↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Brain network analysis traditionally focuses on short-range connections, neglecting crucial long-range interactions that reflect long-distance communication. This limitation hinders a complete understanding of brain-wide information processing and impacts the accuracy of neurological disease diagnosis. Existing graph learning methods struggle to effectively capture these long-range dependencies, limiting their ability to provide a holistic view of brain connectivity.
This paper introduces ALTER, a novel brain graph transformer that specifically addresses this limitation. ALTER utilizes a biased random walk, guided by the correlation between brain regions, to explicitly model long-range dependencies. By integrating both short- and long-range dependencies within a transformer framework, ALTER provides a more comprehensive and accurate representation of brain communication. The extensive experiments on ABIDE and ADNI datasets demonstrate that ALTER surpasses state-of-the-art methods in neurological disease diagnosis, underscoring the importance of considering long-range connections for a more accurate and effective analysis.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in brain network analysis as it directly addresses the limitations of existing methods by incorporating long-range dependencies. It introduces a novel graph transformer model that significantly improves accuracy in neurological disease diagnosis, setting a new benchmark and opening avenues for further research in brain connectivity analysis using graph neural networks and transformers.
Visual Insights#
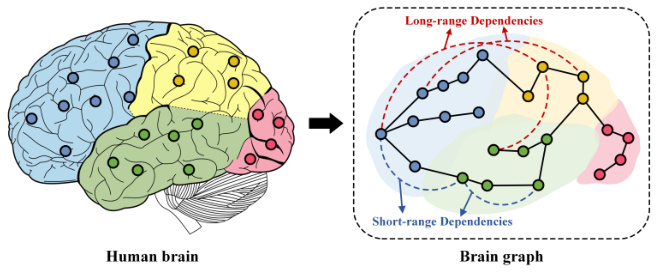
This figure illustrates the concept of short-range and long-range dependencies in brain networks. The left panel shows a simplified representation of the human brain, divided into regions with different colors representing different brain regions of interest (ROIs). Nodes (dots) within each region represent specific areas within that region. The right panel depicts a brain graph where nodes