TL;DR#
Many organizations share pre-trained molecular models to facilitate drug discovery without revealing proprietary training data. However, this practice may pose privacy risks. This paper explores the risk of extracting private training molecular data from these models, which is challenging because they are non-generative and exhibit diverse model architectures.
The researchers introduce a novel, model-independent scoring function for evaluating molecule candidates extracted from molecular pre-trained models and a Molecule Extraction Policy Network to improve the efficiency of molecule extraction. Experimental results demonstrate that there is a considerable risk of extracting training data from pre-trained models. This challenges the assumption that sharing only pre-trained models provides adequate data protection.
Key Takeaways#
Why does it matter?#
This paper is crucial because it reveals a significant vulnerability in the widespread practice of sharing pre-trained molecular models without exposing the underlying data. The research highlights that malicious actors can extract sensitive training data from these models, raising serious concerns about commercial confidentiality and collaborative trust. This necessitates a reassessment of current data sharing practices and motivates the development of robust data protection techniques for pre-trained models in the field.
Visual Insights#
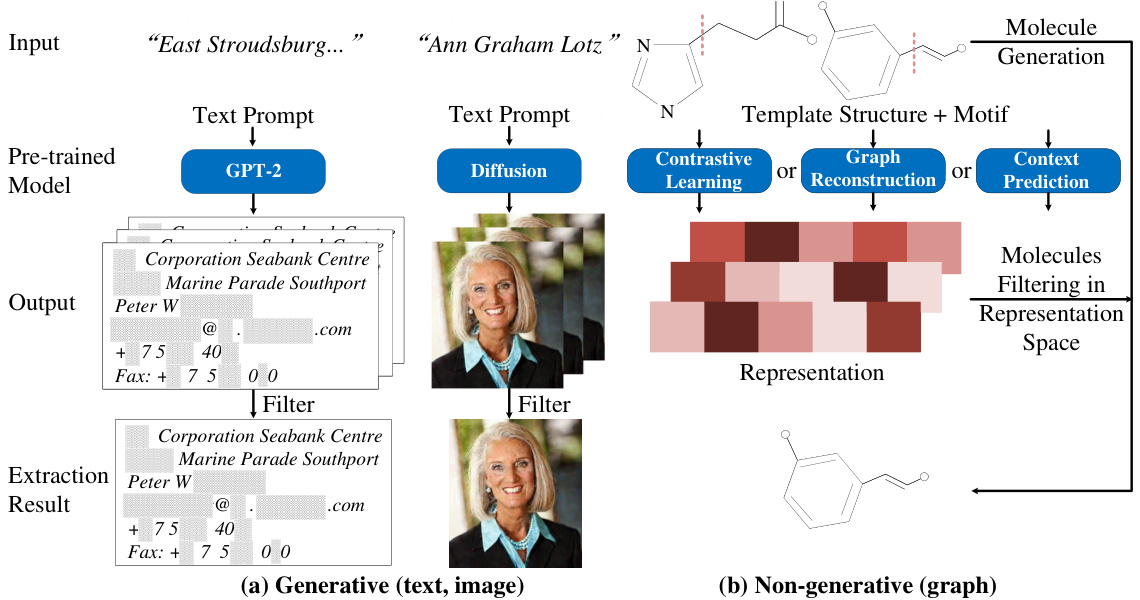
🔼 The figure illustrates data extraction attacks across different data types (text, image, and graph). Panel (a) shows how generative models (like GPT-2 and diffusion models) for text and image data can be directly exploited by inputting specific prompts to extract private training data from their outputs. Panel (b) contrasts this by showing that graph-based models, which are commonly used for molecular data, are often non-generative. They typically use various pre-training techniques (contrastive learning, reconstruction, and context prediction) making direct data extraction more difficult and requiring alternative approaches.
read the caption
Figure 1: Data extraction attacks across text, image, and graph. (a) In domains like text and image, by inputting specific text prompts, private training data can be directly extracted from the outputs generated by models. (b) Conversely, in the graph domain, the pre-trained models are typically non-generative, and exhibit a diversity of pre-training tasks, such as contrastive learning, graph reconstruction, and context prediction.

