↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Whole slide image (WSI) analysis using Multiple Instance Learning (MIL) heavily relies on the quality of extracted features. While foundation models show promise, they often include task-irrelevant information. This paper addresses this by proposing a novel framework.
The proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) framework dynamically calibrates generic image features using task-specific concepts. This is achieved via two interconnected modules: a Concept-guided Information Bottleneck module to enhance relevant characteristics, and a Concept-Feature Interference module to generate discriminative features. Extensive experiments demonstrate CATE significantly enhances performance and generalizability of MIL models, providing a practical and effective solution for improving WSI analysis.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in computational pathology and medical image analysis. It introduces CATE, a novel paradigm for task-specific adaptation of pathology foundation models, addressing a critical limitation in current weakly supervised methods. The ‘free lunch’ approach of CATE, requiring minimal additional resources, offers significant improvements in model performance and generalizability, opening new avenues for developing more effective and adaptable WSI analysis tools.
Visual Insights#
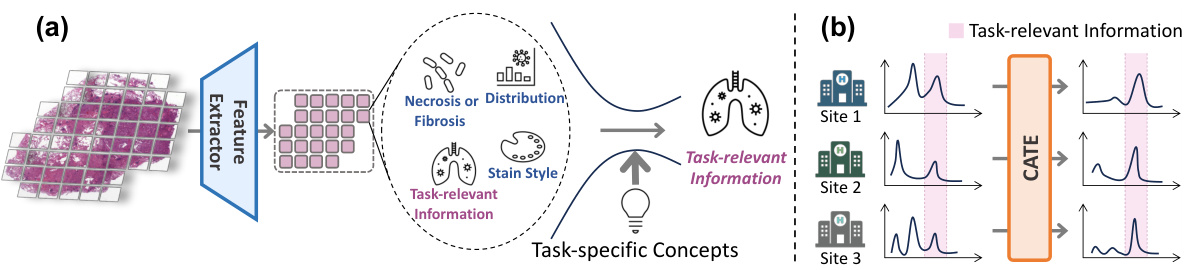
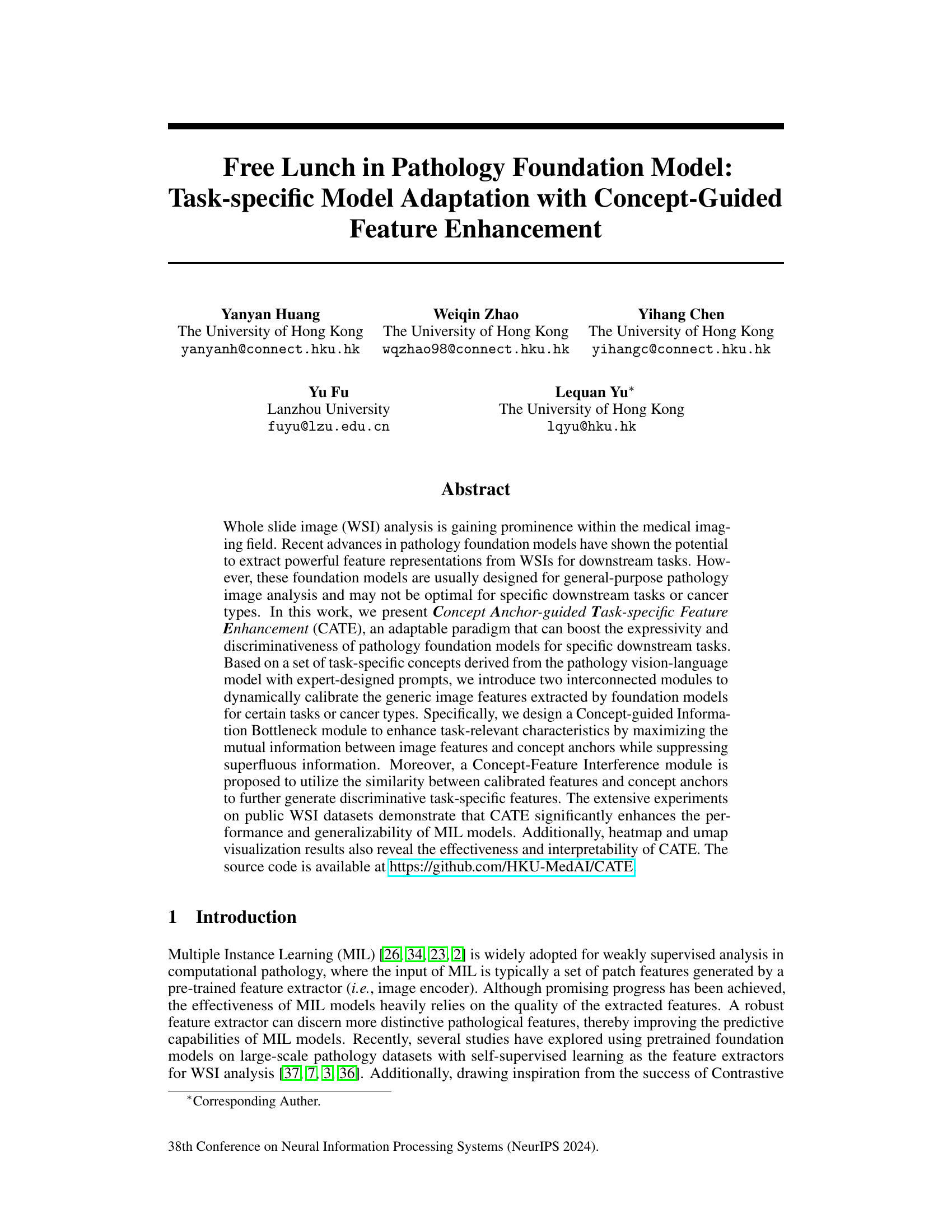
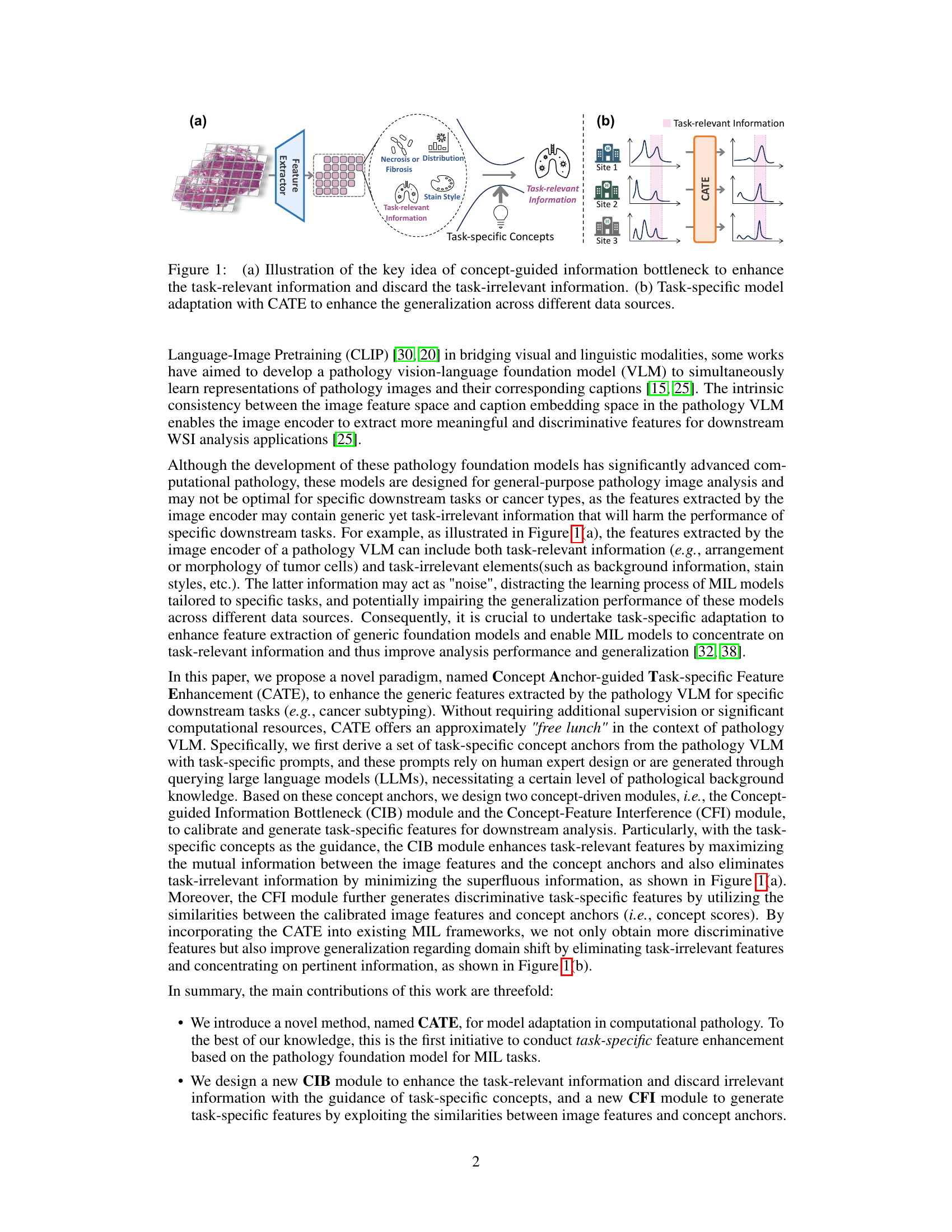
The figure illustrates the core idea of the Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. (a) shows how CATE uses task-specific concepts to filter out task-irrelevant information from generic image features, focusing on the task-relevant aspects. (b) demonstrates how CATE improves the generalizability of models across different datasets (data sources) by enhancing task-relevant features while suppressing irrelevant information, leading to better performance and robustness.