represent ROIs and edges represent the connections between them. The black solid lines represent short-range connections within nearby ROIs (local connectivity). Dashed and colored arcs show long-range connections bridging distant ROIs. This visual representation highlights that brain communication involves both local interactions and extensive long-distance connections crucial for integrating information across the entire brain.
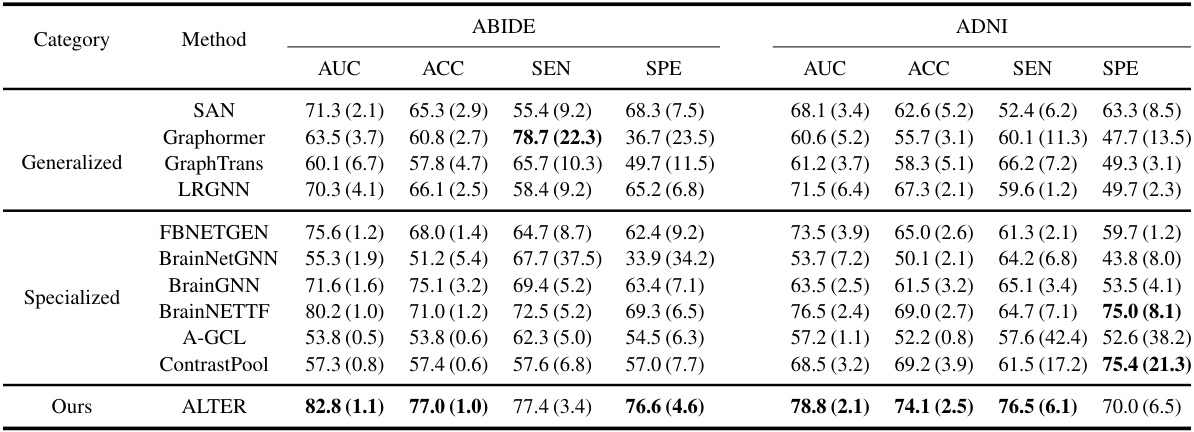
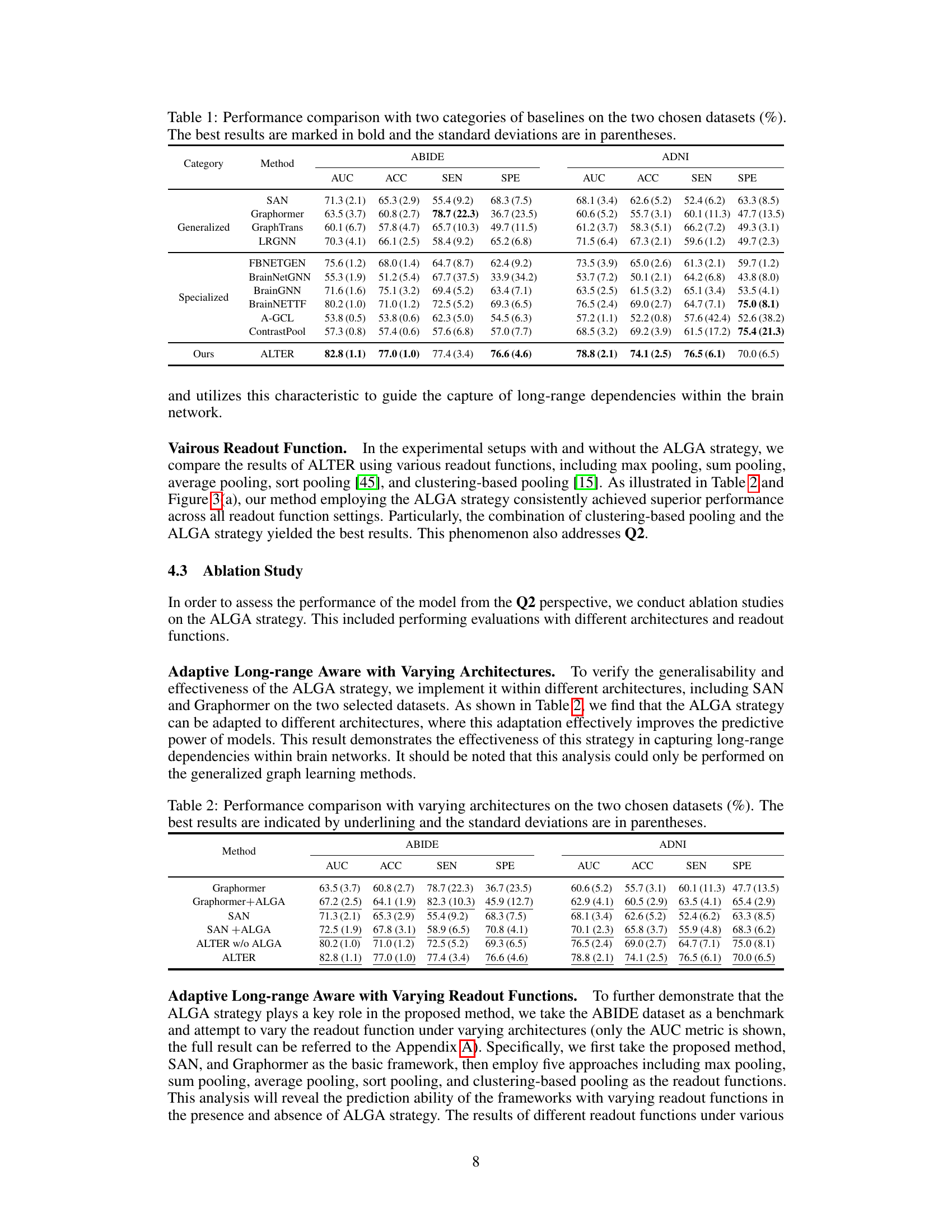
This table compares the performance of the proposed ALTER model against various baseline methods for brain network analysis on two datasets (ABIDE and ADNI). It shows the AUC, ACC, SEN, and SPE for each method, categorized into generalized graph learning methods and specialized brain graph methods. The best results for each metric on each dataset are highlighted in bold, demonstrating the superior performance of ALTER compared to other methods.
In-depth insights#
Long-Range Dependency#
The concept of “Long-Range Dependency” in brain networks is crucial, as it highlights the importance of connections between distant brain regions in information processing. Traditional methods often focus on local connections, neglecting the significant role of long-range interactions in complex brain functions. The existence of long-range dependencies challenges the simplistic view of brain connectivity as solely localized, emphasizing the need for models that can capture both short-range and long-range interactions. Understanding these long-range effects is vital for unraveling the intricacies of brain function and dysfunction, particularly in neurological disorders. Methods employing strategies like biased random walks are promising avenues to model these complex dependencies, helping bridge the gap between observed brain activity and the underlying neural mechanisms. Future research should focus on more sophisticated models that can effectively incorporate these long-range interactions within a holistic framework for a comprehensive understanding of brain dynamics.