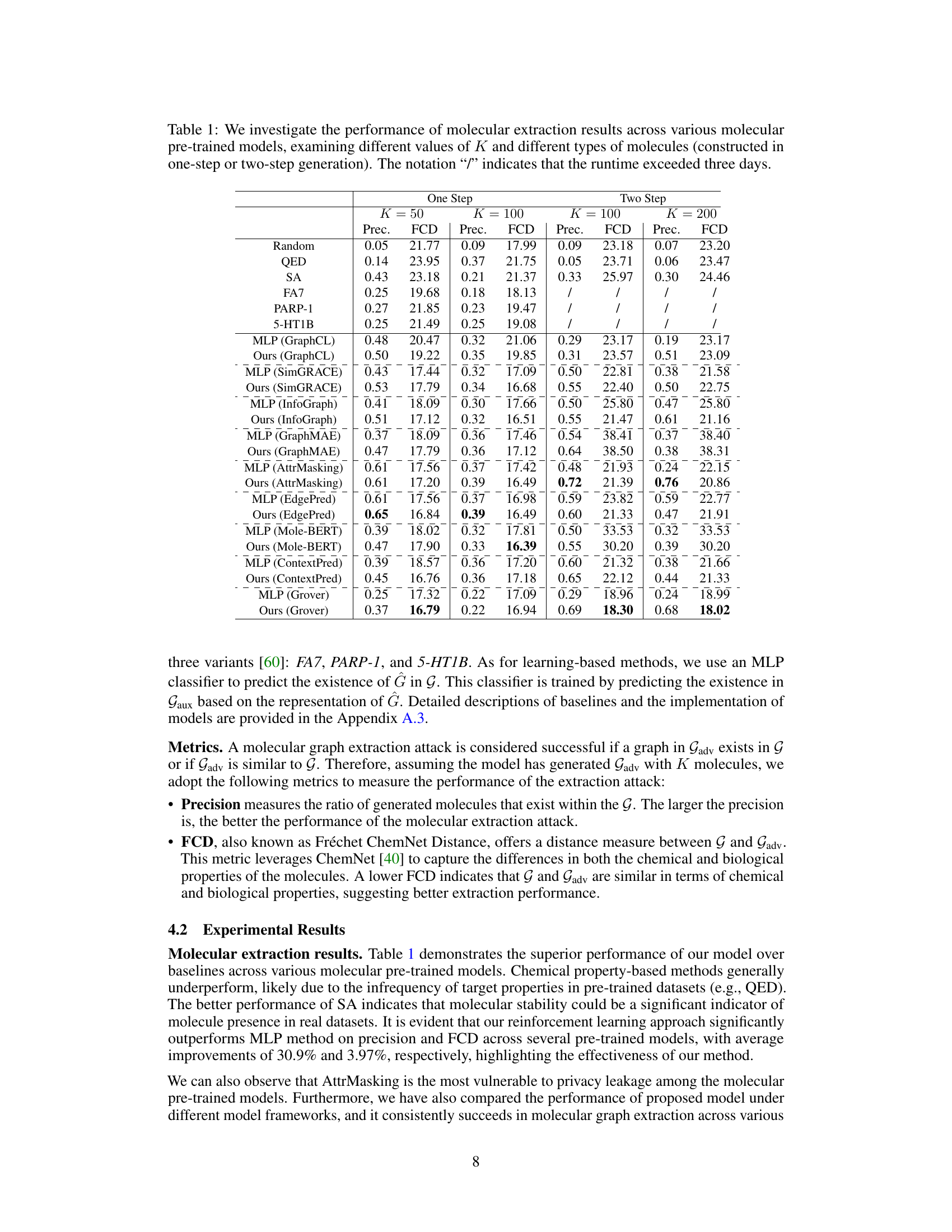
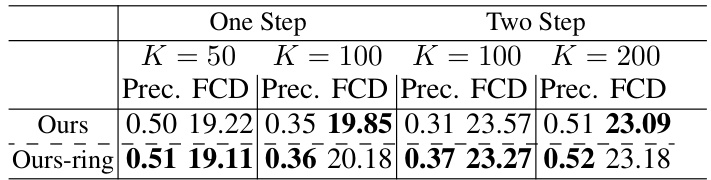
🔼 This table presents the precision and Fréchet ChemNet Distance (FCD) for molecular extraction attacks against various molecular pre-trained models. Different numbers of molecules (K) are tested for both one-step and two-step generation methods. The results are broken down by model type and molecule generation method, showing the success rate (precision) and similarity (FCD) of extracted molecules to the original training data.
