TL;DR#
Antibody design is crucial but challenging due to its complexity. Traditional methods often struggle with producing antibodies possessing both rational structures and high binding affinity, relying on metrics like amino acid recovery (AAR) and root mean square deviation (RMSD) which are insufficient and susceptible to manipulation. This paper addresses these limitations by focusing on energy-based optimization.
The proposed method, ABDPO, uses a pre-trained conditional diffusion model fine-tuned with a residue-level energy preference to generate antibodies. This approach involves fine-tuning a pre-trained diffusion model with residue-level decomposed energy preference, and employing gradient surgery to resolve conflicts. Experiments demonstrate that ABDPO outperforms existing methods, creating antibodies with superior binding affinity and lower total energy, highlighting the importance of energy-based metrics for evaluating antibody design.
Key Takeaways#
Why does it matter?#
This paper is significant because it introduces a novel approach to antibody design, addressing the limitations of existing methods. It offers a more energy-focused and functional design process. This opens avenues for more efficient antibody design in therapeutic and biological research, and offers new techniques for preference optimization in generative models. The residue-level energy-based preference optimization technique is particularly noteworthy, advancing the field of protein design.
Visual Insights#
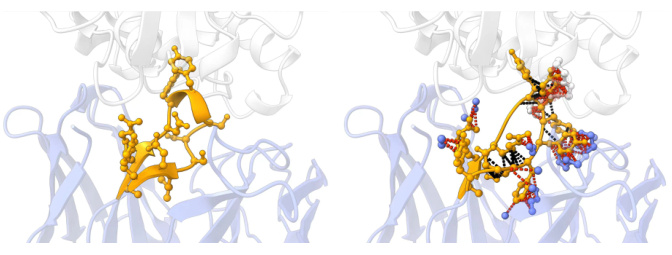