ALTER Model#
The Adaptive Long-range aware TransformER (ALTER) model is a novel brain graph transformer designed to address the limitations of existing methods in brain network analysis by explicitly incorporating long-range dependencies between brain regions of interest (ROIs). ALTER uses a biased random walk strategy, guided by the correlation between ROIs, to capture these long-range dependencies effectively. This is a significant improvement over traditional methods that primarily focus on short-range connections. The integration of these long-range dependencies with short-range ones within the transformer framework provides a more comprehensive and accurate representation of brain-wide communication. Extensive experiments on the ABIDE and ADNI datasets demonstrate that ALTER consistently outperforms state-of-the-art graph learning methods, highlighting the model’s effectiveness in neurological disease diagnosis. The model’s success underscores the importance of considering long-range interactions for a fuller understanding of complex brain processes.
ALGA Strategy#
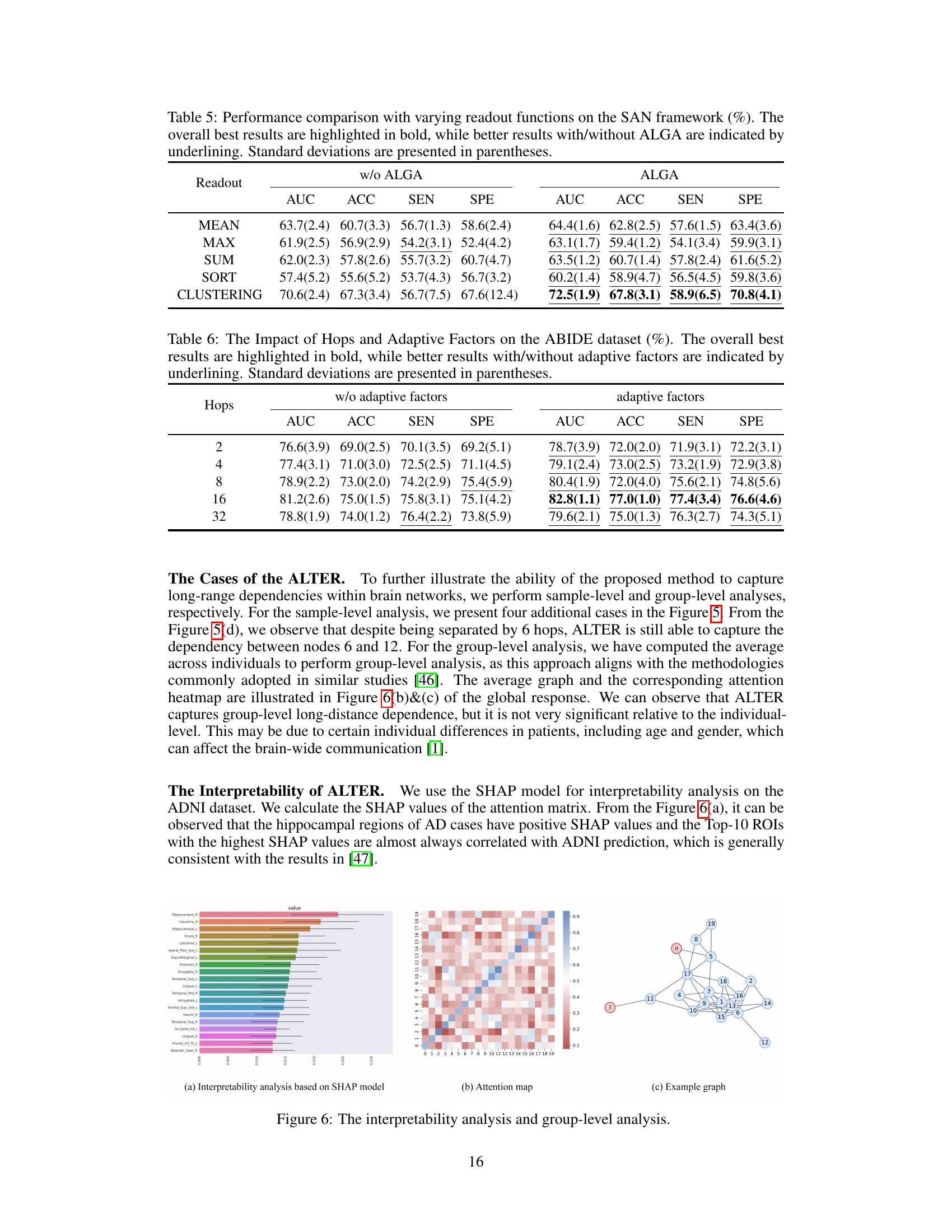
The Adaptive Long-range Aware (ALGA) strategy is a crucial component of the ALTER model, designed to address the limitations of existing brain network analysis methods that primarily focus on short-range dependencies. ALGA leverages biased random walks to explicitly capture long-range dependencies, moving beyond the limitations of uniform probability transitions. By incorporating adaptive factors that reflect varying communication strengths between brain regions of interest (ROIs), the random walk is guided towards next hops with higher correlation values, effectively mimicking real-world brain communication patterns. This approach generates long-range embeddings that effectively encode the long-distance communication connectivity within the brain network. The integration of these embeddings into a transformer framework allows for the adaptive integration of both short- and long-range dependencies, ultimately contributing to a more holistic and accurate representation of brain-wide communication. This innovative approach is shown to significantly improve performance in neurological disease diagnosis tasks.