read the caption
Table 1: We investigate the performance of molecular extraction results across various molecular pre-trained models, examining different values of K and different types of molecules (constructed in one-step or two-step generation). The notation '/' indicates that the runtime exceeded three days.
In-depth insights#
Molecule Data Leakage#
The hypothetical heading ‘Molecule Data Leakage’ in a research paper would likely explore the risk of sensitive molecular data being unintentionally or maliciously revealed through the use of pre-trained molecular models. This is a crucial concern in collaborative drug discovery settings, where model sharing is common but poses a privacy risk. The core issue revolves around the ability of an adversary to extract or reconstruct training data from the model’s output, potentially compromising proprietary information or intellectual property. A thorough analysis under this heading could involve discussing various attack vectors, such as membership inference attacks or reconstruction attacks, and propose defense mechanisms to mitigate such data leakage, emphasizing the need for robust privacy-preserving techniques in the development and deployment of molecular AI models. The investigation might also delve into the legal and ethical implications of model sharing in relation to data privacy concerns, which are increasingly significant in this domain. Furthermore, a discussion of practical solutions to address data leakage, like differential privacy or federated learning techniques, would strengthen the analysis.
Model-Agnostic Scoring#
A model-agnostic scoring function is a crucial element in evaluating the likelihood of a data point belonging to a model’s training set without direct access to the training data or model architecture. Its key advantage lies in its applicability across various model types, making it highly valuable for tasks like data extraction attacks or assessing the memorization capabilities of different pre-trained models. The effectiveness of such a function rests on its ability to identify subtle patterns and relationships within the model’s output that correlate with the presence of specific data points in the training set. A well-designed scoring function needs to be robust, not only considering the model’s output representation of individual data points but also their relationships within the training set. This is especially true for complex data domains, such as molecular graphs, where a single scoring function may require supplementary techniques to sufficiently constrain the search space and enhance performance. Therefore, a robust and efficient method needs to be model-independent, addressing the diverse architectures of pre-trained models and mitigating the inherent challenges related to scalability and complexity.
RL-Based Extraction#
An RL-based extraction approach for molecular data from pre-trained models is a novel and promising direction. Leveraging reinforcement learning (RL) allows for a more efficient and targeted search of the vast chemical space, overcoming the limitations of exhaustive enumeration. The agent learns a policy to guide molecule generation, prioritizing candidates most likely to be present in the original training set. A key innovation is the model-independent scoring function enabling application across diverse model architectures. However, this approach’s effectiveness heavily relies on the quality of the molecule generation process and the scoring function’s ability to discriminate between training and non-training data. The robustness of the method against various pre-trained models and different types of datasets requires thorough investigation. While promising, challenges remain in evaluating and mitigating potential vulnerabilities in this approach.
Privacy Risk Analysis#
A thorough privacy risk analysis of a system employing machine learning models, especially pre-trained ones, necessitates a multi-faceted approach. It should evaluate data leakage risks inherent in model architectures and training processes, examining if sensitive information might be inadvertently memorized or reconstructed from model outputs. The analysis must also consider the potential for membership inference attacks, where an adversary seeks to determine if a specific data point was part of the training set. Furthermore, a robust analysis should account for the attacker’s capabilities and resources, ranging from simple query access to more sophisticated techniques like adversarial attacks. Finally, it’s crucial to assess the impact of different model sharing mechanisms on privacy, considering whether anonymization or other privacy-enhancing techniques are effective in mitigating potential data breaches. Quantifiable metrics for privacy risk are vital, enabling comparison of different systems and the evaluation of mitigation strategies.