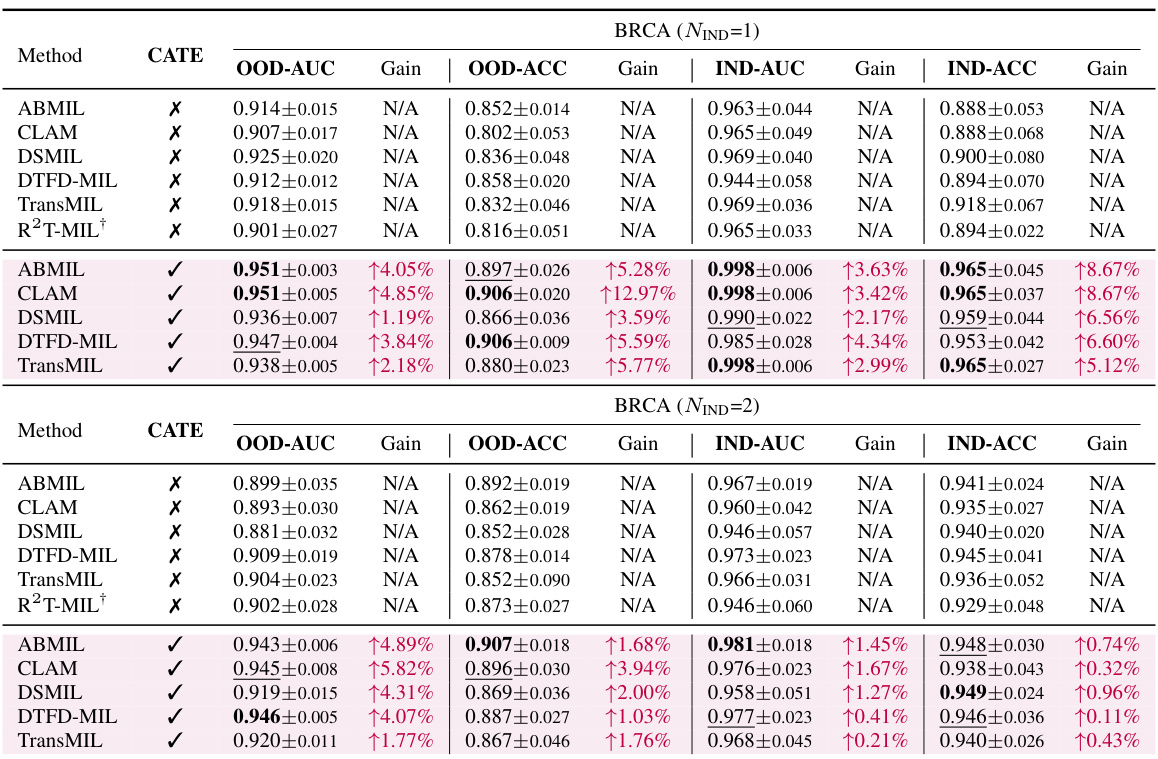
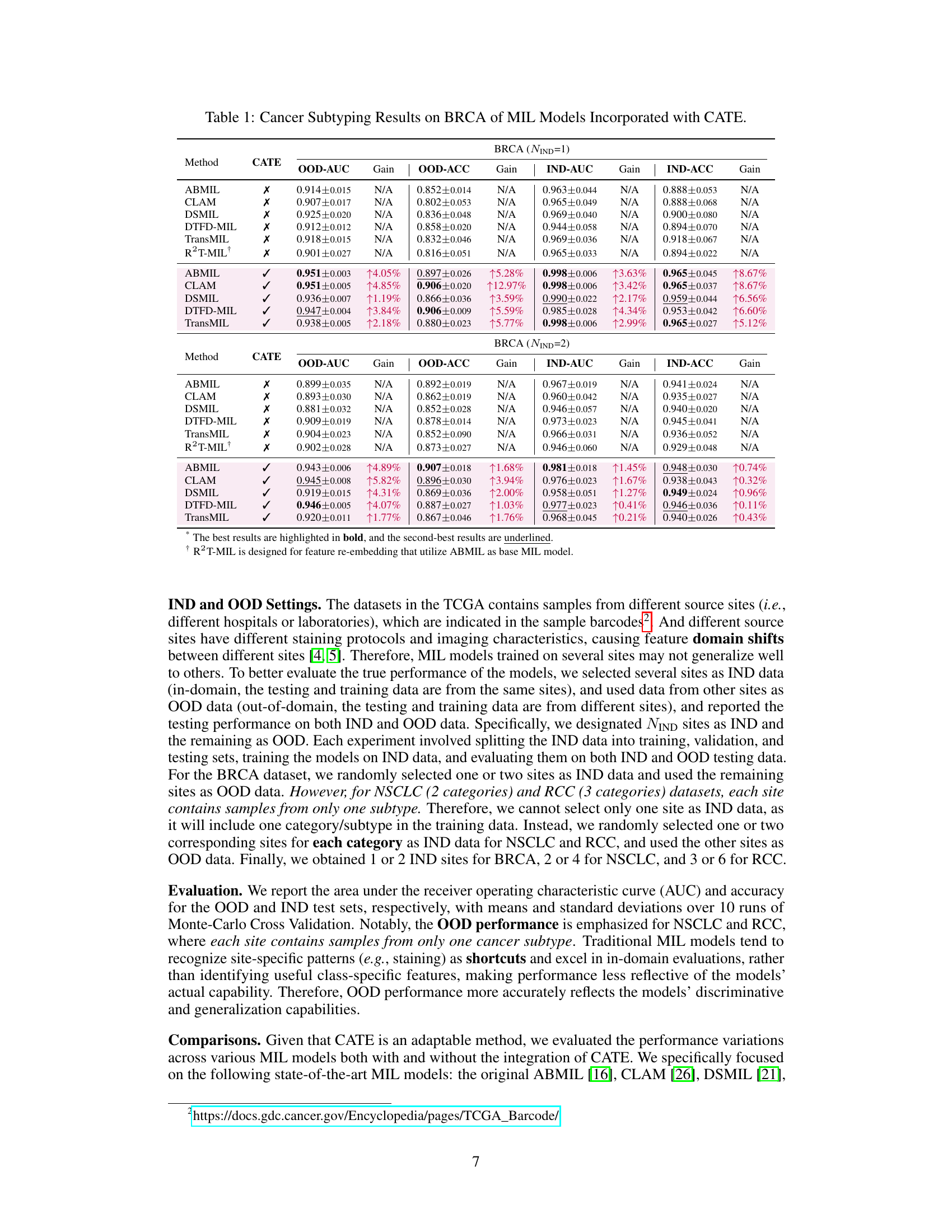
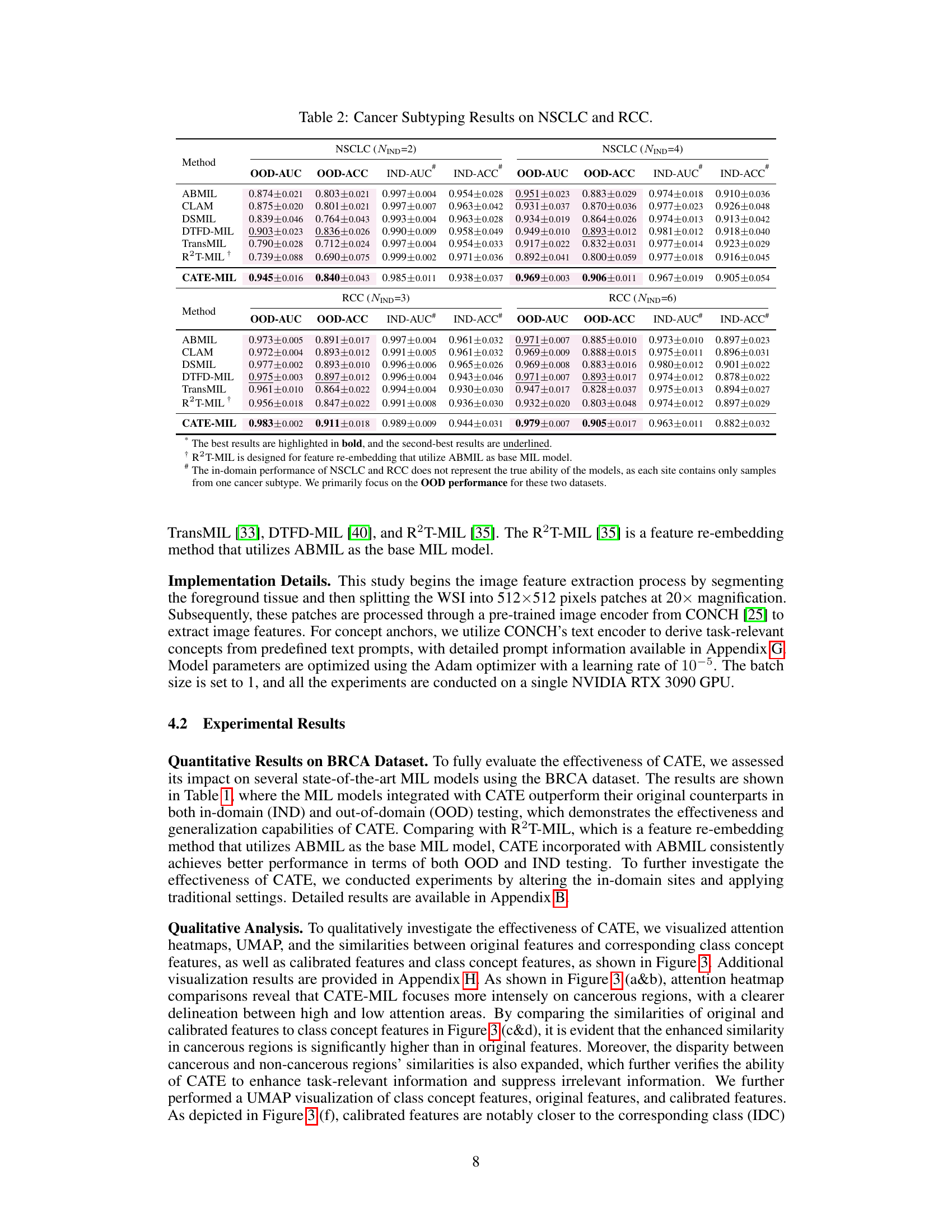
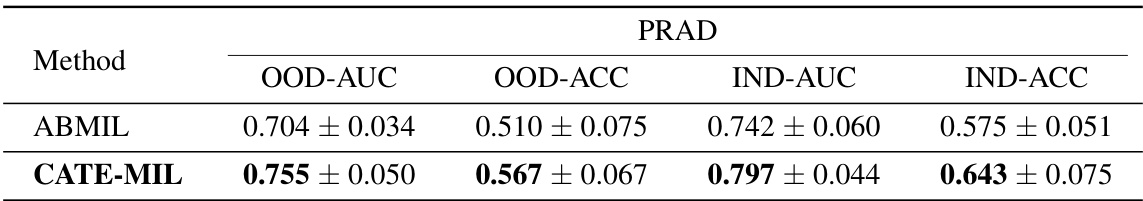
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models. It compares the performance of these models with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The table shows the Area Under the Curve (AUC) and accuracy for both in-domain (IND) and out-of-domain (OOD) settings, highlighting the improvements achieved by incorporating CATE. In-domain refers to testing on the same sites as training, while out-of-domain uses different sites to assess generalization performance. The ‘Gain’ columns indicate the percentage improvement achieved with CATE compared to the baseline model.
In-depth insights#
Concept-driven MIL#
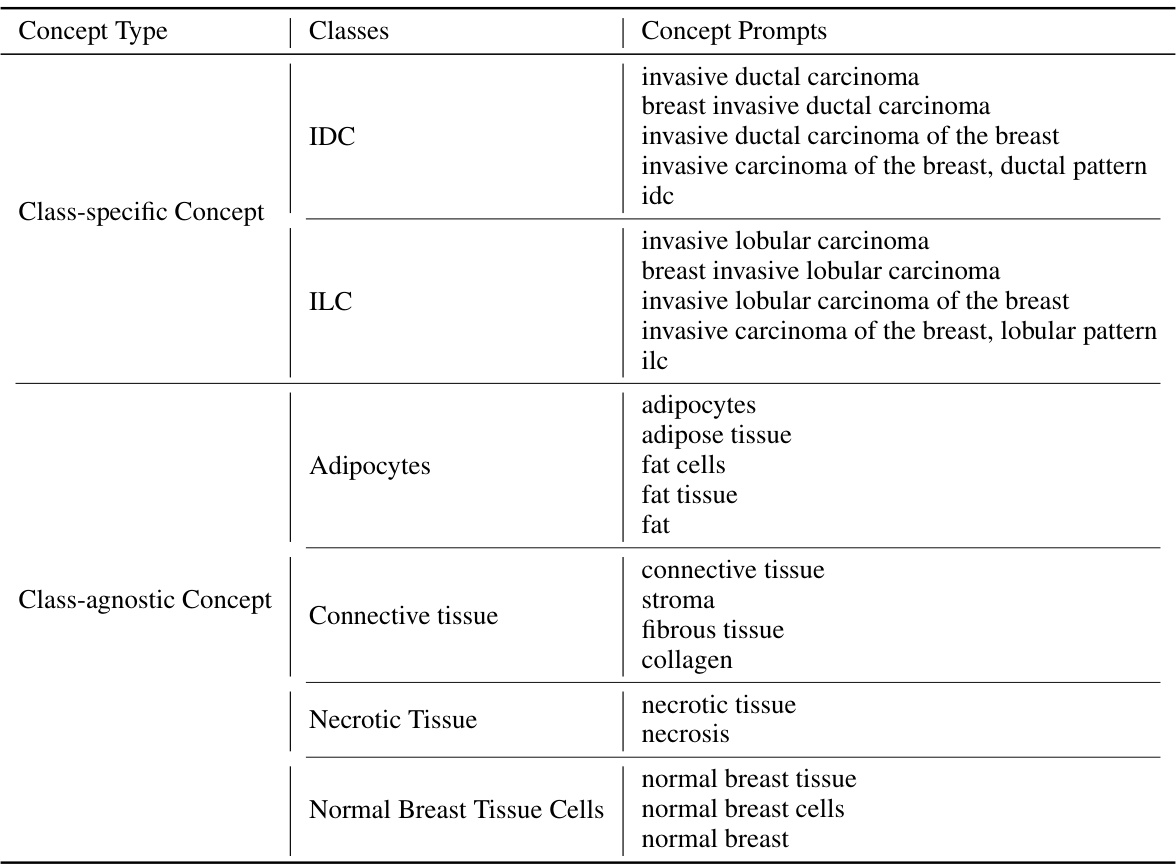
Concept-driven Multiple Instance Learning (MIL) represents a significant advancement in weakly supervised learning for image analysis, particularly within the context of pathology. The core idea revolves around leveraging high-level semantic concepts to guide the learning process, rather than relying solely on low-level image features. This approach addresses a crucial limitation of traditional MIL, which often struggles with the inherent ambiguity and variability in weakly labeled data. By incorporating prior knowledge in the form of concepts, Concept-driven MIL enhances the model’s ability to discern relevant patterns and improve classification accuracy. This is achieved through mechanisms such as concept embedding, which provides a structured representation for incorporating human-interpretable knowledge, resulting in improved generalization across different datasets and better handling of noisy or incomplete annotations. A key advantage lies in its interpretability, as the model’s decisions can be better understood through the lens of these concepts, fostering trust and facilitating better clinical decision-making. However, challenges exist in defining and selecting appropriate concepts, as well as in effectively integrating concept information into the MIL framework. Future research should focus on developing robust methods for concept selection, representation, and integration to further enhance the effectiveness and applicability of concept-driven MIL.