Brain Graph Transformer#
The concept of a ‘Brain Graph Transformer’ represents a significant advancement in neuroimaging analysis. It leverages the power of graph neural networks to model the complex relationships between brain regions, treating the brain as a graph where nodes represent regions of interest (ROIs) and edges represent the strength of connections between them. The integration of transformer architectures allows the model to capture both local and long-range dependencies within this brain graph, something traditional methods often struggle with. This is crucial because brain function relies heavily on communication across both short and long distances. Long-range connections are particularly important for higher-level cognitive functions, and the transformer’s ability to attend to distant nodes in the graph is a key innovation. By using attention mechanisms, this model can prioritize information from the most relevant ROIs, regardless of their physical proximity, and potentially provide a more holistic understanding of brain activity than previous methods. The use of this architecture opens possibilities for improved accuracy in neurological disease diagnosis, prediction of cognitive decline, and for understanding the functional organization of the brain itself.
Future Work#
The authors mention exploring optimal balance between short and long-range dependencies in brain network analysis. This is crucial because an over-reliance on either might hinder the accuracy and generalizability of the model. Further research could investigate the impact of diverse data modalities beyond fMRI, such as DTI, to enrich model understanding and improve robustness. Addressing the potential for bias stemming from individual differences (age, gender) in long-range communication strengths is also a key area. The paper also acknowledges the limitations of solely relying on Pearson correlation coefficient for calculating adaptive factors and suggests exploring alternative methods to refine this process. Expanding the methodology to other brain network analysis tasks, beyond neurological disease diagnosis, such as sex prediction or cognitive function analysis, is another avenue for future work. Ultimately, extending the work into real-world clinical settings, validating the model’s generalizability and ensuring its clinical utility are critical steps towards broad implementation.
More visual insights#
More on figures
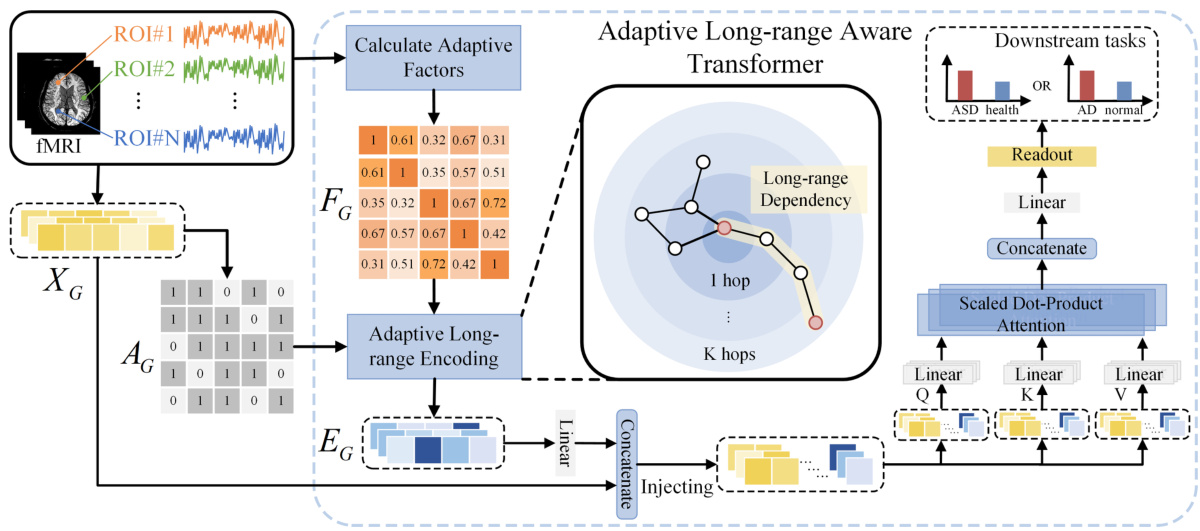
The figure illustrates the overall framework of the proposed ALTER model for brain network analysis. It shows the process of extracting node features and adjacency matrix from fMRI data, calculating adaptive factors, performing adaptive long-range encoding using biased random walks based on the adaptive factors, injecting the long-range embeddings into a transformer framework, integrating short- and long-range dependencies using the self-attention mechanism, and generating the graph-level representation for downstream tasks such as neurological disease diagnosis. The figure visually represents the data flow and processing stages of the ALTER model.