Future Defenses#
Future defenses against molecular data extraction attacks necessitate a multi-pronged approach. Strengthening model architectures to be less susceptible to memorization effects is crucial. This may involve techniques like differential privacy, or the use of more robust training methods that enhance generalization and reduce overfitting. Developing more sophisticated scoring functions that are less vulnerable to manipulation by adversaries is also important, potentially through the incorporation of more nuanced chemical features and increased complexity to make reverse engineering more difficult. Finally, implementing robust detection mechanisms to identify and mitigate data extraction attempts is critical. This requires a deep understanding of attack strategies, and the development of effective monitoring systems that can distinguish legitimate queries from malicious ones. Combining these three strategies will provide a robust defense, making the extraction of training data significantly more challenging.
More visual insights#
More on figures
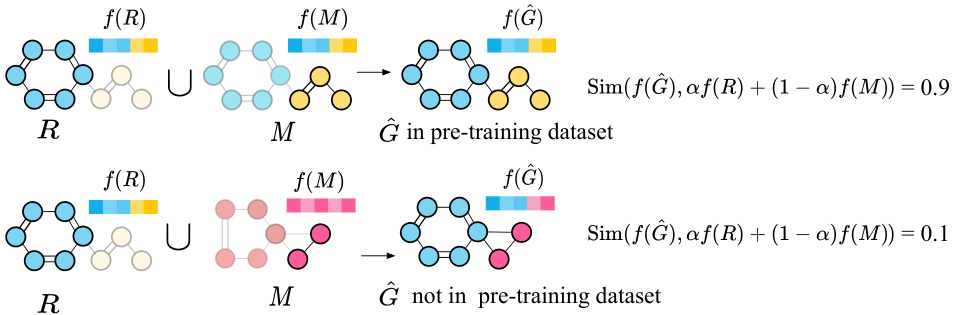
🔼 This figure illustrates different data extraction attack scenarios across various data types (text, image, graph). Panel (a) demonstrates how generative models (text and image) can directly leak training data when prompted with specific inputs. In contrast, panel (b) shows that non-generative models (graphs) require different attack strategies due to their non-generative nature and diverse pre-training tasks. It highlights the unique challenges in extracting training data from molecular pre-trained models.
read the caption