🔼 This figure visualizes three examples of reference antibodies from the RAbD dataset and the antibodies generated by ABDPO for the same antigens. Each pair shows the CDR-H3 region (in yellow) of both the reference and ABDPO-generated antibody, bound to its antigen. The remaining antibody structures are shown in blue. The visualizations highlight the structural differences between the reference and the model-generated antibodies, and the annotated energy values (omitted for brevity in the caption) give a quantitative comparison of their binding energies. The goal is to illustrate how ABDPO produces antibodies with similar structures to the reference antibodies but with improved binding affinity.
read the caption
Figure 3: Visualization of reference antibodies in RAbD and antibodies designed by ABDPO given specific antigens (PDB ID: 1iqd (left), 1ic7 (middle), and 2dd8 (right)). The unit of energy annotated is kcal/mol and omitted here for brevity.
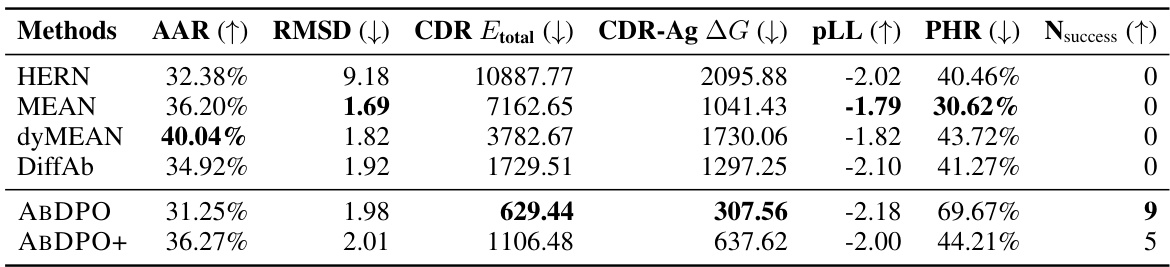
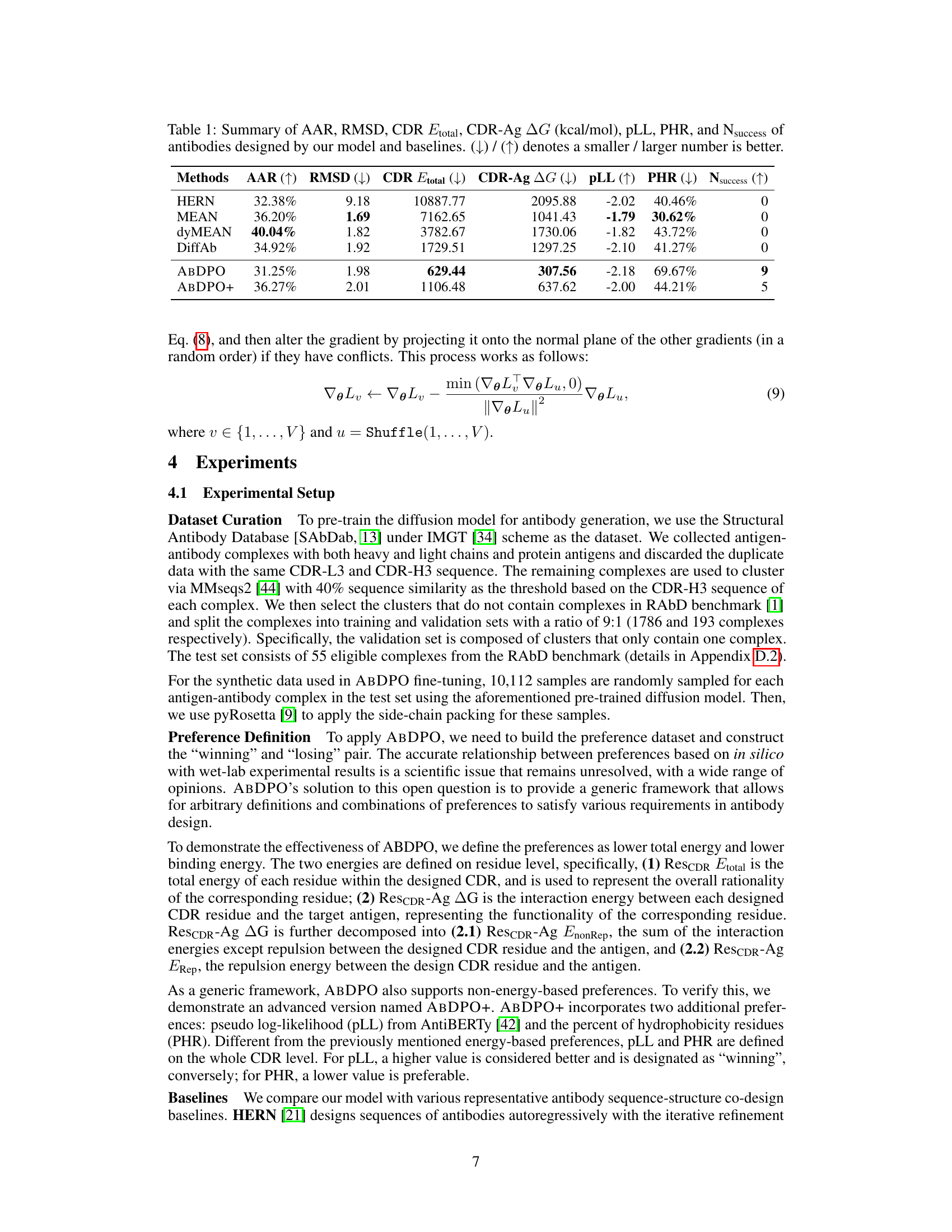
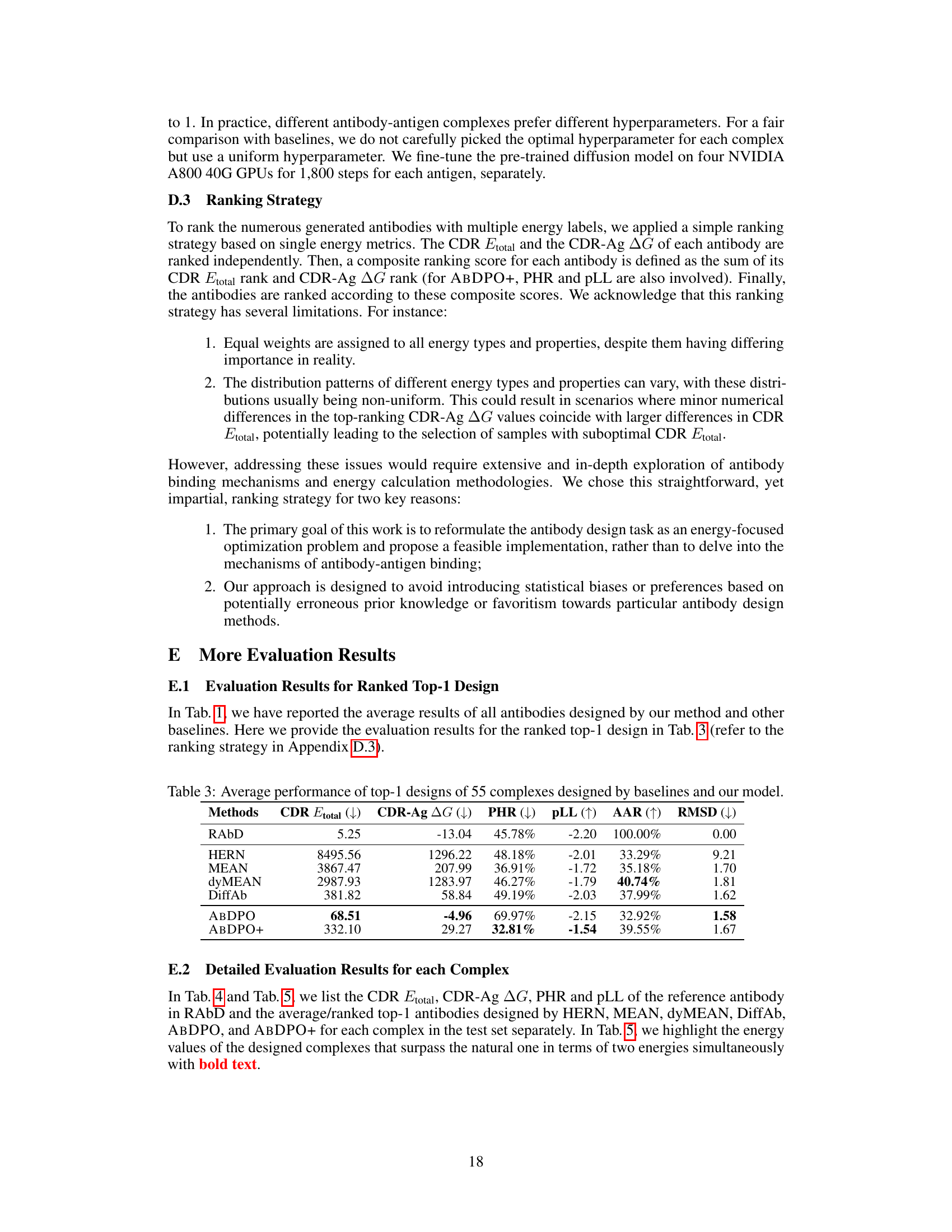
🔼 This table presents a comparison of the performance of different antibody design methods, including the proposed ABDPO and several baselines. The metrics used to evaluate the methods include Amino Acid Recovery (AAR), Root Mean Square Deviation (RMSD), total CDR energy (CDR Etotal), antigen-binding energy (CDR-Ag AG), pseudo log-likelihood (pLL), percent of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). Lower values are preferred for RMSD, CDR Etotal, CDR-Ag AG, and PHR, while higher values are preferred for AAR, pLL, and Nsuccess. The table shows that ABDPO achieves state-of-the-art performance on several of the key metrics.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.
In-depth insights#
Antibody Design#
