TL;DR#
Co-crystallization, combining a drug with a coformer, is a promising technique to enhance drug properties like solubility and tabletability. However, finding suitable coformer-drug combinations is challenging due to the vast chemical space and complex experimental screening processes. This significantly hinders drug development, making efficient design methods crucial.
This research introduces GEMCODE, a novel pipeline that leverages the power of AI to address this challenge. GEMCODE combines deep generative models for exploring chemical space, machine learning models for predicting co-crystal properties (like tabletability), and evolutionary optimization to refine designs. The results demonstrate GEMCODE’s effectiveness in generating numerous potentially useful co-crystals, showing its potential to significantly accelerate the drug discovery process.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in pharmaceutical science and AI. It introduces GEMCODE, a novel AI-driven method to design co-crystals with improved tabletability, a significant challenge in drug development. This opens new avenues for accelerating drug development and offers a valuable tool for researchers working on related problems.
Visual Insights#
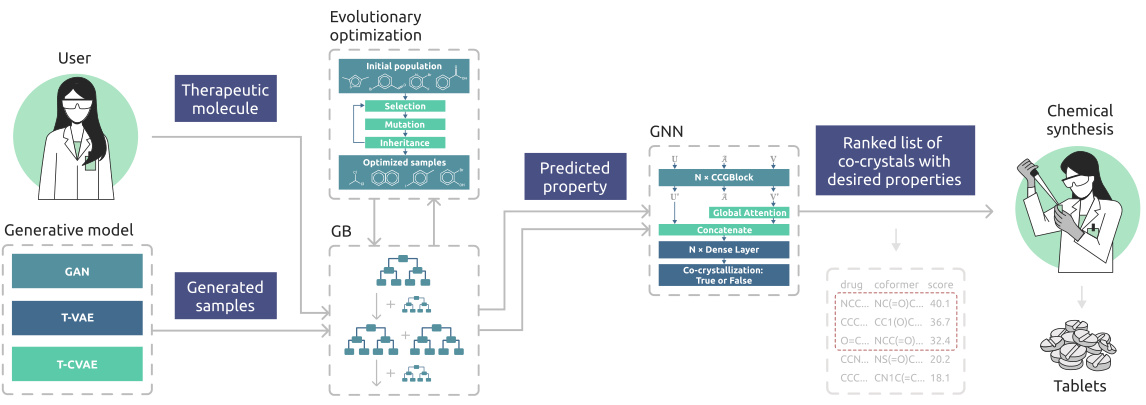
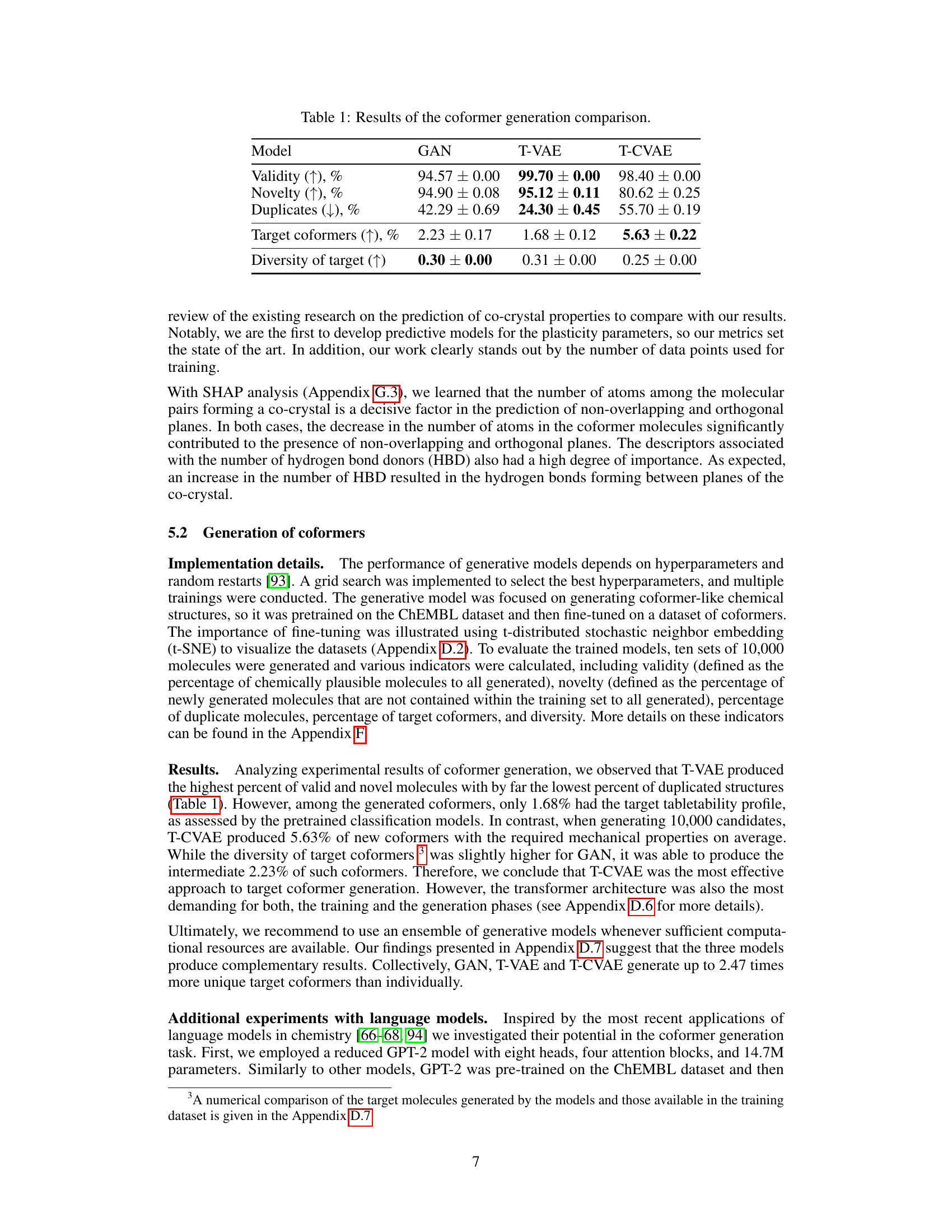
🔼 This figure illustrates the GEMCODE pipeline, which starts with a therapeutic molecule as input. A generative model (GAN, T-VAE, or T-CVAE) generates potential coformers. These coformers are then evaluated using gradient boosting (GB) models to predict their mechanical properties relevant to tabletability. An evolutionary optimization algorithm refines these coformers to improve tabletability, and finally, a graph neural network (GNN) ranks the resulting co-crystals based on their probability of formation and desired properties. The process culminates in a ranked list of co-crystals suitable for chemical synthesis into tablets.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.

