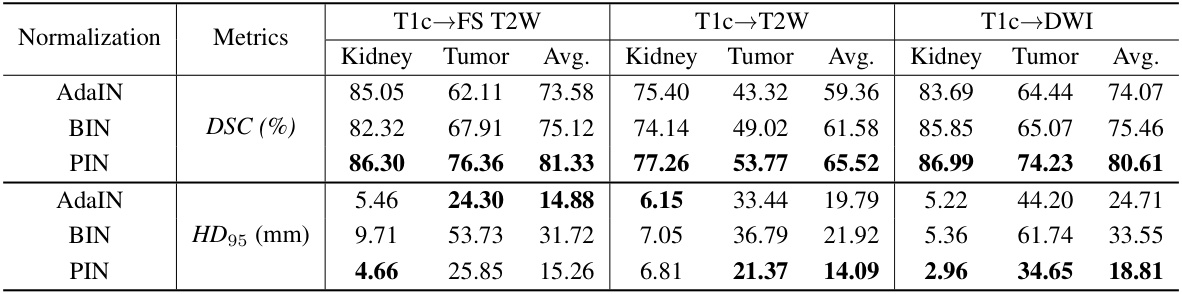
🔼 This table presents a quantitative comparison of the performance of different methods on the MSKT dataset for kidney and tumor segmentation. It shows the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) achieved by various methods across three different MRI sequences (FS T2W, T2W, and DWI) using T1c as the source domain. The results highlight the superior performance of the proposed PSTUDA method compared to other state-of-the-art unsupervised domain adaptation (UDA) techniques. The table also shows the results of a supervised training scenario and a no-adaptation scenario (baseline).
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
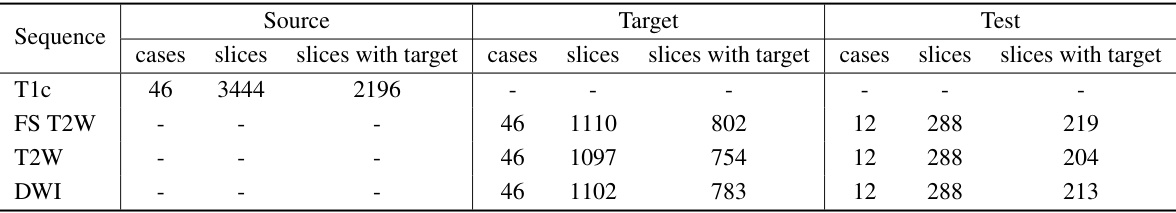
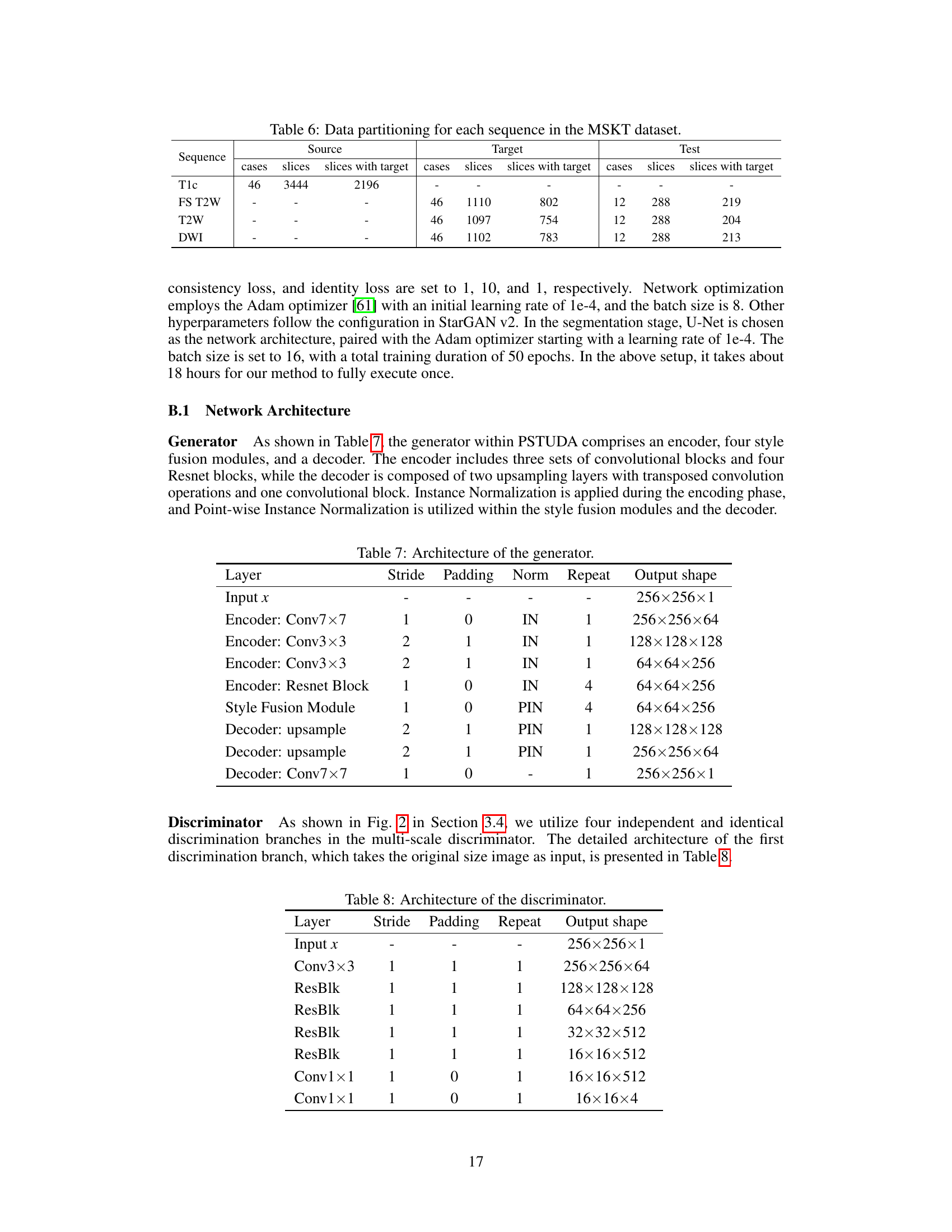
🔼 This table shows the distribution of the MSKT dataset across different MRI sequences (T1c, FS T2W, T2W, DWI) and their splits into training sets (source and target) and testing set. The source domain uses the T1c sequence, while the target domains use the other three sequences. Note the different numbers of cases and slices in each split, with and without tumor targets.
