↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Predicting drug interactions is crucial for safe and effective medicine but challenging due to the complexity of drug mechanisms and the vastness of potential combinations. Existing methods often rely on hand-designed features or generic models that fail to capture subtle nuances in drug interactions. This paper addresses these limitations by introducing a novel approach: customized subgraph selection and encoding (CSSE-DDI). The core problem is addressed by utilizing neural architecture search (NAS) to automatically customize the subgraph selection and encoding processes for improved accuracy and efficiency. This data-driven strategy adapts to the specific characteristics of different drug interaction datasets, allowing for more precise and adaptable predictions.
The CSSE-DDI method shows substantial improvements over state-of-the-art methods across various datasets. The findings highlight the advantages of utilizing a data-specific design rather than relying on generic models. The approach demonstrates its superior performance, especially when dealing with new drugs not present in the training data. The authors also provide visualizations of the discovered subgraphs and encoding functions, providing insights into the model’s decision-making process and offering potential explainability benefits. Furthermore, the successful application of NAS to the DDI prediction problem opens new avenues for research and development in other areas requiring precise multi-relational prediction.
Key Takeaways#
Why does it matter?#
This paper is important because it presents a novel approach to drug-drug interaction (DDI) prediction that significantly improves accuracy and efficiency. The method, CSSE-DDI, uses neural architecture search (NAS) to automatically design data-specific components, overcoming limitations of existing hand-designed methods. This opens avenues for improving DDI prediction, facilitating drug development, and enhancing patient safety. The adaptable nature of CSSE-DDI has significant implications for other multi-relational prediction problems.
Visual Insights#

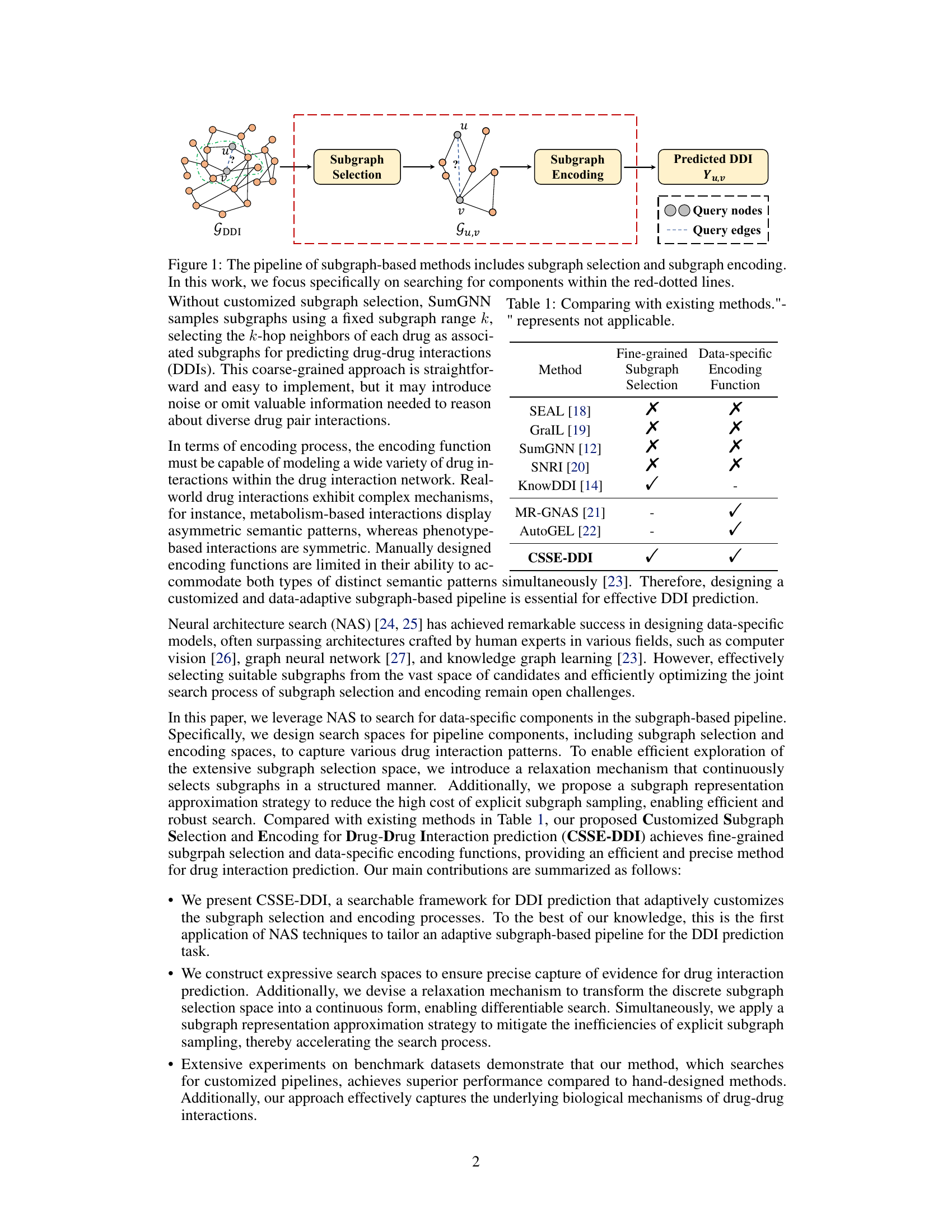
This figure illustrates the general pipeline of subgraph-based methods for drug-drug interaction prediction. It starts with the input drug interaction network (GDDI). A subgraph selection module then extracts a subgraph (Gu,v) relevant to a specific query pair of drugs (u,v). This subgraph is further encoded using a subgraph encoding module that learns a representation. Finally, this representation is used by a predictor to generate the prediction of drug-drug interaction (Yuv). The figure highlights the two key components, subgraph selection and subgraph encoding, that the authors’ proposed method focuses on improving.

This table compares the proposed method, CSSE-DDI, with existing methods for drug-drug interaction prediction. It highlights whether each method uses fine-grained subgraph selection and data-specific encoding functions, showing that CSSE-DDI uniquely incorporates both features, unlike other methods that might use one or neither.
In-depth insights#
DDI Subgraph Search#