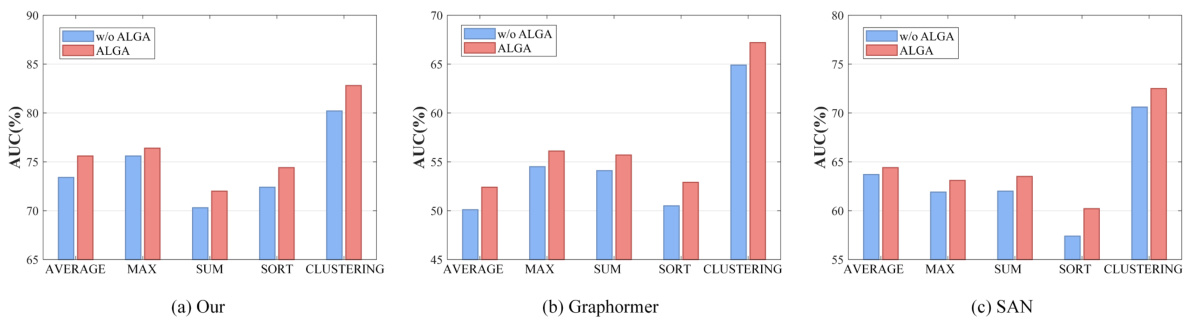
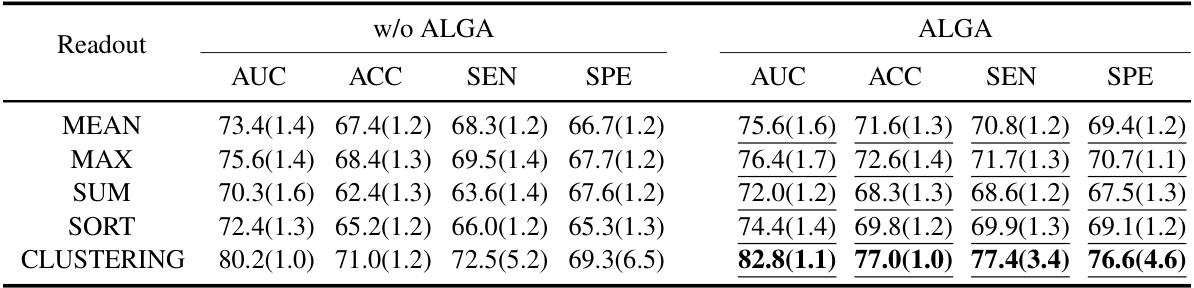
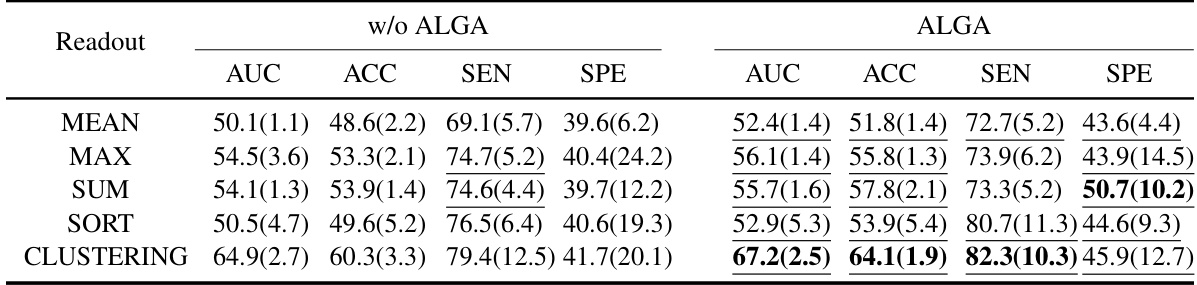
This figure compares the performance of different readout functions (Average, Max, Sum, Sort, Clustering) in three different model architectures (ALTER, Graphormer, and SAN) with and without the Adaptive Long-range Aware (ALGA) strategy. The y-axis represents the AUC (Area Under the Curve), a metric used to evaluate the performance of a classification model. The results show that the ALGA strategy consistently improves the AUC across all readout functions and model architectures, indicating its effectiveness in improving model performance.