CATE Framework#
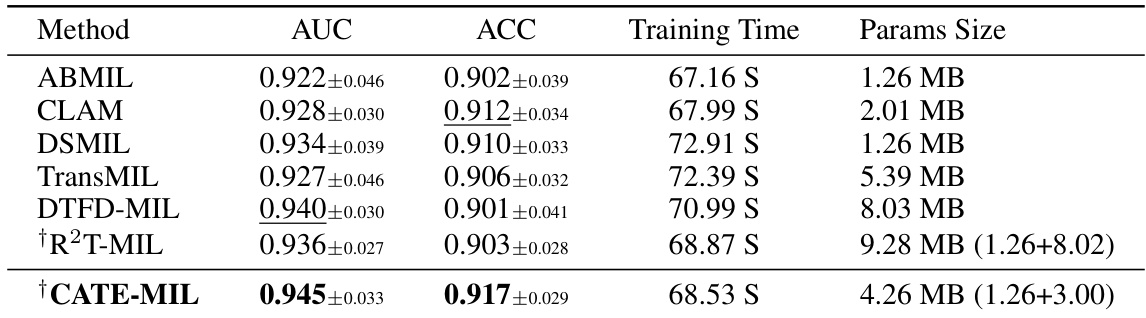
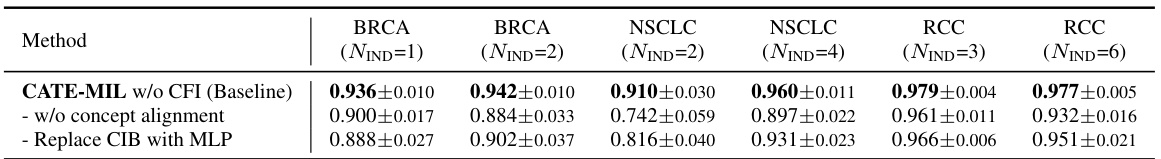
The CATE (Concept Anchor-guided Task-specific Feature Enhancement) framework offers a novel approach to task-specific adaptation of pathology foundation models. It leverages the inherent consistency between image and text modalities in vision-language models to dynamically calibrate generic image features. By introducing two interconnected modules, Concept-guided Information Bottleneck (CIB) and Concept-Feature Interference (CFI), CATE enhances task-relevant characteristics while suppressing superfluous information. The CIB module maximizes mutual information between image features and concept anchors, effectively calibrating the feature space. The CFI module utilizes similarities between calibrated features and concept anchors to generate task-specific features, improving both performance and generalizability. The framework’s adaptability makes it a valuable tool for enhancing MIL models in computational pathology, particularly for specific downstream tasks or cancer types. The reliance on expert-designed prompts or large language models for concept extraction is a notable aspect of the method.
Pathology Feature#
In the context of pathology, features are crucial pieces of information extracted from medical images, specifically whole slide images (WSIs), that provide insights into the condition of the tissue. Effective feature extraction is paramount because it directly impacts the performance of downstream tasks, such as cancer subtyping or disease classification. The concept of ‘pathology features’ encompasses various data representations, ranging from low-level image features like texture or color to high-level semantic features indicative of specific tissue structures or cellular patterns. The quality of these features heavily relies on the strength of the underlying image analysis models, which may leverage techniques such as deep learning and multiple instance learning (MIL). Foundation models in this context can play a critical role in pretraining robust feature extractors capable of generalizing across diverse datasets, while subsequent task-specific adaptations can enhance performance. However, achieving optimal feature extraction often requires careful calibration to ensure that task-relevant information is emphasized, and task-irrelevant or confounding details are suppressed. In essence, the goal is to create a refined representation that boosts the accuracy and robustness of downstream analyses, thus improving diagnostic capabilities and potentially clinical outcomes.
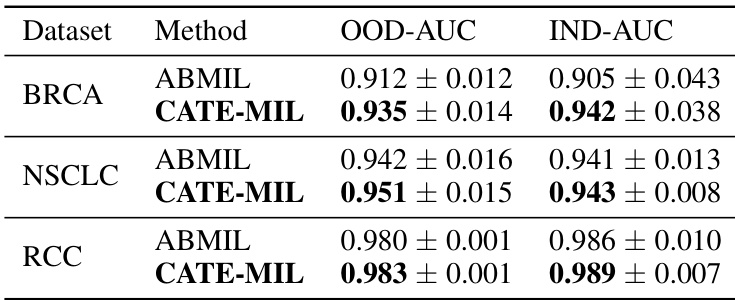
Generalization Gains#
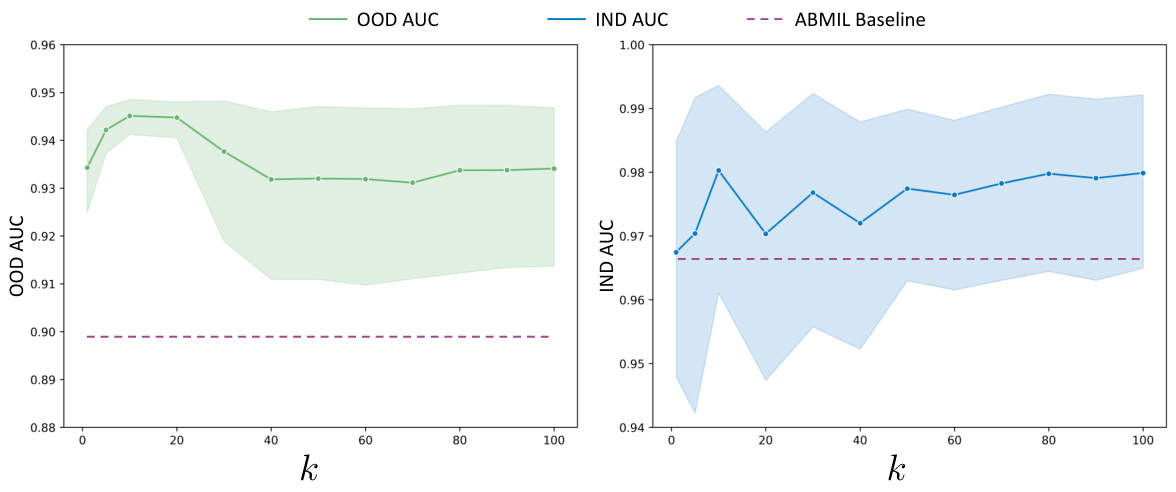
The concept of ‘Generalization Gains’ in a machine learning context, particularly within the realm of medical image analysis, refers to the improvement in a model’s ability to perform well on unseen data that differs from its training data. In this specific scenario of pathology foundation models, generalization gains would be observed if a model trained on one set of whole slide images (WSIs) from a particular hospital or using a specific staining technique, performs well on WSIs from other sources or with different staining methods. This improvement often signifies that the model has learned underlying, transferable features rather than just memorizing specific characteristics of the training data. Factors contributing to significant generalization gains might include robust model architecture, extensive pretraining on diverse datasets, and employing techniques that mitigate overfitting, such as regularization or data augmentation. Measuring these gains usually involves evaluating performance on held-out datasets representing different distributions (out-of-distribution generalization) and comparing the results to models that do not exhibit such robust performance. Strong generalization is crucial in medical applications due to the inherent variability across different sources of medical images and the risk of applying models effectively in practice that have been trained on data from specific sites or patient groups.