Antibody design is a complex process with significant implications for therapeutics and biology. Traditional methods are often inefficient, relying on sampling protein sequences and struggling with local minima. Deep generative models offer advantages by modeling sequence and structure relationships, enabling sequence-structure co-design. However, challenges persist, including insufficient training data due to a scarcity of high-quality data and inadequate evaluation metrics such as amino acid recovery (AAR) and root mean square deviation (RMSD). Therefore, this paper proposes a more rational approach using direct energy-based preference optimization which leverages pre-trained diffusion models and addresses limitations through energy decomposition and conflict mitigation techniques. This approach shows superiority in designing high-quality antibodies with low total energy and high binding affinity, paving the way for more effective antibody engineering.
Energy Optimization#
The concept of ‘Energy Optimization’ within the context of a research paper likely refers to minimizing a system’s energy consumption or maximizing energy efficiency. This could involve various approaches, depending on the specific system. For example, in a study involving protein design, energy optimization might focus on finding protein structures with minimal energy states, indicating high stability and functionality. This process could leverage computational methods to explore various configurations and identify optimal ones. Another example could be within the realm of machine learning, where energy optimization could involve improving the efficiency of deep learning models by reducing energy usage during training and inference. This can be achieved through techniques like model compression, optimized hardware utilization, or novel training algorithms. Regardless of the specific application, energy optimization is a crucial aspect for improving the performance, sustainability, and cost-effectiveness of any system.