🔼 This table presents a comparison of the performance of three different generative models (GAN, T-VAE, and T-CVAE) in generating coformers. The metrics used for comparison include validity (percentage of chemically plausible molecules), novelty (percentage of novel molecules not found in the training set), duplicates (percentage of duplicate molecules), the percentage of target coformers (coformers with desired properties), and diversity of target (diversity among target coformers). The results show that T-CVAE outperforms the other models in generating target coformers, although it is also the most computationally expensive.
read the caption
Table 1: Results of the coformer generation comparison.
In-depth insights#
AI Co-crystal Design#
AI is revolutionizing co-crystal design by automating the traditionally laborious process of screening potential coformers. Generative models, trained on vast chemical databases, are employed to propose novel coformers tailored to specific drug molecules. These models predict physicochemical properties, including crucial tabletability parameters, allowing researchers to focus on promising candidates. Evolutionary algorithms further optimize these designs, refining properties like plasticity and mechanical strength for improved tablet formation. Integrating graph neural networks allows prediction of successful co-crystal formation probabilities, further streamlining the design process. This integrated AI approach significantly accelerates drug development by enabling rapid exploration of a much larger chemical space than previously possible, ultimately leading to faster discovery and optimization of co-crystals with enhanced properties.
GEMCODE Pipeline#
The GEMCODE pipeline is a novel, automated approach for designing pharmaceutical co-crystals with enhanced tabletability. It cleverly integrates several powerful AI techniques, starting with deep generative models (GAN, T-VAE, T-CVAE) to create a diverse pool of potential coformer candidates. These candidates are then evaluated using machine learning models that predict crucial mechanical properties related to tabletability. A crucial step is the evolutionary optimization which refines the coformer candidates to optimize their predicted properties. Finally, a graph neural network (GNN) ranks the resulting co-crystal pairs based on their probability of formation. This multi-stage pipeline effectively explores a vast chemical space to identify promising co-crystals, accelerating the drug development process and addressing a significant challenge in pharmaceutical formulation.
ML Property Prediction#
Machine learning (ML) techniques are increasingly used to predict the properties of co-crystals, reducing the need for extensive and costly experimental screening. The accuracy of these predictions is crucial for the success of co-crystal design and development. Current ML models primarily focus on predicting properties such as lattice energy, density, melting point, solubility, and stability. However, a limitation is often the limited size of available datasets, limiting the ability to train robust predictive models with broader applicability. A significant challenge lies in effectively incorporating complex structural information into ML models, as this is key to accurately predicting co-crystal properties and accurately modeling the impact of coformers on the drug molecule. Future work should explore larger, more diverse datasets and improved ML architectures that incorporate detailed structural information to improve predictive accuracy and expand the range of predictable properties. This could include mechanical properties, which are particularly important for pharmaceutical applications, and other physicochemical properties such as bioavailability and permeability.
Evolutionary Approach#
An evolutionary approach in the context of a research paper on co-crystal design using AI would likely involve employing genetic algorithms or similar methods to optimize the properties of generated co-crystals. The core idea is to treat the design process as a form of natural selection, where a population of potential co-crystal structures is generated, evaluated based on desired properties (e.g., tabletability, solubility), and iteratively improved through processes mirroring mutation, crossover, and selection. This approach allows for exploration of a vast chemical space beyond the capabilities of traditional methods and could be especially valuable when dealing with complex interactions between drug molecules and coformers, leading to better identification of superior co-crystals.