DDI subgraph search focuses on efficiently identifying informative subgraphs within a drug-drug interaction (DDI) network to predict interactions. Effective subgraph selection is crucial as it directly impacts prediction accuracy. The challenge lies in the vast search space of possible subgraphs and the computational cost of exploring them. Methods typically involve designing a search strategy, such as those inspired by neural architecture search (NAS), to navigate this space efficiently. Relaxation techniques might be employed to transform a discrete search problem into a continuous one, making the search process differentiable and more tractable. Furthermore, approximation methods are often used to reduce the computational burden by estimating the properties of large subgraphs. The ultimate goal is to find subgraphs that capture the relevant contextual information for accurate DDI prediction, allowing for more efficient and accurate model training, while maintaining model interpretability.
NAS for DDIs#
Utilizing Neural Architecture Search (NAS) for Drug-Drug Interaction (DDI) prediction is a novel approach with significant potential. NAS automates the design of optimal DDI prediction models, eliminating the need for manual design and potentially leading to improved accuracy and efficiency. By searching through a vast space of possible architectures, NAS can identify models specifically tailored to the complexities and nuances of DDI data, such as the diverse types of interactions, asymmetric relationships, and the often dense nature of the interaction network. A key advantage is the potential for increased interpretability. By examining the components of the discovered architectures, researchers gain valuable insights into the features and relationships most relevant for DDI prediction, making the process more transparent. However, challenges remain, including the computational cost of NAS, the need for effective search strategies to navigate the vast architecture space, and the necessity for careful consideration of evaluation metrics in the context of DDI prediction. Future research should focus on developing efficient NAS techniques, incorporating domain expertise to guide the search process, and thoroughly validating the robustness of NAS-discovered models across various DDI datasets and prediction tasks.
DDI Relax & Approx#
The heading ‘DDI Relax & Approx’ likely refers to methods within a drug-drug interaction (DDI) prediction model that address challenges posed by the complexity of subgraph selection and encoding. Relaxation techniques likely involve transforming a discrete search space (e.g., choosing from a finite set of subgraphs) into a continuous space, enabling the use of gradient-based optimization methods which are more efficient than discrete search strategies. Approximation methods likely focus on simplifying the representation of subgraphs to reduce computational cost, such as using lower-dimensional embeddings to capture important structural information. These combined techniques would aim to enable efficient exploration of a large search space without sacrificing predictive accuracy, achieving a balance between exploration and exploitation during model training. The success hinges on the design of appropriate relaxation and approximation functions; poor choices could lead to significant performance loss. Therefore, the effectiveness of ‘DDI Relax & Approx’ heavily depends on carefully designed strategies that preserve essential information while significantly reducing computational burdens.