Future of CATE#
The future of Concept Anchor-guided Task-specific Feature Enhancement (CATE) looks promising, given its demonstrated ability to improve the performance and generalizability of MIL models in computational pathology. Further research could explore expanding CATE’s application beyond cancer subtyping to other downstream tasks, such as survival prediction or treatment response. Improving the efficiency of concept anchor generation is crucial; exploring alternative methods like larger language models (LLMs) or active learning could streamline the process and enhance scalability. Investigating CATE’s performance on diverse WSI datasets is also vital to ascertain its robustness across different imaging modalities, staining protocols, and acquisition settings. A deeper understanding of the interplay between the CIB and CFI modules will help fine-tune the model architecture and potentially unlock improved performance. Finally, exploring the integration of CATE with other feature enhancement techniques will determine the limits of performance improvement and identify promising avenues for future enhancement.
More visual insights#
More on figures
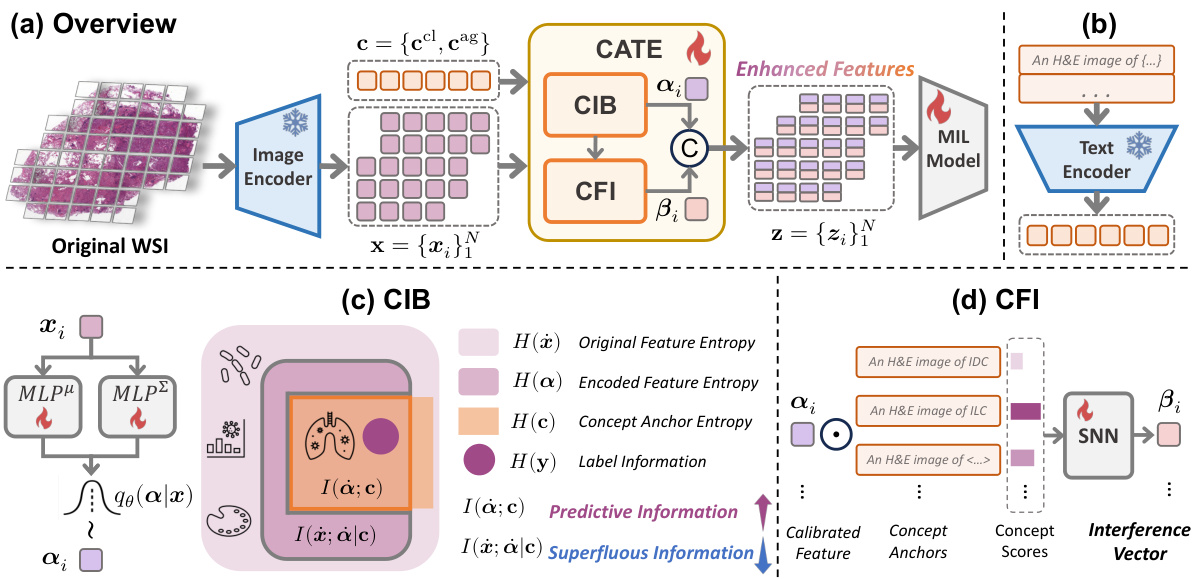
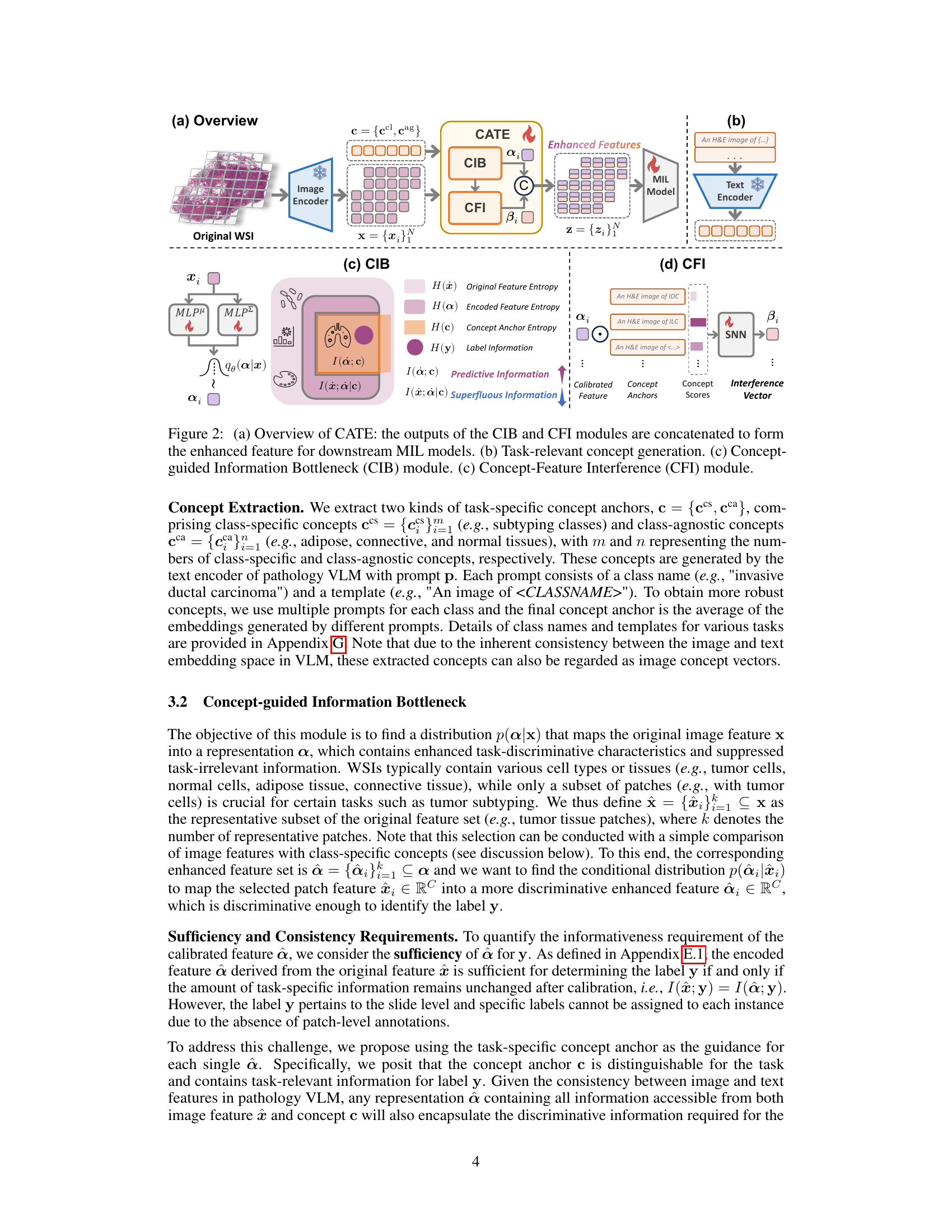
This figure illustrates the overall architecture of the Concept Anchor-guided Task-specific Feature Enhancement (CATE) framework. Panel (a) shows the flow of the process: original WSI patches are passed through an image encoder, then the CIB and CFI modules process the features before being fed into a MIL model. Panel (b) shows the generation of task-relevant concepts using a text encoder. Panels (c) and (d) detail the inner workings of the CIB and CFI modules, respectively, showing how they process features and concept anchors to generate enhanced, task-specific features.