Future Directions#
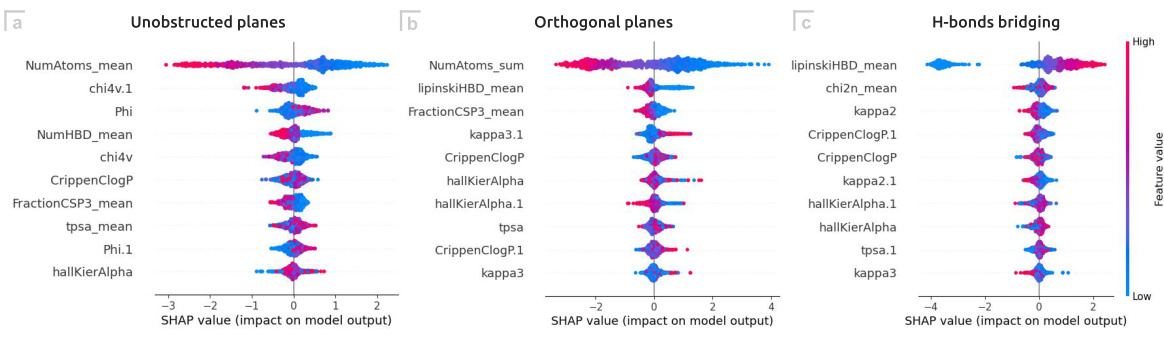
Future research should focus on expanding the GEMCODE dataset to encompass a wider range of drug molecules and coformers, thus enhancing the model’s generalizability and predictive capabilities. Addressing the current bias toward predicting the absence of orthogonal planes in the GB model is crucial, potentially through exploring alternative model architectures or feature engineering techniques. Investigating the impact of polymorphism on the predicted mechanical properties of co-crystals is essential, requiring the development of new methods to handle the complexities of polymorphic forms and their different physical properties. Finally, further exploration of language models for coformer generation, focusing on higher-capacity models with appropriate fine-tuning and data curation, could significantly improve the efficiency and diversity of the generated molecules, ultimately leading to the discovery of innovative co-crystals for pharmaceutical applications.
More visual insights#
More on figures
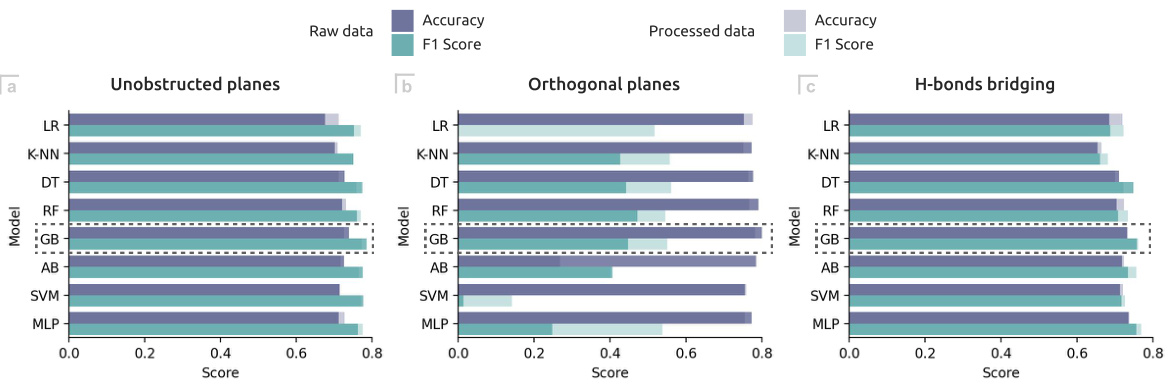
🔼 This figure shows the performance of various machine learning models in predicting three mechanical properties related to co-crystal tabletability: Unobstructed planes, Orthogonal planes, and H-bonds bridging. The performance is measured using Accuracy and F1-score, both before and after feature engineering and selection steps. This allows comparison of model performance with and without data preprocessing.
read the caption
Figure 2: Accuracy and F1 score metrics for the ML models predicting three mechanical properties of co-crystals. (a) Unobstructed planes. (b) Orthogonal planes. (c) H-bonds bridging. The performance of each model is shown before ('Raw data') and after ('Processed data') the feature engineering and feature selection steps.
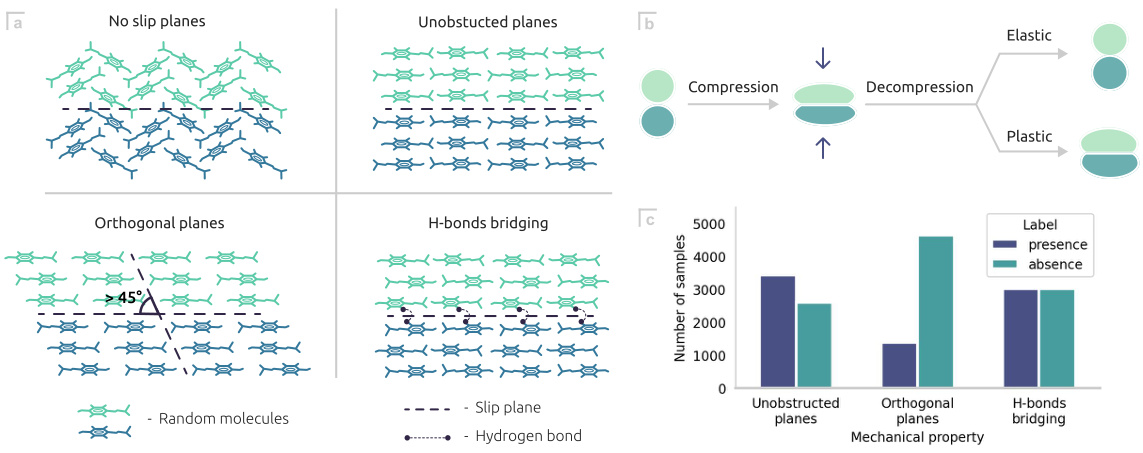
🔼 This figure shows a schematic representation of the mechanical properties of co-crystals, illustrating how the presence or absence of slip planes and hydrogen bonds affects tabletability. It also includes a schematic of particle deformation during powder compression and a bar chart showing the number of coformer samples for each mechanical property.