CSSE-DDI: S0/S1#
The heading ‘CSSE-DDI: S0/S1’ likely refers to a comparative analysis of the CSSE-DDI model’s performance across two distinct experimental settings: S0 and S1. S0 probably represents a standard, well-established benchmark setting, perhaps involving known drug interactions within a complete dataset. S1 likely introduces a more challenging, novel scenario, potentially using a dataset with newly discovered or less-well-understood drug interactions or involving predicting interactions where one or more drugs are new or not fully represented in the existing knowledge graph. The comparison is crucial because it evaluates the model’s generalizability and robustness. A strong performance in both S0 and S1 would suggest that CSSE-DDI is not only effective at replicating known results but can also generalize well to unseen data, a key aspect of robust DDI prediction models. The contrast allows researchers to gauge the impact of data novelty and missing information on the algorithm’s accuracy, offering insights into its real-world applicability and limitations.
Future of CSSE-DDI#
The future of CSSE-DDI (Customized Subgraph Selection and Encoding for Drug-Drug Interaction prediction) looks promising, building upon its success in achieving superior performance compared to existing methods. Further research could focus on expanding the search space to encompass more intricate interaction patterns and incorporate diverse data modalities beyond network structures. Integrating external knowledge sources, such as pathway information and protein-protein interactions, could enhance the model’s reasoning capabilities. Exploring transfer learning techniques to leverage knowledge from related domains (e.g., drug repurposing) would further improve efficiency and prediction accuracy, especially when dealing with limited data for certain drugs. Additionally, developing more robust evaluation metrics that consider the multifaceted nature of DDIs is crucial to accurately assess the model’s performance in real-world settings. Finally, investigating explainability methods will be vital to enhance interpretability and build trust in the predictions, facilitating better clinical decision-making.
More visual insights#
More on figures

This figure compares the training time and performance of CSSE-DDI against two other methods: KnowDDI and CompGCN. The left panel shows the F1 scores on the DrugBank dataset over training time in hours, while the right panel shows the ROC-AUC scores on the TWOSIDES dataset over training time in minutes. CSSE-DDI demonstrates superior convergence speed and ultimately achieves higher performance in both datasets, showcasing the benefits of its adaptive subgraph selection and encoding strategies.
This figure compares the training efficiency and performance of the proposed CSSE-DDI method against existing human-designed methods (CompGCN and KnowDDI). The plots show the learning curves for F1 score, accuracy, and Cohen’s Kappa on the DrugBank dataset (Figure 2a) and ROC-AUC, PR-AUC, and AP@50 on the TWOSIDES dataset (Figure 2b). CSSE-DDI demonstrates faster convergence and superior performance compared to baselines, highlighting the benefits of automated search for model architecture.

This figure visualizes two example subgraphs identified by the CSSE-DDI model for specific drug pairs from the DrugBank dataset. Each subgraph highlights the relationships between drugs, illustrating the model’s ability to identify relevant contextual information for accurate prediction. The subgraphs reveal domain concepts such as pharmacokinetics and metabolic interactions, demonstrating the model’s capacity to capture nuanced details that impact drug interactions.

This figure visualizes the encoding functions searched by the CSSE-DDI model across different datasets (DrugBank and TWOSIDES). It shows the specific combinations of operations (CORR, MULT, MLP, etc.) and aggregation functions (SUM, MEAN, CONCAT) discovered through the search process. The variation in the encoding functions highlights the model’s ability to adapt to the diverse interaction patterns in different datasets. For example, the DrugBank dataset, which features asymmetric interactions, predominantly employs CORR operations, while the symmetric TWOSIDES dataset shows a preference for MULT operations. This visualization illustrates the data-specific nature of the learned encoding functions.