read the caption
Table 6: Data partitioning for each sequence in the MSKT dataset.

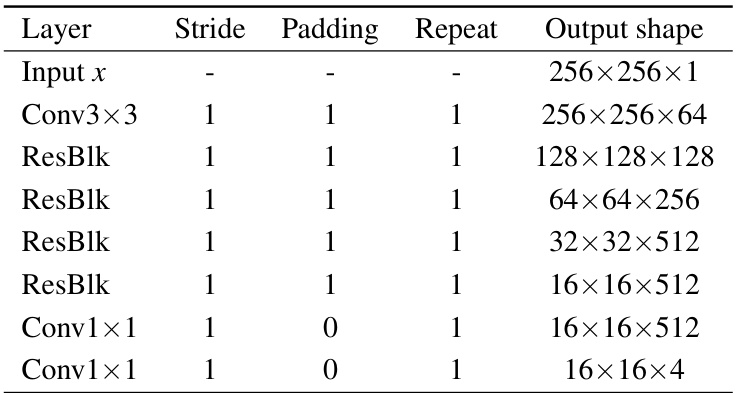
🔼 This table details the architecture of the generator used in the PSTUDA framework. It shows the different layers, including convolutional layers, residual blocks, and upsampling layers, along with the stride, padding, normalization method (Instance Normalization or Point-wise Instance Normalization), number of repetitions, and output shape for each layer. The table provides a comprehensive overview of the generator’s structure, which is crucial for understanding the image generation process within the PSTUDA framework.
read the caption
Table 7: Architecture of the generator.
🔼 This table details the architecture of the discriminator used in the PSTUDA framework. It shows the layers, stride, padding, repeat count, and output shape for each layer in the first (original size image input) discrimination branch. The discriminator uses multiple residual blocks (ResBlk) to enhance feature extraction and a multi-scale mechanism for more comprehensive image assessment.
read the caption
Table 8: Architecture of the discriminator.