read the caption
Figure 3: (a) Schematic representation of the mechanical properties of co-crystals. No slip plane and H-bond bridging are associated with low tabletability. The other two properties positively correlate with tabletability. (b) Schematic representation of the particle deformation during powder compression. (c) Number of coformer samples of each category per mechanical property.

🔼 This figure illustrates three different ways of representing molecules: SMILES (Simplified Molecular Input Line Entry System), molecular fingerprints, and molecular descriptors. SMILES notation is a string-based representation of a molecule’s structure; molecular fingerprints are vectors representing the presence or absence of certain substructures; and molecular descriptors are numerical values describing various physicochemical properties. The figure uses caffeine (C8H10N4O2) as an example to show how each method represents the molecule.
read the caption
Figure 4: Molecular representation using the chemical structure of caffeine as an example in the form of SMILES, molecular fingerprints, and molecular descriptors.

🔼 This figure shows the results of training a Generative Adversarial Network (GAN) on two datasets: ChEMBL and a smaller dataset of coformers. Panel (a) is a plot showing the increase in the percentage of valid chemical structures generated by the GAN over the course of training. The validity increases significantly after the GAN is fine-tuned on the smaller coformer dataset. Panel (b) uses t-SNE to visualize the distribution of the molecules from both datasets in a lower-dimensional space. The plot shows a clear separation between the ChEMBL and coformer molecules, highlighting the distinct features and chemical spaces represented by each dataset.
read the caption
Figure 5: GAN training results on ChEMBL datasets and coformers: (a) plot of the growth of the valid chemical structures share in a batch, (b) t-SNE visualization of molecules from the ChEMBL dataset and coformers.
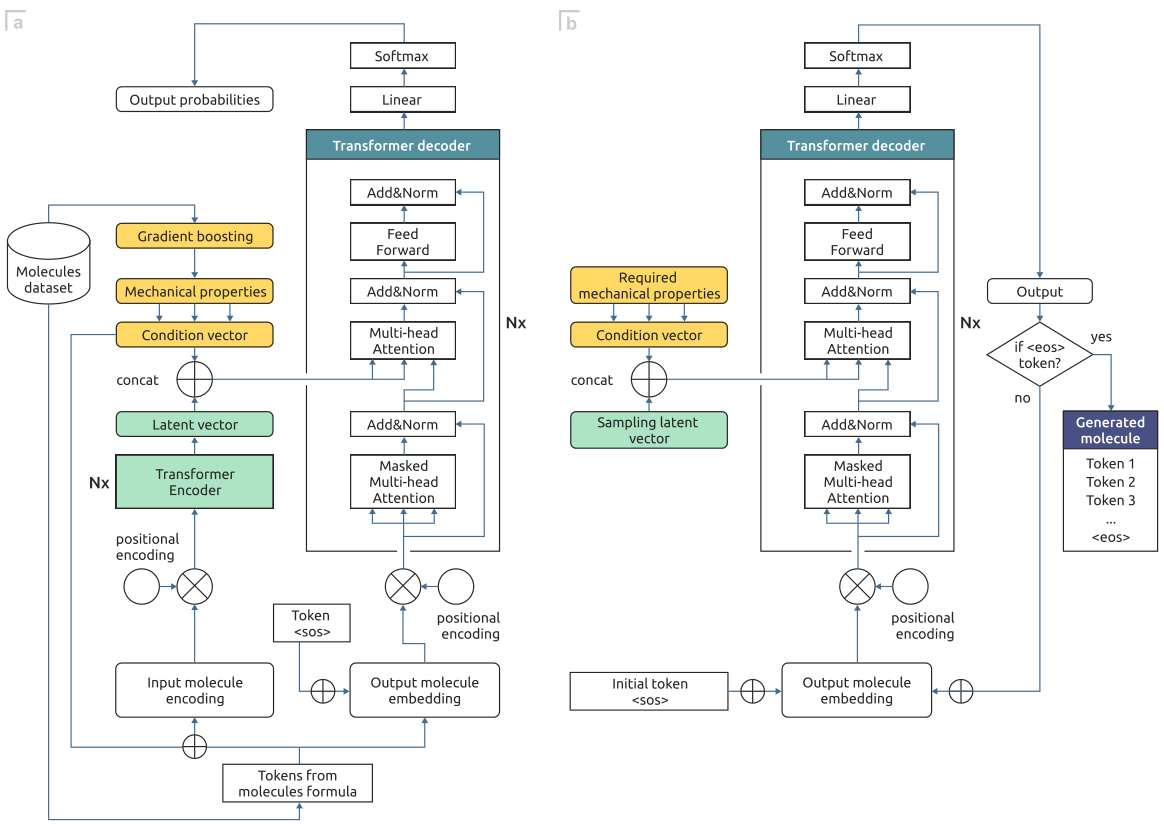
🔼 This figure illustrates the GEMCODE pipeline, which is a hybrid approach for designing cocrystals. It consists of several stages: The first is the generation of coformer candidates using generative models (LSTM-based GAN, T-VAE, T-CVAE). Next, gradient boosting models predict the mechanical properties of potential cocrystals using the generated coformers. The pipeline then uses an evolutionary algorithm to improve the tabletability of the coformers. Finally, a graph neural network ranks the cocrystals based on the likelihood of formation, ultimately guiding chemical synthesis and tablet creation.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.