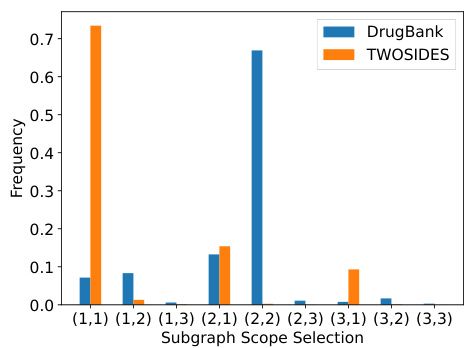
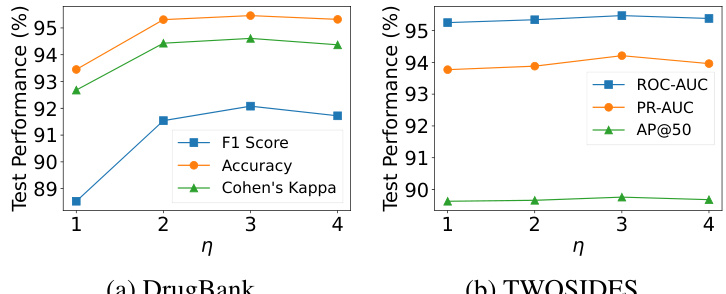
The figure shows a bar chart visualizing the distribution of subgraph scopes selected by the CSSE-DDI model across two benchmark datasets, DrugBank and TWOSIDES. The x-axis represents different subgraph scope combinations (e.g., (1,1), (1,2), etc.), indicating the number of hops considered from each node of a drug pair in the subgraph sampling process. The y-axis represents the frequency of each subgraph scope being selected by the model. The chart visually compares the scope selection preferences between the two datasets, showing variations in the model’s subgraph sampling strategies depending on the characteristics of the data.
More on tables
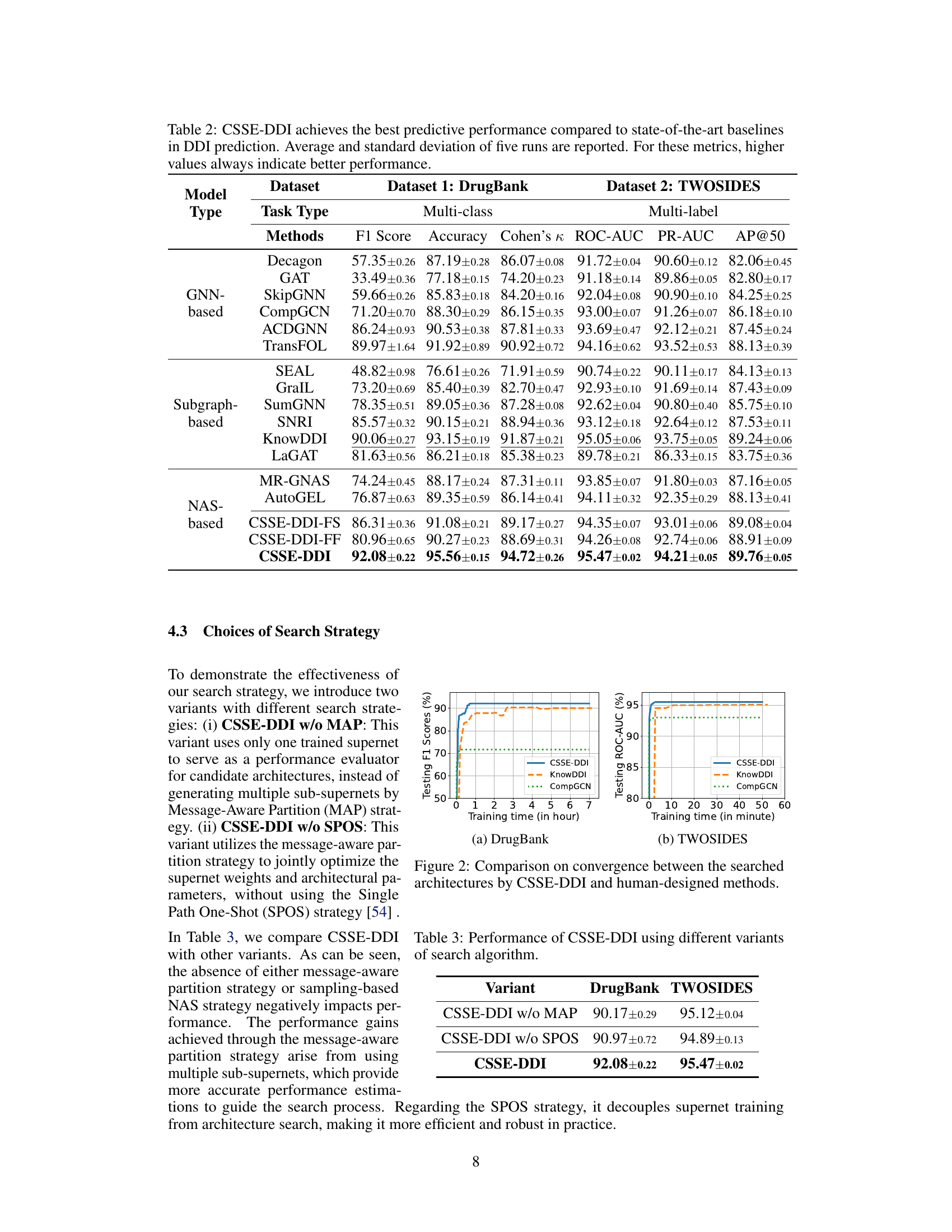
This table compares the performance of the proposed CSSE-DDI model against various baseline methods for drug-drug interaction (DDI) prediction on two benchmark datasets: DrugBank (multi-class) and TWOSIDES (multi-label). The metrics used for evaluation include F1 score, accuracy, Cohen’s kappa, ROC-AUC, PR-AUC, and AP@50, depending on the dataset. The table shows that CSSE-DDI consistently outperforms all baselines across different evaluation metrics, demonstrating its effectiveness in DDI prediction.