Figure 1: Data extraction attacks across text, image, and graph. (a) In domains like text and image, by inputting specific text prompts, private training data can be directly extracted from the outputs generated by models. (b) Conversely, in the graph domain, the pre-trained models are typically non-generative, and exhibit a diversity of pre-training tasks, such as contrastive learning, graph reconstruction, and context prediction.
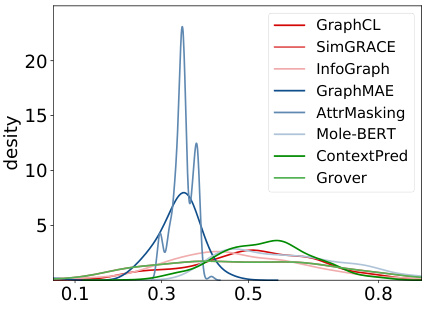
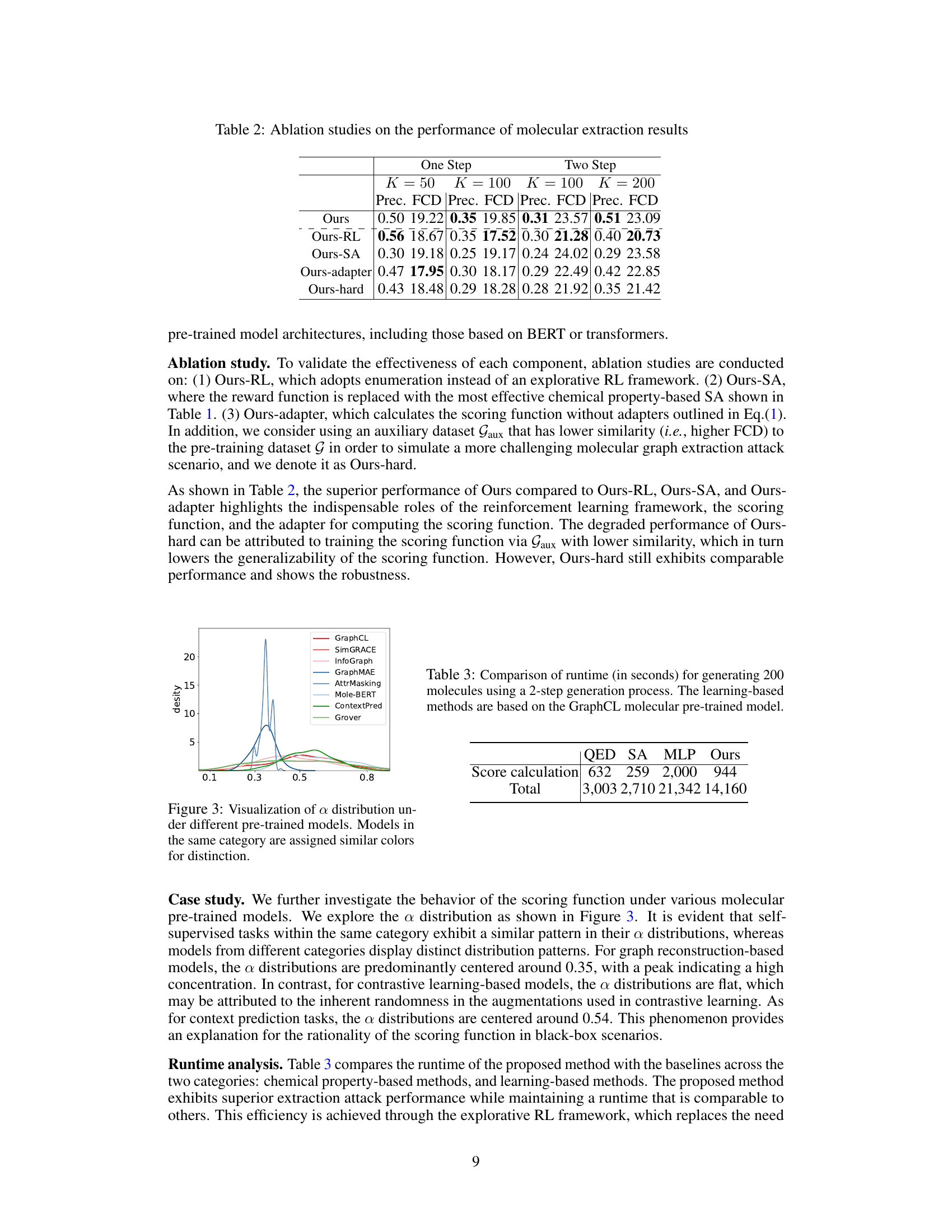
🔼 This figure visualizes the distribution of the hyperparameter α in the scoring function across various molecular pre-trained models. The x-axis represents the value of α, and the y-axis shows the density of α values. Different colors represent different categories of models (Contrastive Learning, Graph Reconstruction, Context Prediction). The distributions are quite distinct across different model categories suggesting that the scoring function is effective despite being model-independent.
read the caption
Figure 3: Visualization of α distribution under different pre-trained models. Models in the same category are assigned similar colors for distinction.
More on tables
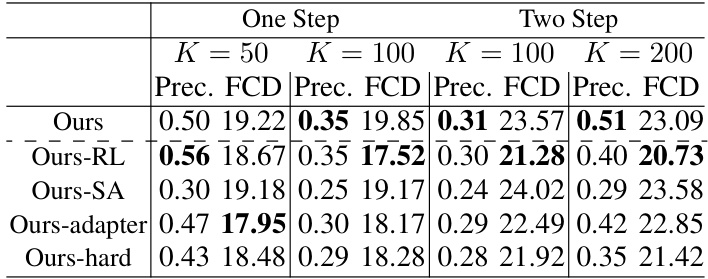
🔼 This table presents the results of ablation studies conducted to evaluate the impact of different components of the proposed molecular extraction attack method. It shows the precision and Fréchet ChemNet Distance (FCD) achieved by the full model and several variations where components such as the reinforcement learning (RL) framework, the scoring function, the adapter, and the auxiliary dataset were modified. This allows for a quantitative assessment of the contribution of each component to the overall performance of the attack.