This figure presents a comparative analysis of CATE-MIL and ABMIL using attention heatmaps and UMAP visualizations. It highlights how CATE-MIL improves the focus on cancerous regions (as seen in the heatmaps (a) and (b)). The similarity plots (c) and (d) demonstrate that CATE-MIL aligns features more effectively with concept anchors, particularly for cancerous areas, leading to a clearer separation of cancerous and non-cancerous regions in the UMAP visualization (f).

This figure shows a comparison of attention heatmaps and UMAP visualizations for CATE-MIL and ABMIL models applied to breast cancer WSIs. The heatmaps highlight the attention given to different regions of the image by each model, while the UMAP visualization shows how the models cluster features in a lower-dimensional space. Specifically, the figure illustrates how CATE-MIL enhances focus on cancerous regions and generates more discriminative features. Comparing (c) and (d) shows that CATE-MIL improves similarity between calibrated features and class-specific concept anchors, indicating better task-relevant feature extraction.
This figure shows a comparison of attention heatmaps and UMAP visualizations for CATE-MIL and the original ABMIL model. It highlights how CATE-MIL focuses attention on cancerous regions more effectively than ABMIL. The similarity plots (c, d) demonstrate that CATE-MIL aligns calibrated features more closely with the corresponding class concept features. The UMAP visualization shows that CATE-MIL produces feature embeddings that are more discriminative and closer to the respective class concept features than those produced by ABMIL.
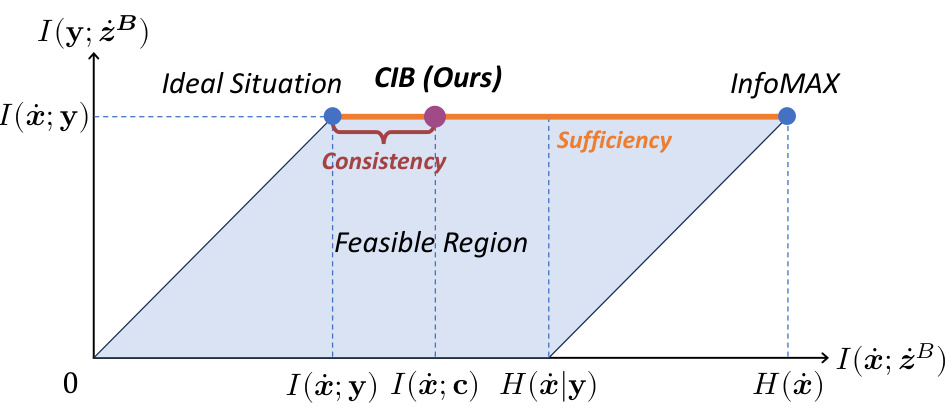
This figure illustrates the CATE framework, showing the two main modules: CIB and CFI. The CIB module calibrates the original image features using concept anchors, while the CFI module generates discriminative task-specific features by leveraging similarities between calibrated features and concept anchors. The final enhanced features are then fed into a downstream MIL model.
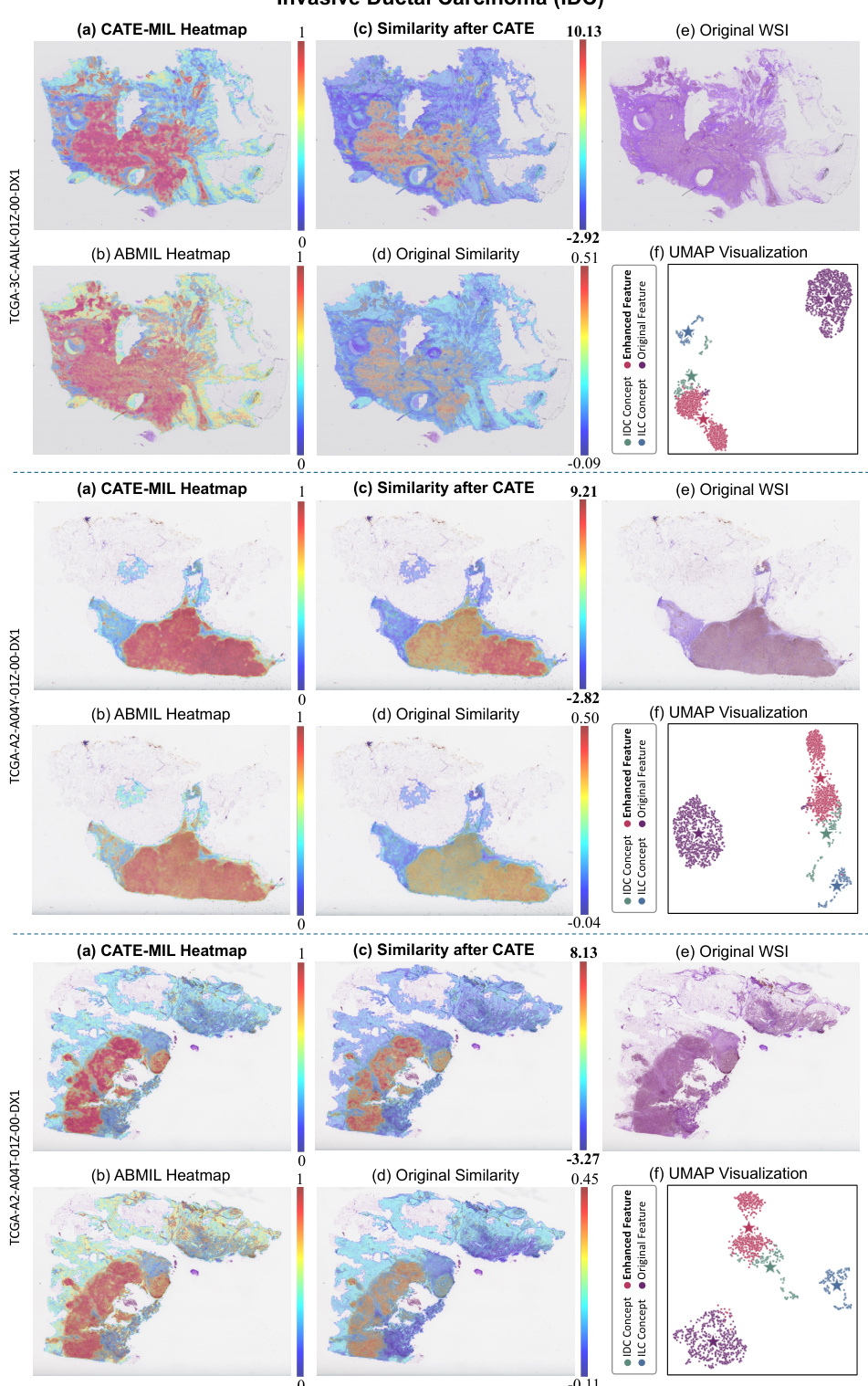
This figure shows a comparison of heatmaps and UMAP visualizations for CATE-MIL and ABMIL models applied to IDC and ILC cancer subtypes. The heatmaps illustrate the attention mechanisms in each model, highlighting areas of interest for classification. The UMAP visualizations show the clustering of feature representations from both models, helping to understand how well they distinguish between the subtypes. The similarity plots quantify the relationship between image features and concept anchors before and after calibration using CATE.

This figure provides a visual comparison of the attention heatmaps generated by CATE-MIL and ABMIL, highlighting the differences in focus on relevant image regions. It also shows the similarity scores (cosine similarity) between image features and class concept features, both before and after the CATE calibration process. Finally, it includes a UMAP visualization that shows how CATE enhances the clustering of features in the feature space, improving the separation of different classes.