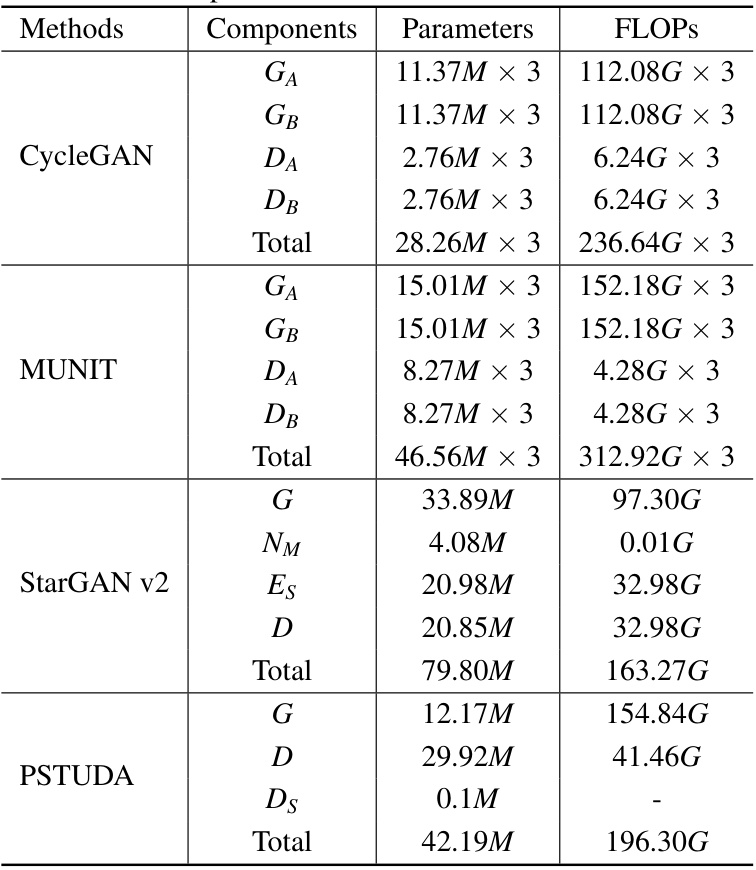
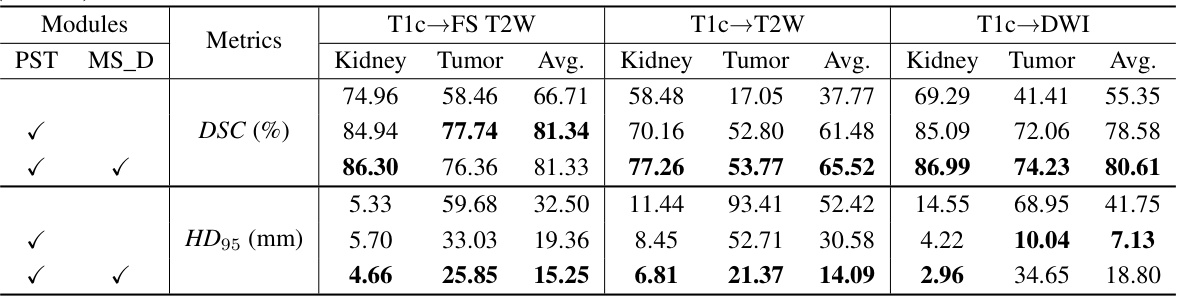
🔼 This table presents a quantitative comparison of the proposed PSTUDA method against several other state-of-the-art unsupervised domain adaptation (UDA) methods for kidney and tumor segmentation on the MSKT dataset. The comparison is based on the Dice Similarity Coefficient (DSC) and the 95% Hausdorff Distance (HD95), two common metrics used to evaluate segmentation performance. The table shows the average results across different MRI sequences (T1c-FS T2W, T1c-T2W, and T1c-DWI) for each method, allowing for a comprehensive assessment of their relative performance and ability to handle the variability in multi-sequence MRI data.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
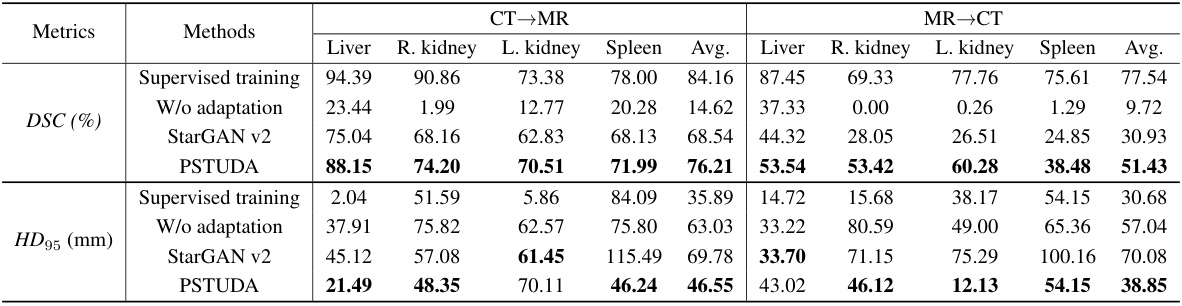
🔼 This table presents a quantitative comparison of different methods for kidney and tumor segmentation on the MSKT dataset. The metrics used are Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95). The results are broken down by kidney and tumor for each of the four MRI sequences (T1c, FS T2W, T2W, DWI), and for each method (CycleGAN, MUNIT, SIFA, DEPL, StarGAN v2, and PSTUDA). The table allows for a comparison of the performance of each method across different sequences and metrics, highlighting the superior performance of PSTUDA.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
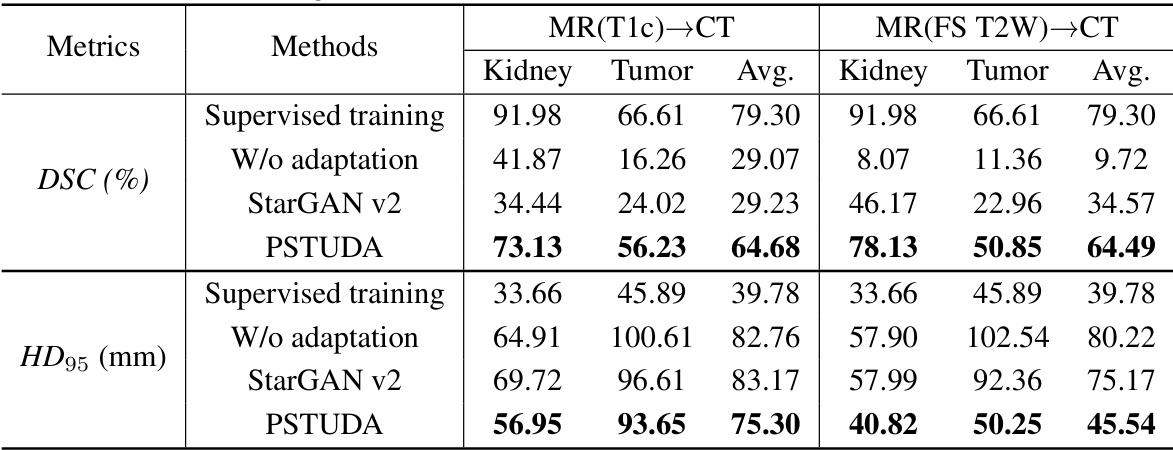
🔼 This table presents a quantitative comparison of different domain adaptation methods’ performance on kidney and tumor segmentation using the MSKT dataset. The metrics used are Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95), calculated for the kidney and tumor separately in three different MRI sequences (FS T2W, T2W, and