read the caption
Table 2: Ablation studies on the performance of molecular extraction results
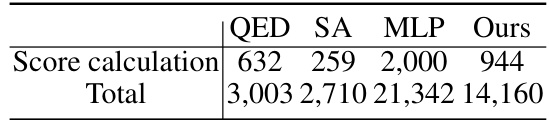
🔼 This table compares the computation time in seconds for generating 200 molecules using a 2-step generation process. It shows a breakdown of the time spent on score calculation versus the total runtime for different methods (QED, SA, MLP, and the proposed ‘Ours’ method). All methods use the GraphCL molecular pre-trained model.
read the caption
Table 3: Comparison of runtime (in seconds) for generating 200 molecules using a 2-step generation process. The learning-based methods are based on the GraphCL molecular pre-trained model.
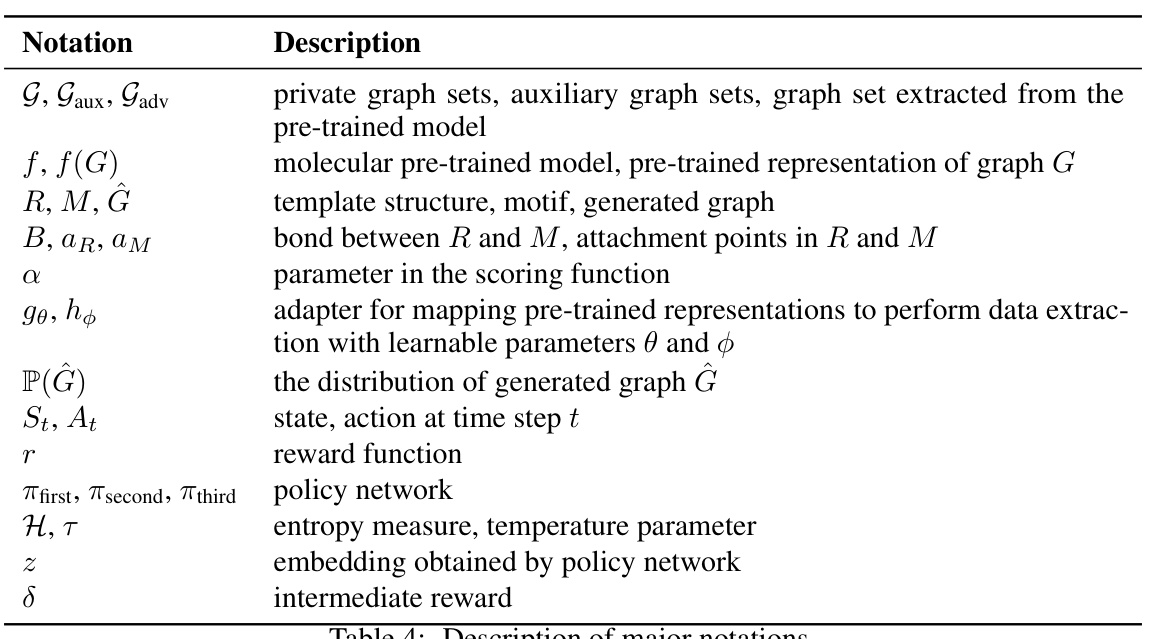
🔼 This table lists the notations used throughout the paper along with their corresponding descriptions. The notations cover various aspects of the proposed molecular graph extraction attack, including the datasets used (private, auxiliary, and extracted), the model itself, the molecular graph components, the scoring function parameters, the policy network, reward and other crucial elements of the reinforcement learning framework employed for the attack.
read the caption
Table 4: Description of major notations.