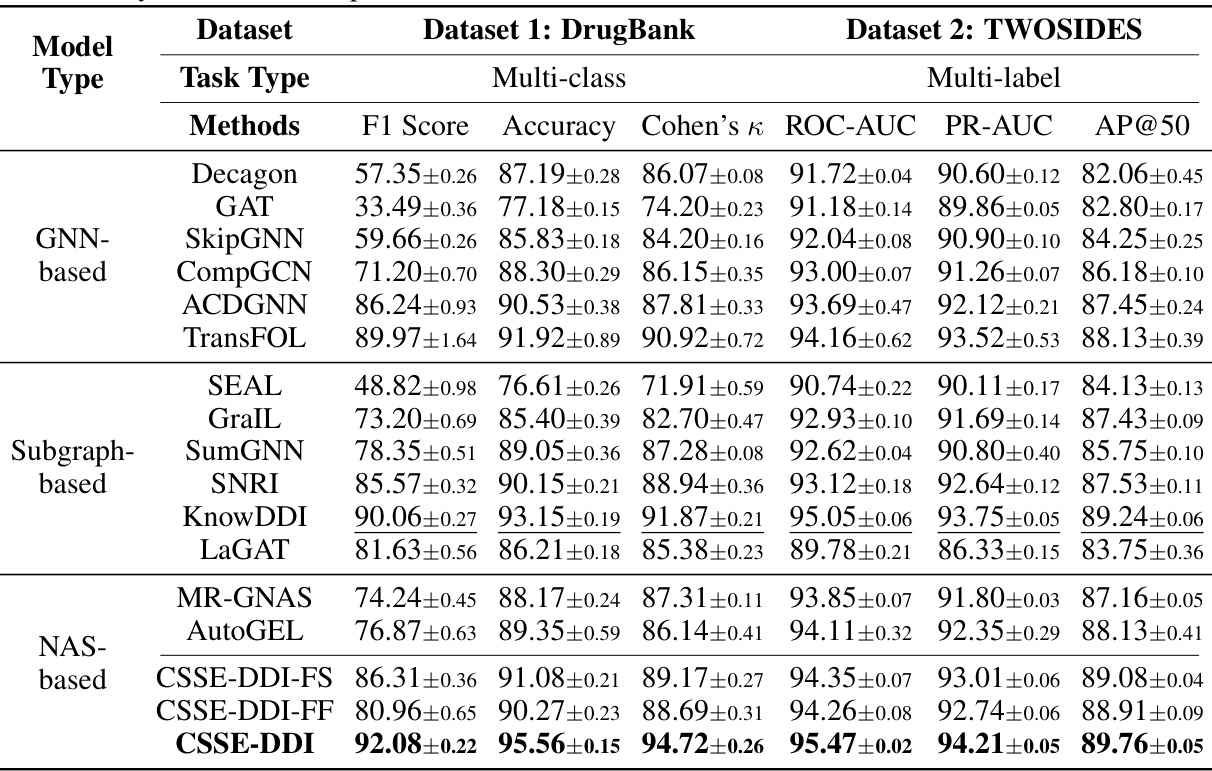
This table presents a comparison of the performance of the proposed CSSE-DDI model against various existing methods for drug-drug interaction (DDI) prediction on two benchmark datasets, DrugBank and TWOSIDES. The table shows performance metrics like F1 Score, Accuracy, Cohen’s Kappa, ROC-AUC, PR-AUC, and AP@50 for multi-class and multi-label classification tasks, respectively. It highlights the superiority of CSSE-DDI across different model types (GNN-based, Subgraph-based, and NAS-based) and variations of CSSE-DDI itself. Higher values indicate better performance.