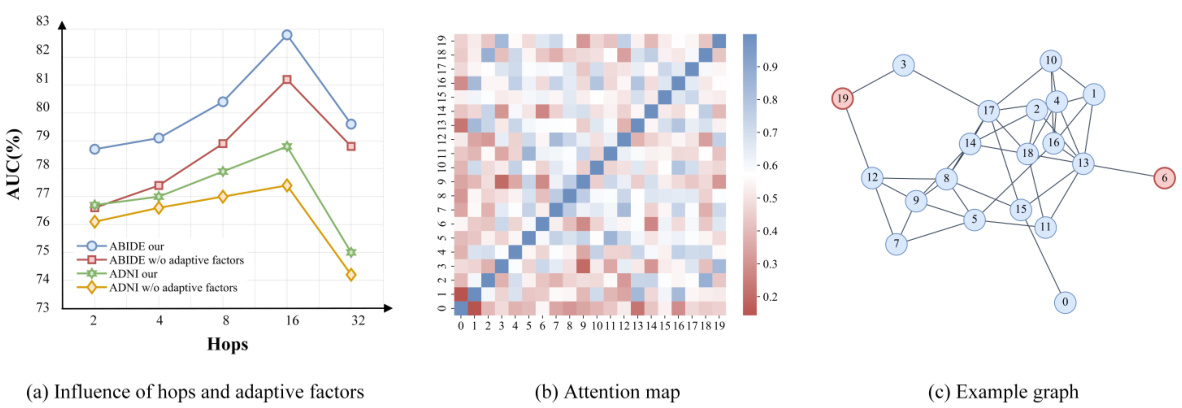
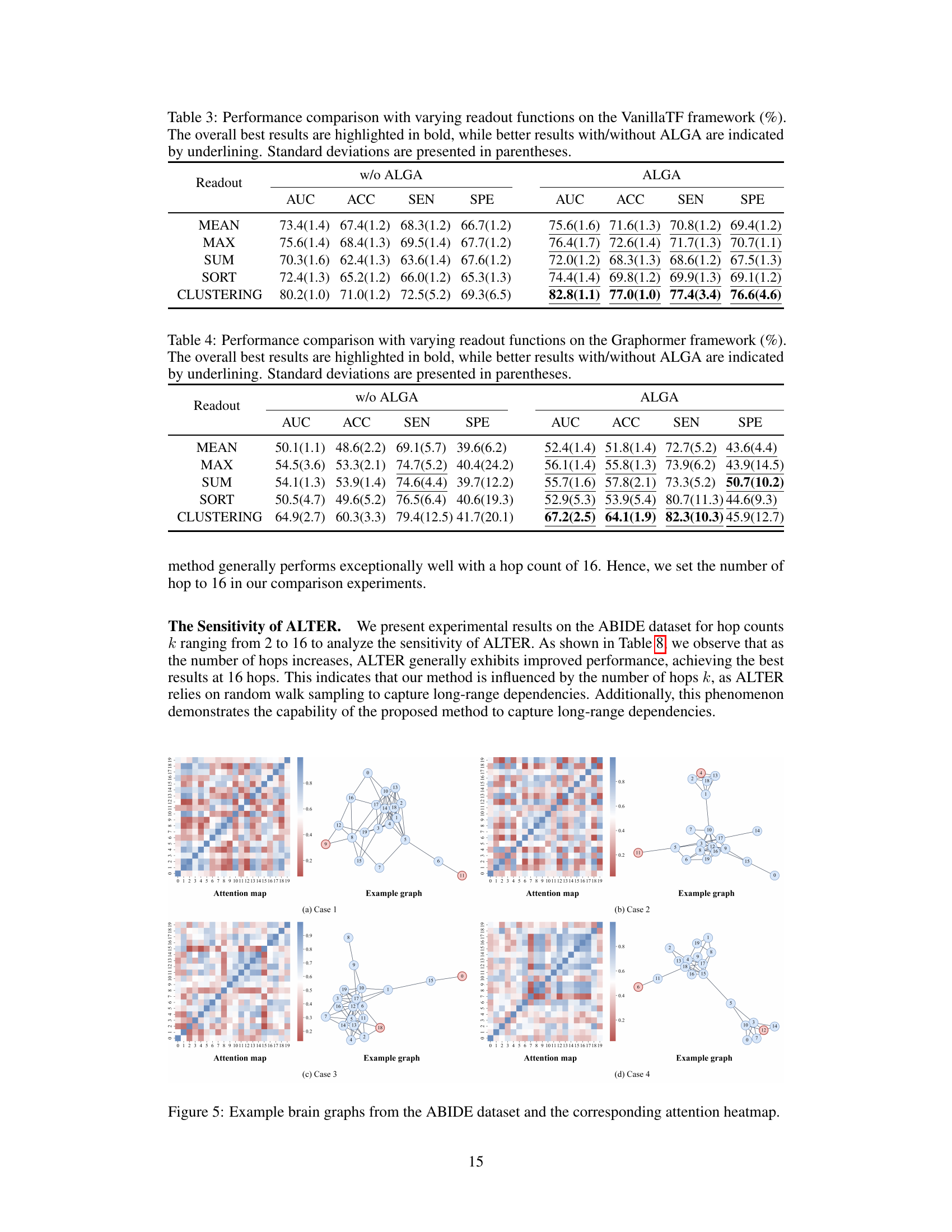
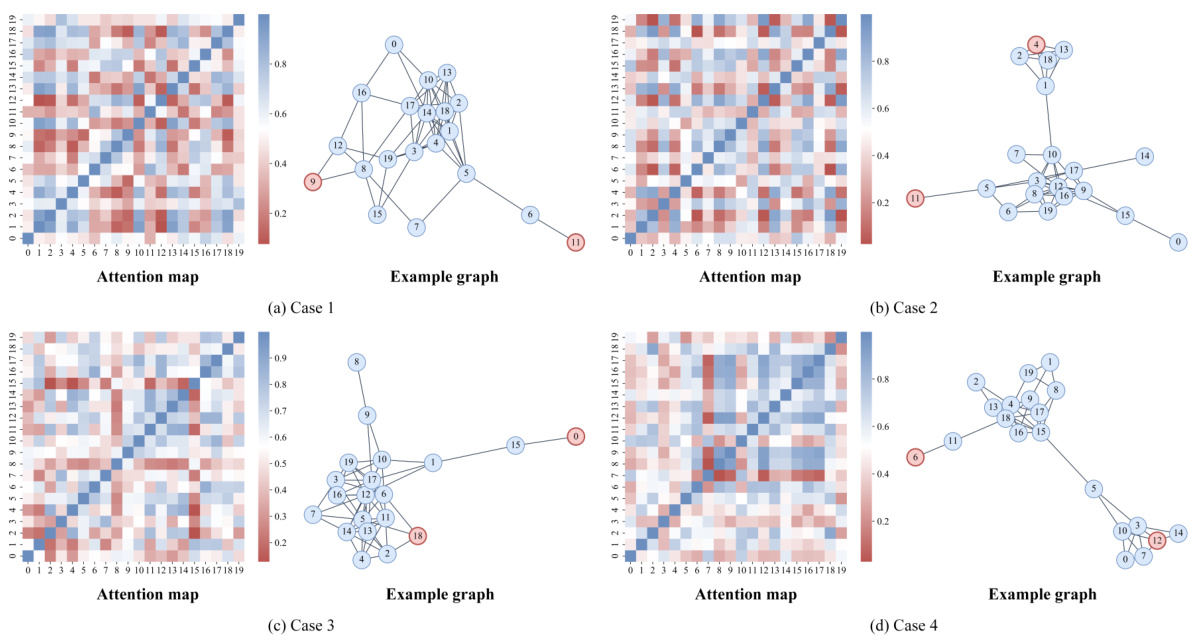
This figure presents an in-depth analysis of the ALTER model and its adaptive long-range aware (ALGA) strategy. Subfigure (a) shows the impact of the number of hops in the ALGA strategy on the model’s performance, demonstrating that increasing the number of hops generally improves the model’s predictive power for both ABIDE and ADNI datasets. Subfigure (b) displays an attention heatmap illustrating the communication patterns between brain regions of interest (ROIs). Subfigure (c) shows an example brain graph used in the analysis, highlighting long-range dependencies captured by the ALTER model.
This figure shows the overall framework of the proposed ALTER model. It details the process, starting with fMRI data as input, through the calculation of adaptive factors, the adaptive long-range encoding using a biased random walk, injecting those long-range embeddings into a transformer framework, and finally using a readout function to produce the final output for downstream tasks such as neurological disease diagnosis. The diagram visually represents the flow of information and the key components of the ALTER model.

This figure illustrates the overall framework of the Adaptive Long-range aware TransformER (ALTER) model. It shows the process flow, starting with fMRI data as input. Adaptive factors (FG) are calculated from the fMRI data to represent communication strength between brain Regions of Interest (ROIs). These factors are used in an adaptive long-range encoding process to generate long-range dependency embeddings (EG). These embeddings are concatenated with the original node features (XG) and fed into a long-range aware transformer. The transformer incorporates both short- and long-range dependencies and produces a graph-level representation (ZG) used for downstream tasks (e.g., disease prediction).