DWI). The table compares the proposed PSTUDA method against several state-of-the-art unsupervised domain adaptation techniques (CycleGAN, MUNIT, SIFA, DEPL, and StarGAN v2). The results show the average DSC and HD95 across all sequences for each method, and also provide a breakdown for each sequence, allowing for a detailed analysis of performance across different image modalities.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
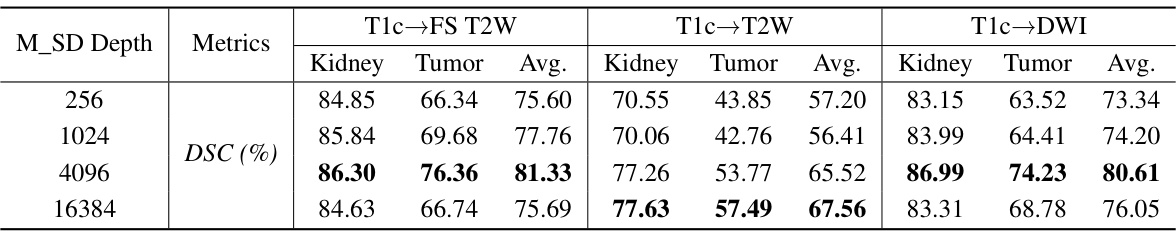
🔼 This table presents a quantitative comparison of the proposed PSTUDA method against several state-of-the-art unsupervised domain adaptation (UDA) methods for kidney and tumor segmentation on the MSKT dataset. It shows the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) for each method across three different MRI sequences (FS T2W, T2W, and DWI) and for both kidney and tumor segmentation. The results highlight the superior performance of PSTUDA compared to the other methods, demonstrating improved accuracy and efficiency in multi-sequence kidney and tumor segmentation.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.

🔼 This table presents a quantitative comparison of different domain adaptation methods on the MSKT dataset for kidney and tumor segmentation. It shows the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) for each method across different MRI sequences (T1c, FS T2W, T2W, DWI). The table allows for a direct comparison of the performance of the proposed method (PSTUDA) against other state-of-the-art unsupervised domain adaptation (UDA) techniques, highlighting its superior performance in multi-sequence kidney and tumor segmentation.
read the caption
Table 1: Quantitative segmentation results of different comparative methods on the MSKT dataset.
Full paper#