🔼 This Venn diagram shows the overlap between the sets of unique molecules generated by three different generative models (GAN, T-VAE, and T-CVAE). Each circle represents the number of unique molecules generated by a single model. The overlapping areas show the number of molecules common to two or three models. The numbers in each section of the diagram indicate the count of unique molecules in that specific region.
read the caption
Figure 7: How unique molecules created in different models intersect.

🔼 This figure presents two Tanimoto similarity histograms. The first histogram (a) shows the distribution of Tanimoto similarity scores between generated molecules and real coformers, providing insight into the novelty of the generated molecules. The second histogram (b) displays the distribution of Tanimoto similarity scores among all generated molecules, indicating the diversity of the generated molecules. Both histograms are broken down by GAN, VAE, and CVAE models, enabling a comparison of the different generative model’s performance regarding novelty and diversity.
read the caption
Figure 8: Tanimoto Similarity Histograms: (a) for generated molecules and real coformers, (b) for all generated molecules.
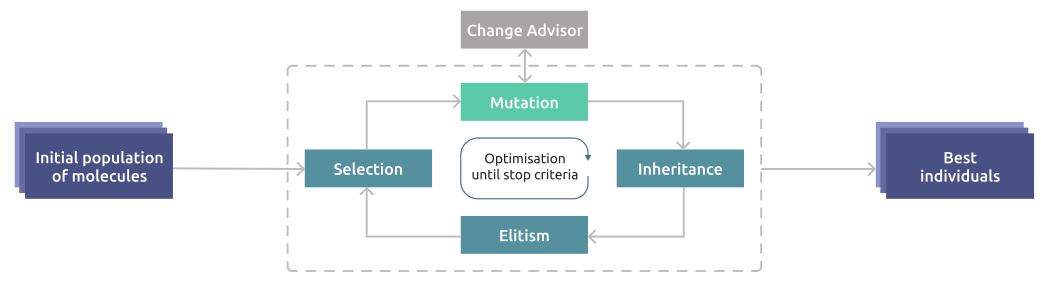
🔼 This figure shows the flowchart of the evolutionary algorithm used to optimize the generated coformers. It starts with an initial population of molecules. These molecules undergo selection based on their fitness. The selected molecules are mutated by applying various mutation operators guided by a change advisor. The mutated molecules are then inherited to form a new population, undergoing further optimization until the stop criteria are met. During the process, the best individuals are selected and used for elitism. This iterative process improves the tabletability properties of the generated coformers.
read the caption
Figure 9: Scheme of the evolutionary algorithm that is used for fine-tuning of solutions.
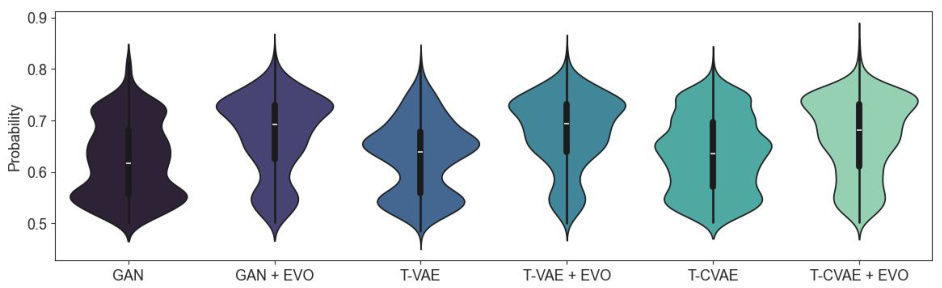
🔼 This violin plot shows the probability distributions of H-bond bridging between planes for coformers generated using different methods (GAN, T-VAE, T-CVAE) with and without evolutionary optimization. The plot visually demonstrates how evolutionary optimization impacts this specific mechanical property of the generated coformers, enhancing the probability of hydrogen bond bridging.
read the caption
Figure 10: Comparison of probability distributions for the presence of hydrogen bonds between the planes (H-bond bridging) for coformers generated by the neural models and optimized by evolution.

🔼 This figure illustrates the GEMCODE pipeline, a multi-stage process for designing co-crystals with enhanced tabletability. It starts with generating coformer candidates using various generative models (LSTM-based GAN, T-VAE, T-CVAE). These candidates are then evaluated by gradient boosting (GB) models to predict mechanical properties. An evolutionary algorithm optimizes these properties, improving tabletability profiles. Finally, a graph neural network (GNN) ranks the co-crystals based on their probability of formation, leading to a final ranked list of potential co-crystals with the desired properties.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.