More on tables
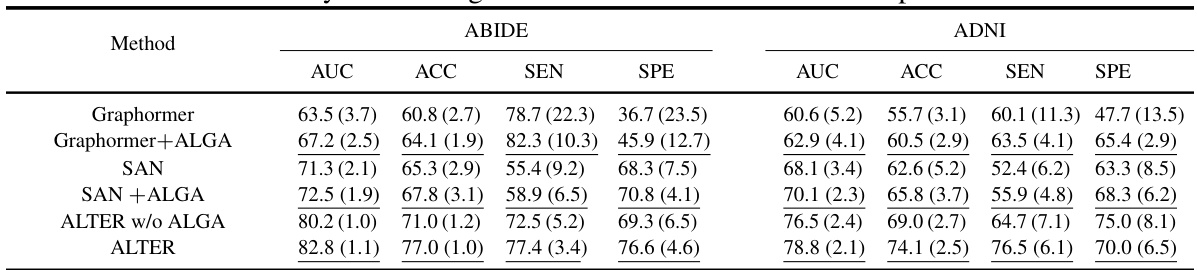
This table compares the performance of the proposed ALTER method with two sets of baseline methods: generalized graph learning methods and brain graph-based methods. The comparison is done on two datasets, ABIDE and ADNI, using metrics AUC, ACC, SEN, and SPE to evaluate the performance of each method in neurological disease diagnosis. The best results for each metric on each dataset are highlighted in bold, and standard deviations are shown in parentheses to indicate the variability in performance across multiple runs.
This table compares the performance of the proposed ALTER model against two categories of baseline methods on two datasets (ABIDE and ADNI). The first category includes generalized graph learning methods applicable to various graph types, while the second category consists of methods specifically designed for brain network analysis. The table shows the AUC, ACC, SEN, and SPE metrics for each method on each dataset. Higher values in bold indicate better performance. Parenthetical values show standard deviations.
This table presents a comparison of the proposed ALTER model’s performance against various baseline methods on two datasets: ABIDE and ADNI. The baselines are categorized into generalized graph learning methods (not specific to brain networks) and specialized brain graph-based methods. The table shows the AUC, ACC, SEN, and SPE for each method on each dataset, highlighting the superior performance of ALTER in bold. Parentheses indicate standard deviations.

This table presents a comparison of the proposed ALTER model against other state-of-the-art methods for two brain network analysis datasets (ABIDE and ADNI). It shows the performance of different models, grouped into generalized graph learning methods and specialized brain graph methods, across various evaluation metrics (AUC, ACC, SEN, SPE). The best performing model for each metric and dataset is highlighted in bold, with standard deviations reported in parentheses to show the variability of the results. The table helps to demonstrate the superior performance of ALTER.
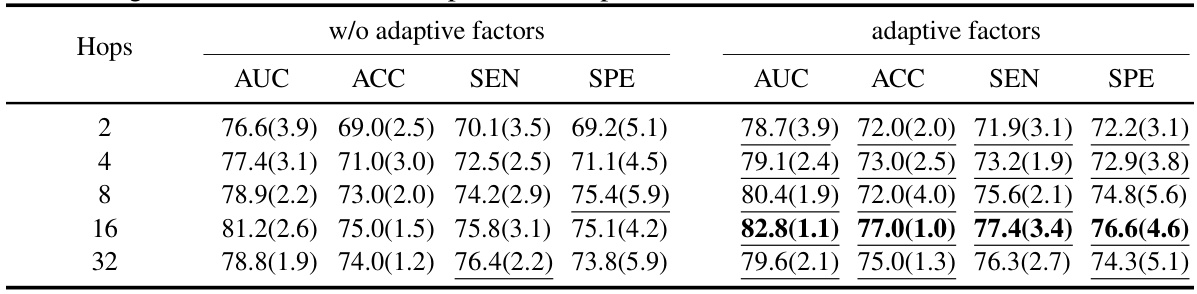
This table compares the performance of the proposed ALTER model against two groups of baseline methods: generalized graph learning methods and brain graph-based methods. The comparison is performed on two datasets, ABIDE and ADNI, using several metrics: AUC, ACC, SEN, and SPE. The best-performing model for each metric in each dataset is highlighted in bold, and standard deviations are included to illustrate variability in the results.

This table compares the performance of the proposed ALTER model against other state-of-the-art graph learning methods for brain network analysis. It shows the Area Under the Curve (AUC), Accuracy (ACC), Sensitivity (SEN), and Specificity (SPE) for two datasets: ABIDE and ADNI. The results are categorized into ‘Generalized’ and ‘Specialized’ methods. The ‘Generalized’ methods are general graph learning methods, not specifically designed for brain networks. The ‘Specialized’ methods are those explicitly designed for brain network analysis. ALTER consistently outperforms all other methods in terms of AUC, ACC, SEN, and SPE across both datasets.

This table presents the performance comparison of the proposed ALTER model against various baseline methods on two datasets, ABIDE and ADNI. It shows the AUC, ACC, SEN, and SPE for each method across both datasets. The best-performing method for each metric on each dataset is highlighted in bold, allowing for a direct comparison of ALTER’s performance against existing state-of-the-art and specialized brain network analysis methods. Standard deviations are included to give a sense of the variability in results.
Full paper#