More on tables
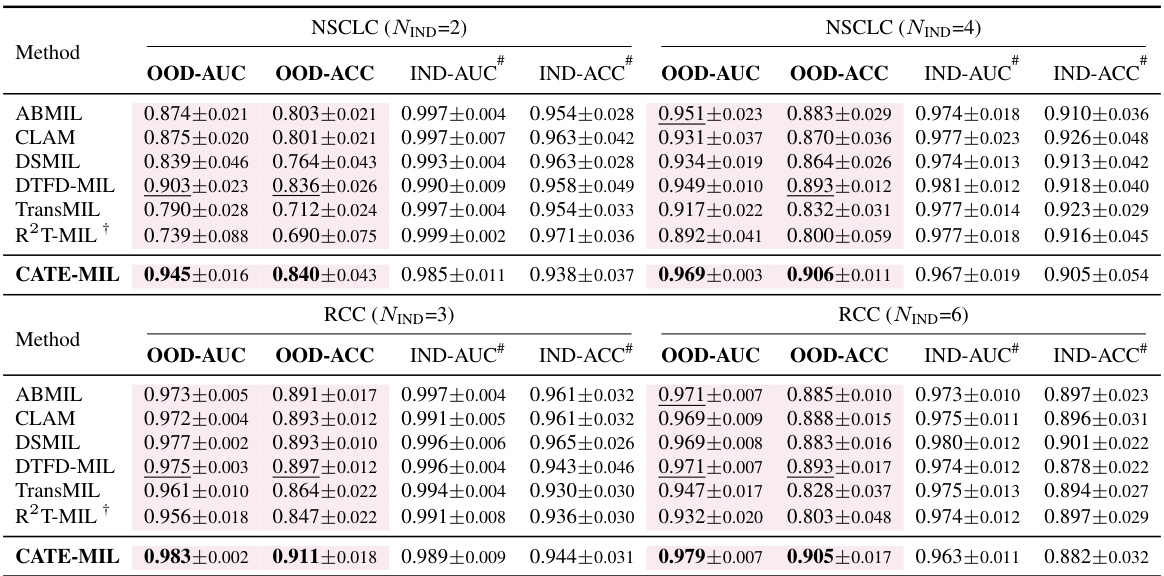
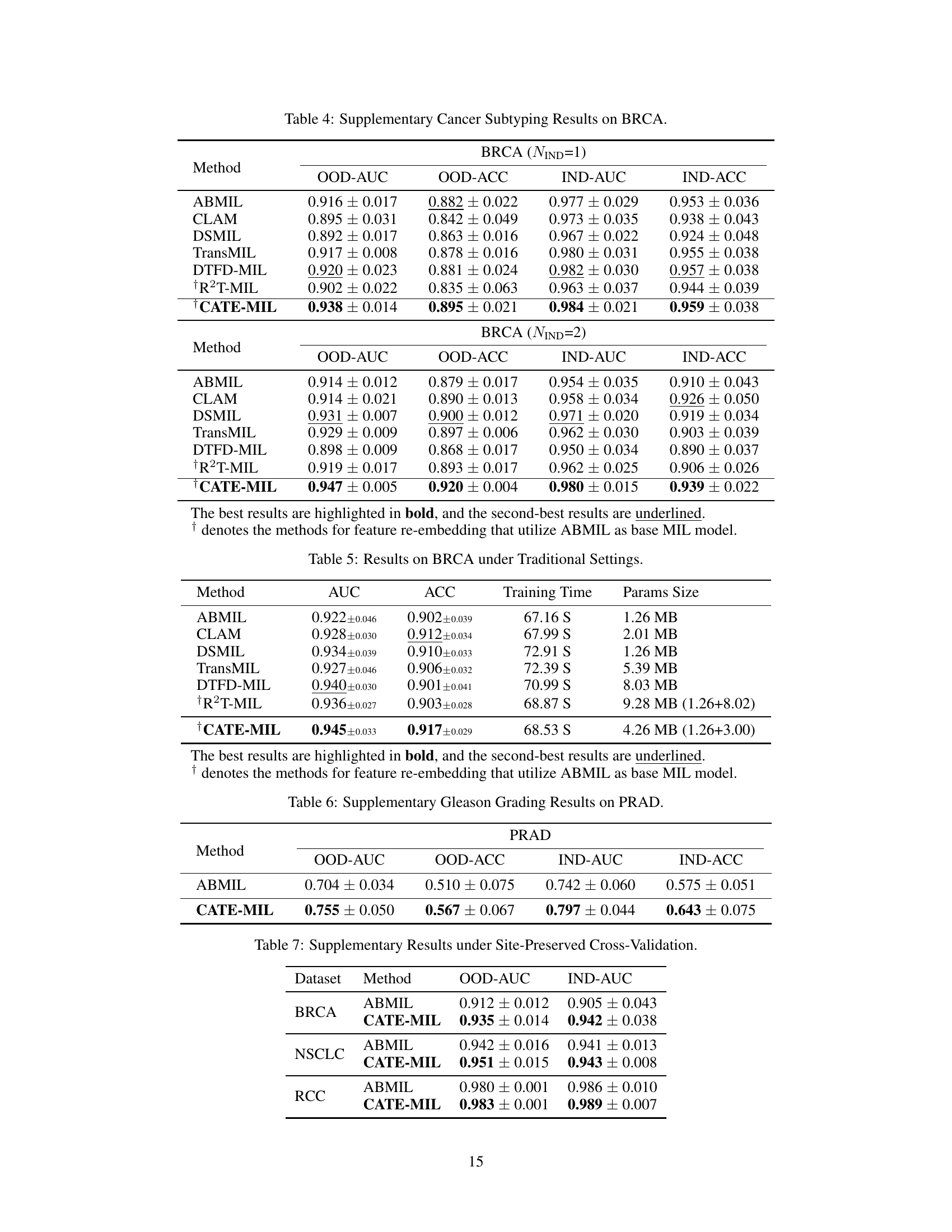
This table presents the results of cancer subtyping experiments using the BRCA dataset. Multiple Instance Learning (MIL) models, both with and without the Concept Anchor-guided Task-specific Feature Enhancement (CATE) method, were evaluated. The table shows the Area Under the Receiver Operating Characteristic curve (AUC) and accuracy metrics for both in-domain (IND) and out-of-domain (OOD) settings. The ‘Gain’ columns highlight the performance improvement achieved by incorporating CATE. In-domain samples are from the same sites, whereas out-of-domain samples represent different sites to assess model generalizability. The experiment was repeated with 1 and 2 in-domain sites (NIND=1, NIND=2).
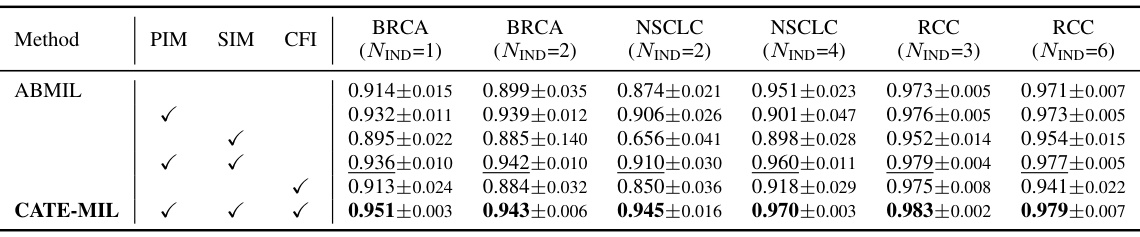
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models, both with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. It shows the Area Under the ROC Curve (AUC) and accuracy for both in-domain (IND) and out-of-domain (OOD) settings. The ‘Gain’ columns show the performance improvement achieved by incorporating CATE. Different numbers of IND sites (NIND=1 and NIND=2) are explored to assess the impact of domain shift on model generalization.