🔼 The figure shows a flowchart of the GEMCODE pipeline. It starts with a therapeutic molecule as input. Three different generative models (LSTM-based GAN, T-VAE, T-CVAE) generate coformer candidates. Gradient boosting models predict the mechanical properties of the co-crystals formed by the therapeutic molecule and each coformer candidate. An evolutionary algorithm optimizes the coformers to improve their tabletability profiles. Finally, a graph neural network ranks the co-crystals according to the probability of formation. The top-ranked co-crystals are then synthesized and tested.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.
🔼 The figure shows a schematic overview of GEMCODE, a pipeline for designing co-crystals. It starts with a user providing a therapeutic molecule. Then, several generative models (LSTM-based GAN, T-VAE, and T-CVAE) create potential coformer candidates. A gradient boosting model and an evolutionary algorithm are used to predict and improve the mechanical properties (tabletability) of the co-crystals. Finally, a graph neural network ranks the co-crystals based on the likelihood of formation. The most promising candidates are then synthesized and tested to form tablets.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.
🔼 The figure illustrates the GEMCODE pipeline, a multi-stage process for designing co-crystals with enhanced tabletability. It starts with several generative models (GAN, T-VAE, T-CVAE) that propose coformer candidates. These are then evaluated using gradient boosting models to predict the mechanical properties of the resulting co-crystals. An evolutionary algorithm is used to optimize these candidates for improved tabletability. Finally, a graph neural network ranks the co-crystals based on their likelihood of formation. The successful candidates are then synthesized and tested experimentally.
read the caption
Figure 1: GEMCODE: a pipeline for generative co-crystal design consisting of models (LSTM-based GAN, T-VAE, T-CVAE) generating coformer candidates, gradient boosting (GB) classification models predicting the mechanical properties of co-crystals based on the generated coformers, an evolutionary algorithm producing additional coformer candidates with improved tabletability profiles, and a graph neural network (GNN) ranking co-crystals according to the probability of formation.
More on tables
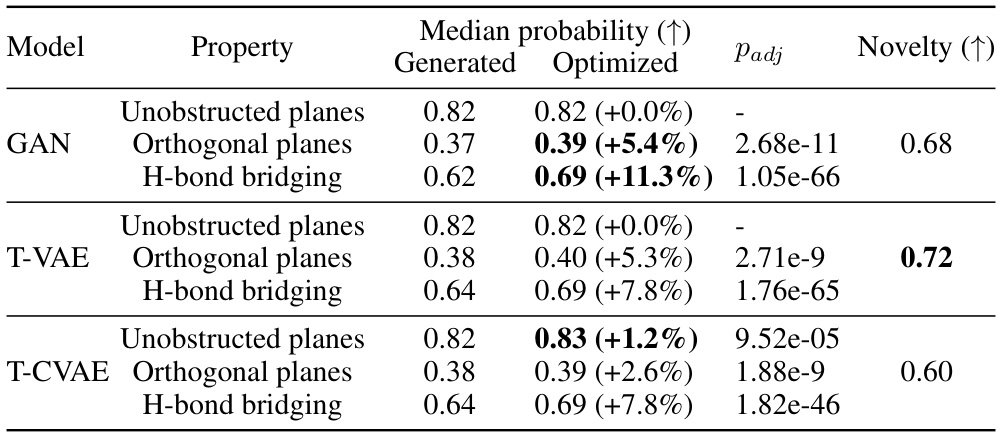
🔼 This table presents the results and statistical significance of the evolutionary optimization process applied to three different generative models: GAN, T-VAE, and T-CVAE. For each model and each of the three mechanical properties (Unobstructed planes, Orthogonal planes, H-bond bridging), the median probability before and after optimization is shown, along with the adjusted p-value from a statistical test and the novelty (increase in unique molecules) introduced by the process. The results indicate that evolutionary optimization significantly improved the median probability for the target mechanical properties, particularly for H-bond bridging.