This table compares the performance of the proposed method, CSSE-DDI, against various baselines on two datasets (DrugBank and TWOSIDES) for two prediction tasks (multi-class and multi-label). It shows that CSSE-DDI outperforms other methods across multiple metrics (F1 Score, Accuracy, Cohen’s Kappa, ROC-AUC, PR-AUC, AP@50) indicating its superior performance in DDI prediction. The table also includes variants of CSSE-DDI to showcase the impact of different design choices.
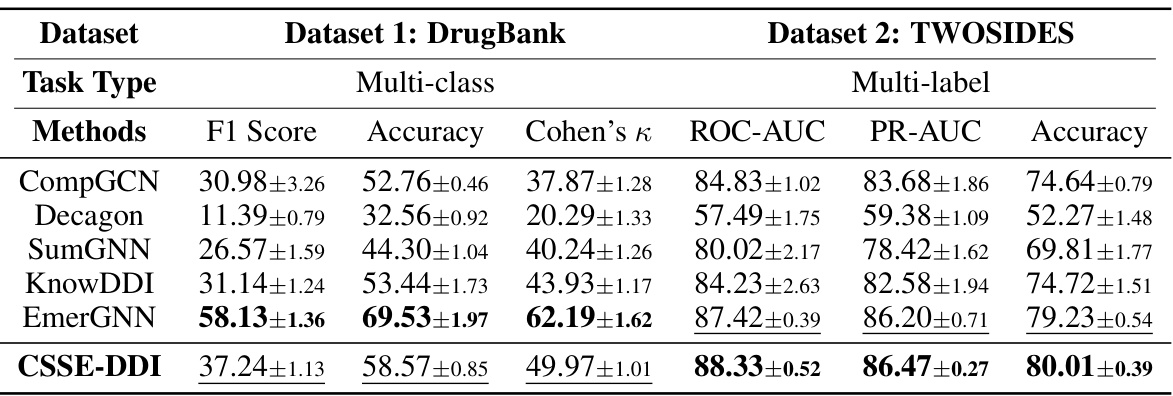
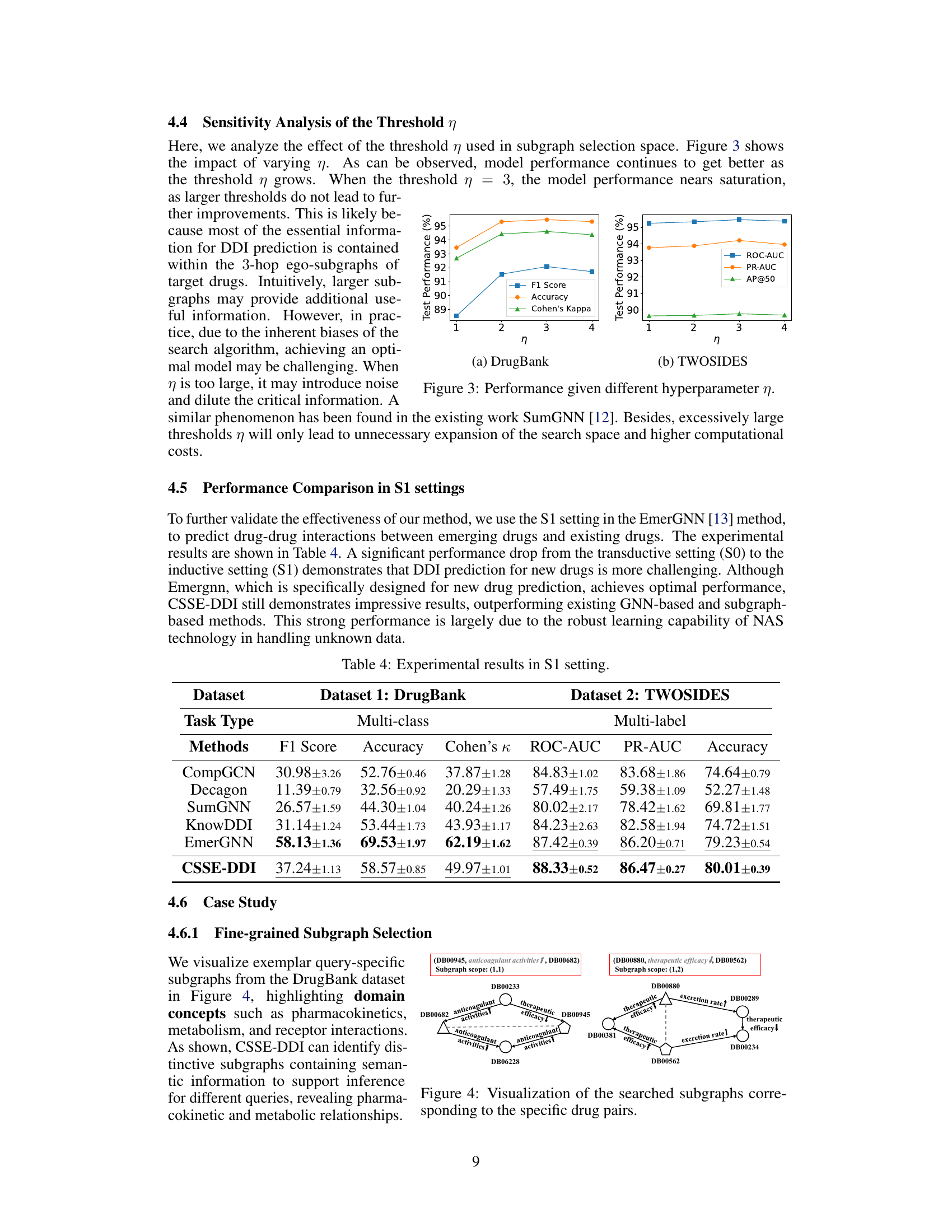
This table presents the performance comparison of different methods in the S1 setting of the drug-drug interaction (DDI) prediction task. The S1 setting is more challenging than the S0 setting because it involves predicting interactions where one drug is novel (not in the training data). The table shows the results on two benchmark datasets, DrugBank and TWOSIDES, using metrics appropriate for multi-class (DrugBank) and multi-label (TWOSIDES) classification. The methods compared include various GNN and subgraph-based methods, and the proposed CSSE-DDI method. The metrics reported include F1 score, Accuracy, Cohen’s Kappa (DrugBank), and ROC-AUC, PR-AUC, and Accuracy (TWOSIDES). Higher values indicate better performance.

This table presents a comparison of the performance of the proposed CSSE-DDI method against various baselines on two datasets, DrugBank and TWOSIDES. For each dataset and method, it lists the F1 Score, Accuracy, Cohen’s Kappa (for DrugBank), ROC-AUC, PR-AUC, and AP@50 (for TWOSIDES). The results demonstrate that CSSE-DDI outperforms all other methods across these metrics, highlighting its superior performance in predicting drug-drug interactions.
This table presents a comparison of the performance of the proposed CSSE-DDI model against various baseline methods on two benchmark datasets (DrugBank and TWOSIDES) for two different prediction tasks (SO and S1). The metrics used for evaluation are F1 score, accuracy, Cohen’s kappa, ROC-AUC, PR-AUC, and AP@50, depending on the dataset and task. The results highlight the superior performance of CSSE-DDI across different evaluation metrics.
This table compares the performance of the proposed CSSE-DDI model against various baseline models for drug-drug interaction (DDI) prediction on two benchmark datasets, DrugBank and TWOSIDES. It shows the F1 score, accuracy, Cohen’s Kappa, ROC-AUC, PR-AUC and AP@50 for different models across two types of DDI prediction tasks. The results demonstrate CSSE-DDI’s superior performance.
This table presents a comparison of the performance of the proposed CSSE-DDI model against various baseline models for drug-drug interaction (DDI) prediction on two benchmark datasets (DrugBank and TWOSIDES). It shows that CSSE-DDI outperforms other methods in terms of F1 score, accuracy, Cohen’s Kappa, ROC-AUC, PR-AUC, and AP@50, demonstrating its superior performance in DDI prediction tasks. The results are averaged over five runs with standard deviation included.
Full paper#