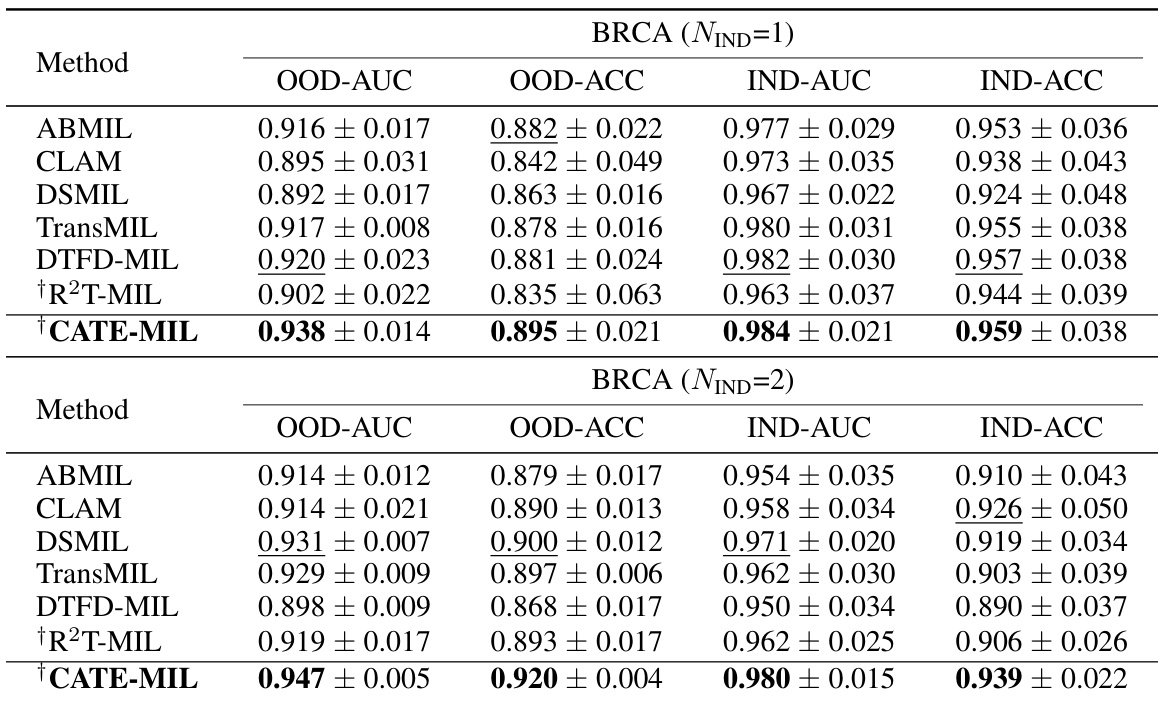
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models. It compares the performance of standard MIL models (ABMIL, CLAM, DSMIL, DTFD-MIL, TransMIL, R2T-MIL) to the same models enhanced with the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. Results are shown for both in-domain (IND) and out-of-domain (OOD) settings, illustrating the improvement in performance and generalization achieved by CATE. The metrics used are AUC (Area Under the Receiver Operating Characteristic Curve) and accuracy (ACC). NIND represents the number of in-domain sites used for training. Gain values indicate percentage improvements with CATE.
This table presents the results of cancer subtyping experiments on the BRCA dataset using several Multiple Instance Learning (MIL) models. The models were tested with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The table shows the Area Under the Receiver Operating Characteristic Curve (AUC) and Accuracy metrics for both in-domain (IND) and out-of-domain (OOD) settings. The performance gain achieved by using CATE with each model is also quantified. The table is split into two parts. The first part displays results when only one site is used as in-domain (IND) data for training, while the second part shows results when two sites are used as IND data. This allows for comparison of the model’s performance under different levels of domain diversity.
This table presents the results of cancer subtyping experiments using the BRCA dataset. It compares various Multiple Instance Learning (MIL) models, both with and without the Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The table shows the Area Under the Curve (AUC) and Accuracy scores for both in-domain (IND) and out-of-domain (OOD) settings. The OOD performance is particularly important as it indicates generalization ability. Results are shown for different numbers of in-domain sites (NIND=1 and NIND=2).
This table presents the results of cancer subtyping experiments on the BRCA dataset using multiple instance learning (MIL) models. It compares the performance of several MIL models (ABMIL, CLAM, DSMIL, DTFD-MIL, TransMIL, R2T-MIL) both with and without the proposed CATE method. Results are shown for both in-domain (IND) and out-of-domain (OOD) settings, with metrics including AUC and accuracy. The ‘Gain’ columns show the improvement in performance achieved by incorporating CATE. The table also shows the results with different numbers of in-domain sites (NIND=1 and NIND=2), illustrating the impact of domain adaptation on model generalizability.
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models. It compares the performance of several state-of-the-art MIL models with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The results are shown for both in-domain (IND) and out-of-domain (OOD) settings, with the gains in Area Under the Curve (AUC) and accuracy reported. The ‘NIND’ column indicates the number of sites used for in-domain training.

This table presents the results of cancer subtyping experiments on the BRCA dataset using multiple instance learning (MIL) models. It compares the performance of several MIL models (ABMIL, CLAM, DSMIL, DTFD-MIL, TransMIL, R2T-MIL) both with and without the integration of the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The results are shown for both in-domain (IND) and out-of-domain (OOD) settings, indicating the performance gains achieved by using CATE in terms of AUC and accuracy. The number of in-domain sites (NIND) is varied (NIND=1 and NIND=2) to evaluate the generalizability of the models across different data sources.
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models. It compares the performance of these models with and without the Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The results are shown separately for in-domain (IND) and out-of-domain (OOD) settings, reflecting the model’s ability to generalize to unseen data. Performance metrics include AUC (Area Under the ROC Curve) and accuracy (ACC). The table helps illustrate how CATE improves the performance and generalizability of the MIL models across various subtypes of breast cancer.
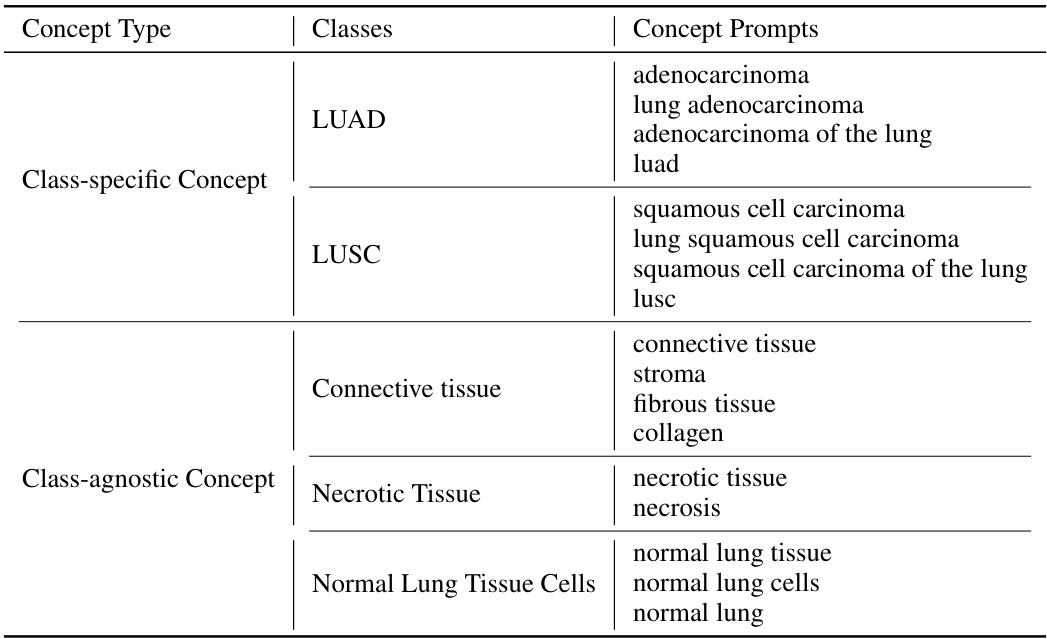
This table presents the results of cancer subtyping experiments on the BRCA dataset using various Multiple Instance Learning (MIL) models. The models are tested both with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. The results are compared using OOD-AUC (out-of-domain Area Under the ROC Curve), OOD-ACC (out-of-domain accuracy), IND-AUC (in-domain AUC), and IND-ACC (in-domain accuracy). The table shows the performance gain achieved by incorporating CATE for each MIL model, highlighting the effectiveness of the method in improving both in-domain and out-of-domain generalization.

This table presents the cancer subtyping results on the BRCA dataset for multiple instance learning (MIL) models with and without the proposed Concept Anchor-guided Task-specific Feature Enhancement (CATE) method. It shows the area under the receiver operating characteristic curve (AUC) and accuracy for both in-domain (IND) and out-of-domain (OOD) settings, highlighting the performance gains achieved by incorporating CATE. The results are broken down for different numbers of in-domain sites (NIND=1 and NIND=2) to demonstrate the impact of domain generalization.
Full paper#