read the caption
Table 2: Results and statistical significance of the evolutionary optimization. Median probability (↑) Property Generated Optimized Padj Novelty (↑)

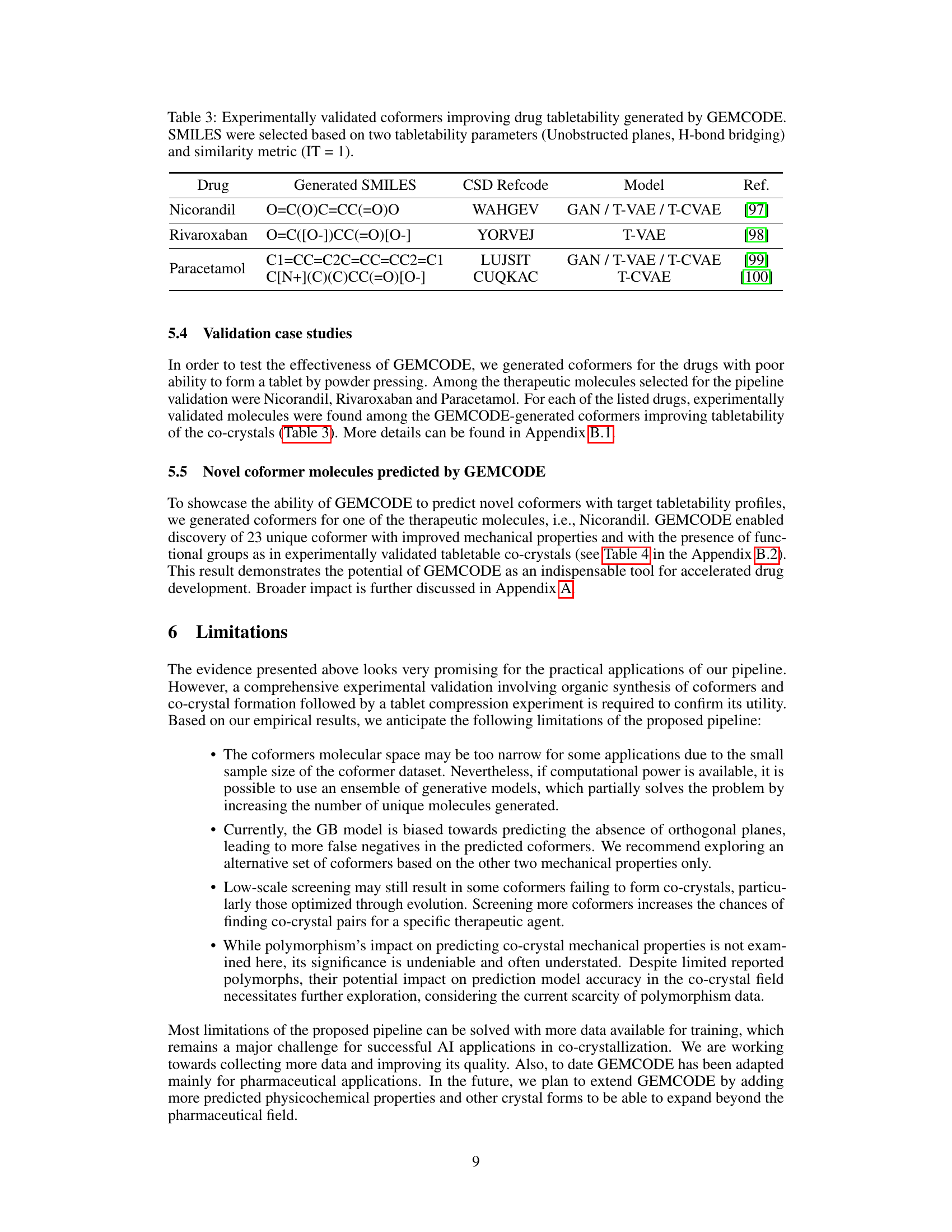
🔼 This table presents three experimentally validated coformers (Nicorandil, Rivaroxaban, Paracetamol) that improve drug tabletability. For each drug, the table shows the generated SMILES (Simplified Molecular Input Line Entry System) notation representing the coformer’s molecular structure, the CSD (Cambridge Structural Database) refcode for the experimentally determined crystal structure, the model used to generate the coformer, and the reference to the experimental validation study.
read the caption
Table 3: Experimentally validated coformers improving drug tabletability generated by GEMCODE. SMILES were selected based on two tabletability parameters (Unobstructed planes, H-bond bridging) and similarity metric (IT = 1).

🔼 This table presents 23 novel coformers for Nicorandil predicted by the GEMCODE pipeline. For each coformer, the SMILES notation, target properties (Unobstructed Planes, Orthogonal Planes, and H-bond bridging), and the CCGNet score (predicting co-crystallization probability) are provided. The coformers were selected based on a similarity metric (Tanimoto Index) of at least 0.7, indicating structural similarity to known effective coformers.
read the caption
Table 4: Previously unknown novel coformers generated using GEMCODE to improve the tabletability of the drug Nicorandil. SMILES are selected based on a similarity metric (IT ≥ 0.7). Target properties abbreviated as follows: Unobstructed planes (U), Orthogonal planes (O), H-bond bridging (H).
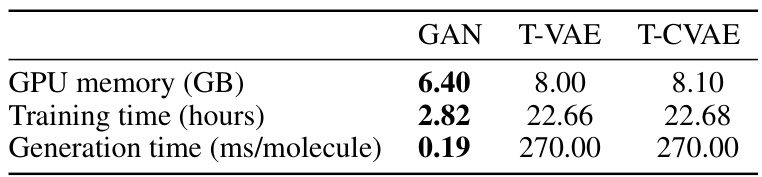
🔼 This table compares the GPU memory usage (in GB), training time (in hours for 10 epochs for GAN and 30 epochs for T-VAE/T-CVAE), and the time required to generate a single molecule (in milliseconds) for three different generative models: GAN, T-VAE, and T-CVAE. It highlights the significant difference in computational cost between GAN and the transformer-based models (T-VAE and T-CVAE), particularly in terms of training and generation time. The GAN model is considerably more efficient in terms of both training and generating molecules, although this is at the cost of lower diversity in generated molecules.
read the caption
Table 5: Comparison of GPU memory usage, training and generation times.