🔼 This table presents the precision and Fréchet ChemNet Distance (FCD) scores achieved by different molecular extraction methods across various molecular pre-trained models, using various values of K (number of molecules extracted). The methods are compared against baseline methods and tested on one-step and two-step generated molecules. The results show the effectiveness of the proposed method across different model architectures and varying data complexities.
read the caption
Table 1: We investigate the performance of molecular extraction results across various molecular pre-trained models, examining different values of K and different types of molecules (constructed in one-step or two-step generation). The notation '/' indicates that the runtime exceeded three days.
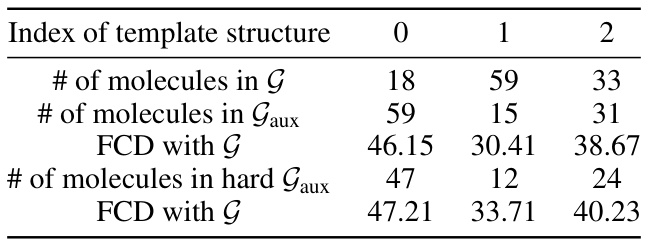
🔼 This table presents the results of ablation studies conducted to evaluate the effectiveness of each component of the proposed molecular extraction attack method. It compares the performance (Precision and FCD) of the full model against variations where reinforcement learning (RL), the scoring function, the adapter, and a harder auxiliary dataset are removed or modified. This helps to understand the contribution of each component to the overall effectiveness of the attack.
read the caption
Table 2: Ablation studies on the performance of molecular extraction results
🔼 This table presents the results of ablation studies conducted to evaluate the impact of different components on the molecular extraction attack’s performance. It compares the performance of the main method (‘Ours’) against variations that remove or modify key elements, such as the reinforcement learning component (‘Ours-RL’), the scoring function (‘Ours-SA’), the adapter used in the scoring function (‘Ours-adapter’), and a variation using a more challenging auxiliary dataset (‘Ours-hard’). The results show the contribution of each component and the robustness of the overall method under more difficult conditions.
read the caption
Table 5: Ablation studies on the performance of molecular extraction results

🔼 This table presents the precision and Fréchet ChemNet Distance (FCD) of molecular extraction attacks across different molecular pre-trained models (GraphCL, SimGRACE, InfoGraph, GraphMAE, AttrMasking, Mole-BERT, EdgePred, Grover, ContextPred) using different numbers (K=50, 100, 200) of generated molecules for both one-step and two-step molecule generation methods. It also includes baseline results using random selection and other chemical property-based methods (QED, SA, FA7, PARP-1, 5-HT1B). The ‘/’ symbol indicates that the runtime exceeded three days.
read the caption
Table 1: We investigate the performance of molecular extraction results across various molecular pre-trained models, examining different values of K and different types of molecules (constructed in one-step or two-step generation). The notation '/' indicates that the runtime exceeded three days.

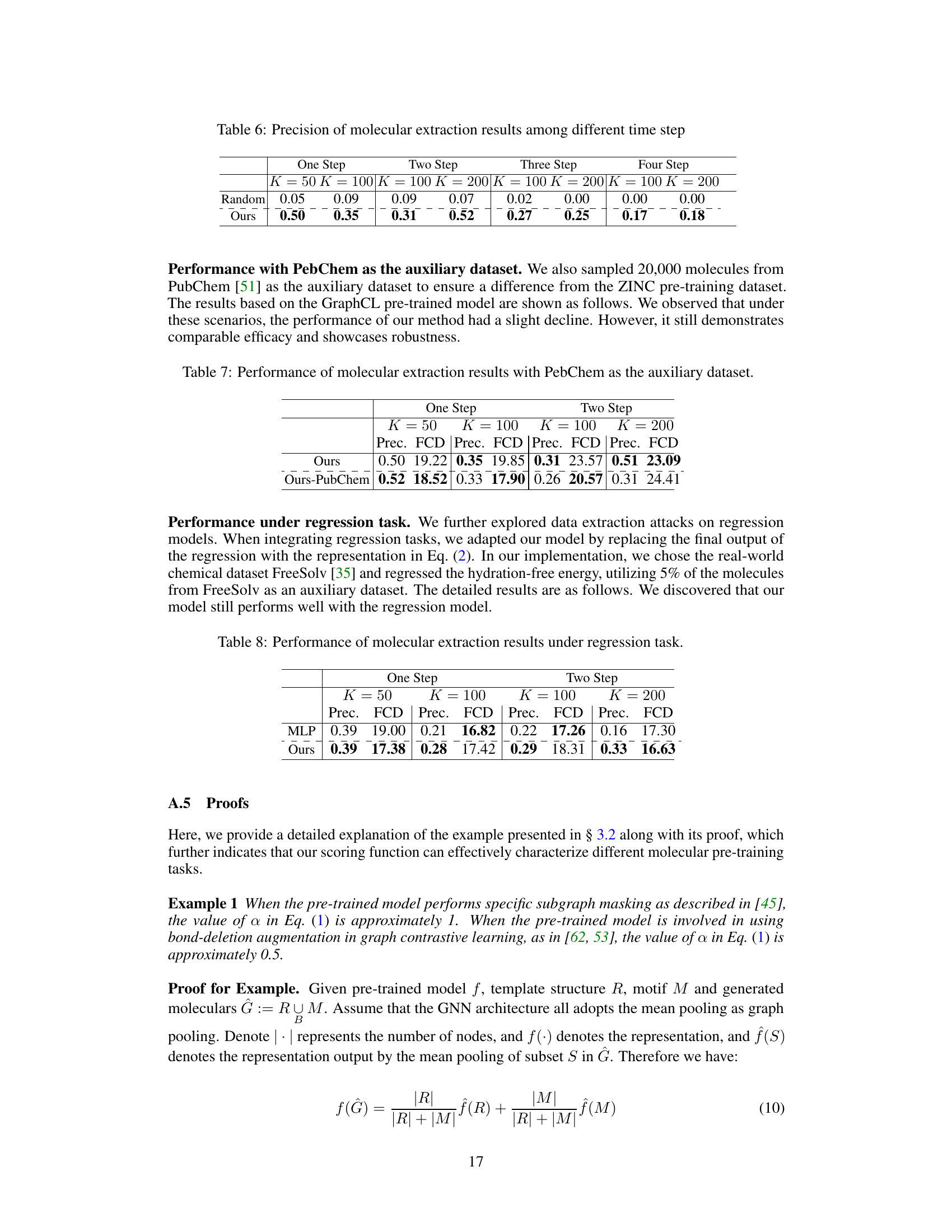
🔼 This table presents the results of molecular extraction attacks using PubChem as the auxiliary dataset, showing the precision and Fréchet ChemNet Distance (FCD) for both one-step and two-step molecular generation methods. It compares the performance of the proposed model (‘Ours’) against a baseline where PubChem is used as the auxiliary dataset (‘Ours-PubChem’). The results demonstrate the model’s robustness across different datasets, although a slight decrease in performance is observed when using PubChem.
read the caption
Table 7: Performance of molecular extraction results with PubChem as the auxiliary dataset.
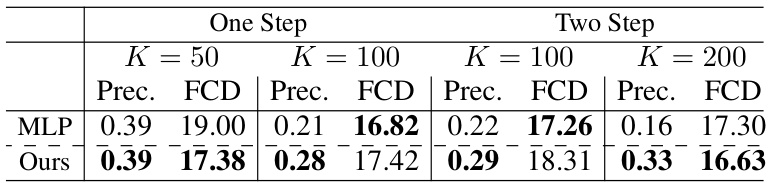
🔼 This table presents the performance of molecular extraction attacks under a regression task using different models. It shows the precision and Fréchet ChemNet Distance (FCD) for both one-step and two-step molecule generation approaches using MLP and the proposed ‘Ours’ method. The results demonstrate the effectiveness of the proposed approach in extracting molecular data from regression models.
read the caption
Table 8: Performance of molecular extraction results under regression task.
Full paper#