🔼 This table compares the performance of GEMCODE with two large language models (GPT-2 and Llama-3-8B) for the task of generating coformers. The metrics used for comparison include validity (percentage of chemically valid molecules), novelty (percentage of novel molecules not found in the training dataset), duplicates (percentage of duplicate molecules), target coformers (percentage of generated molecules with the desired properties for co-crystallization with Nicorandil), and diversity of target (diversity of the generated target coformers). The results show that GEMCODE outperforms both language models in terms of generating target coformers, indicating its superior ability for this specific task.
read the caption
Table 6: Comparison of coformer generation: GEMCODE vs. language models.

🔼 This table presents the results of comparing two evolutionary optimization schemes: SPEA-2 and MOEA/D. The median probabilities and p-values for three mechanical properties (Unobstructed planes, Orthogonal planes, and H-bond bridging) are shown for each scheme and for three different generative models (GAN, T-VAE, and T-CVAE). The p-values indicate the statistical significance of the differences in median probability between the two evolutionary schemes for each property and model.
read the caption
Table 7: Results and statistical significance (non-parametric one-sided Mann-Whitney test) for 10 runs of evolutionary algorithms based on SPEA-2 and MOEA/D selections.
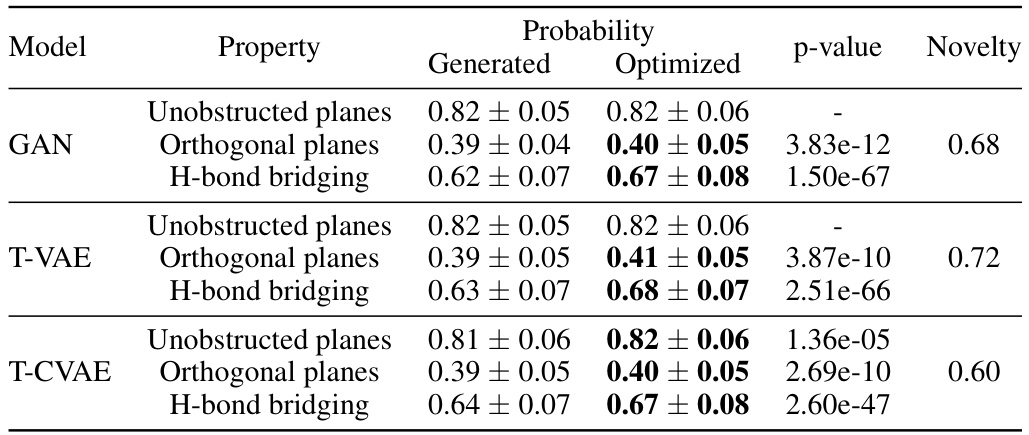
🔼 This table presents the results of applying evolutionary optimization to improve the mechanical properties of generated co-crystals. It shows the median probability of achieving target properties (Unobstructed planes, Orthogonal planes, and H-bond bridging) before and after optimization for different generative models (GAN, T-VAE, and T-CVAE). The p-value indicates the statistical significance of the improvement, and Novelty shows the percentage of new molecules generated after the optimization process.
read the caption
Table 2: Results and statistical significance of the evolutionary optimization. Median probability (↑) Model Property Generated Optimized Padj Novelty (↑)
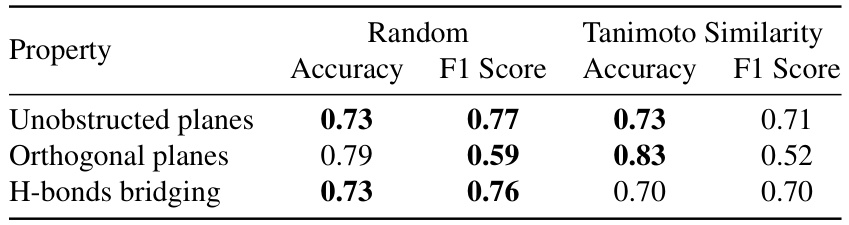
🔼 This table presents the performance metrics (Accuracy and F1 Score) of the Gradient Boosting model used for predicting the mechanical properties of co-crystals. The performance is evaluated using two different data splitting strategies: random splitting and splitting based on Tanimoto similarity. The results are shown for three different mechanical properties: Unobstructed planes, Orthogonal planes, and H-bonds bridging.
read the caption
Table 9: Metrics of the Gradient Boosting model for predicting the mechanical properties of co-crystals upon changing the data splitting strategy.

🔼 This table compares the performance of various machine learning models in predicting different physicochemical properties of co-crystals, including crystal density, melting temperature, melting enthalpy, melting entropy, ideal solubility, lattice energy, and mechanical properties (unobstructed planes, orthogonal planes, and H-bonds bridging). It highlights the best metric achieved by each model and whether or not a generative design approach was used. The table shows that the authors’ work is state-of-the-art in predicting the mechanical properties.
read the caption
Table 10: Comparative table with model metrics on prediction of various co-crystals properties.

🔼 This table compares the performance of the proposed machine learning (ML) models with those obtained using AutoML (Automated Machine Learning) for predicting three mechanical properties of co-crystals: Unobstructed planes, Orthogonal planes, and H-bond bridging. For each property, the table shows the precision, recall, and F1-score achieved by both the proposed models and the AutoML models. The results indicate the performance of the proposed models against a state-of-the-art automated machine learning approach.
read the caption
Table 11: Comparison of the proposed ML models with AutoML. Best achieved metrics are given.
Full paper#