Residue-Level DPO#
Residue-Level Direct Preference Optimization (DPO) represents a significant advancement in antibody design. Instead of considering the entire antibody sequence at once, Residue-Level DPO focuses on individual amino acid residues, evaluating their energetic contributions separately. This granular approach offers several key advantages. Firstly, it allows for a more nuanced understanding of the interactions between the antibody and antigen, identifying specific residues crucial for binding. This detailed understanding is essential for optimizing antibody-antigen interactions. Secondly, the fine-grained evaluation allows for more precise adjustments to the antibody sequence during optimization, potentially leading to higher-quality antibodies with improved binding affinity. It mitigates the issue of ‘shortcuts’ that can arise when using coarser-grained optimization methods. Lastly, residue-level DPO is computationally more efficient than whole-sequence methods as it avoids the high dimensionality of the whole-sequence space, speeding up the overall design process.
Gradient Surgery#
Gradient surgery, as described in the context of the research paper, is a technique to mitigate the interference between conflicting energy gradients during the optimization process. In essence, it addresses situations where multiple objectives, such as attraction and repulsion forces, pull the optimization process in opposing directions. The core idea is to decompose the overall energy into its constituent components and then selectively adjust the gradient updates for each component, preventing a single component from dominating and potentially leading to suboptimal solutions. This involves a careful recalibration of gradients, effectively “surgical” intervention, to achieve a balance between the conflicting forces. This method is crucial in complex systems with multiple interacting elements, ensuring convergence to high-quality solutions that wouldn’t be possible with standard gradient-descent alone. The method’s effectiveness hinges on its ability to maintain equilibrium between opposing forces, directing the optimization towards the desirable characteristics, and preventing the algorithm from being trapped in local minima or unsatisfactory solutions due to the dominance of a single objective.
Future Directions#
Future research directions stemming from this antigen-specific antibody design work could involve improving the accuracy and efficiency of the energy calculations, perhaps through incorporating more sophisticated physics-based models or refining the existing energy decomposition strategies. Another key area is exploring novel ways to define and optimize preferences, moving beyond simplistic energy metrics and incorporating additional criteria relevant to therapeutic antibody development such as stability, immunogenicity, and manufacturability. Integrating machine learning with more advanced experimental techniques, such as high-throughput screening, could also significantly enhance the process of designing and validating antibodies. Finally, expanding the application of this methodology to other areas of protein design, such as enzyme engineering and materials science, warrants further investigation.
More visual insights#
More on figures

🔼 This figure illustrates the workflow of the ABDPO method. Starting with a pre-trained diffusion model, antibodies are generated. Their residue-level energies (attraction, repulsion, total) are calculated and used to create preference data for fine-tuning. Gradient surgery addresses conflicts between energy types. The model is updated iteratively, improving antibody design.
read the caption
Figure 2: Overview of ABDPO. This process can be summarized as: (a) Generate antibodies with the pre-trained diffusion model; (b) Evaluate the multiple types of residue-level energy and construct preference data; (c) Compute the losses for energy-based preference optimization and mitigate the conflicts between losses of multiple types of energy; (d) Update the diffusion model.
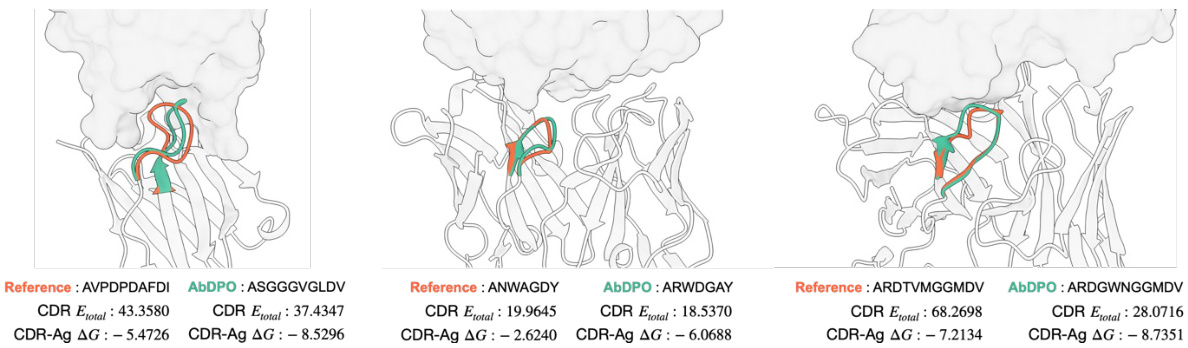
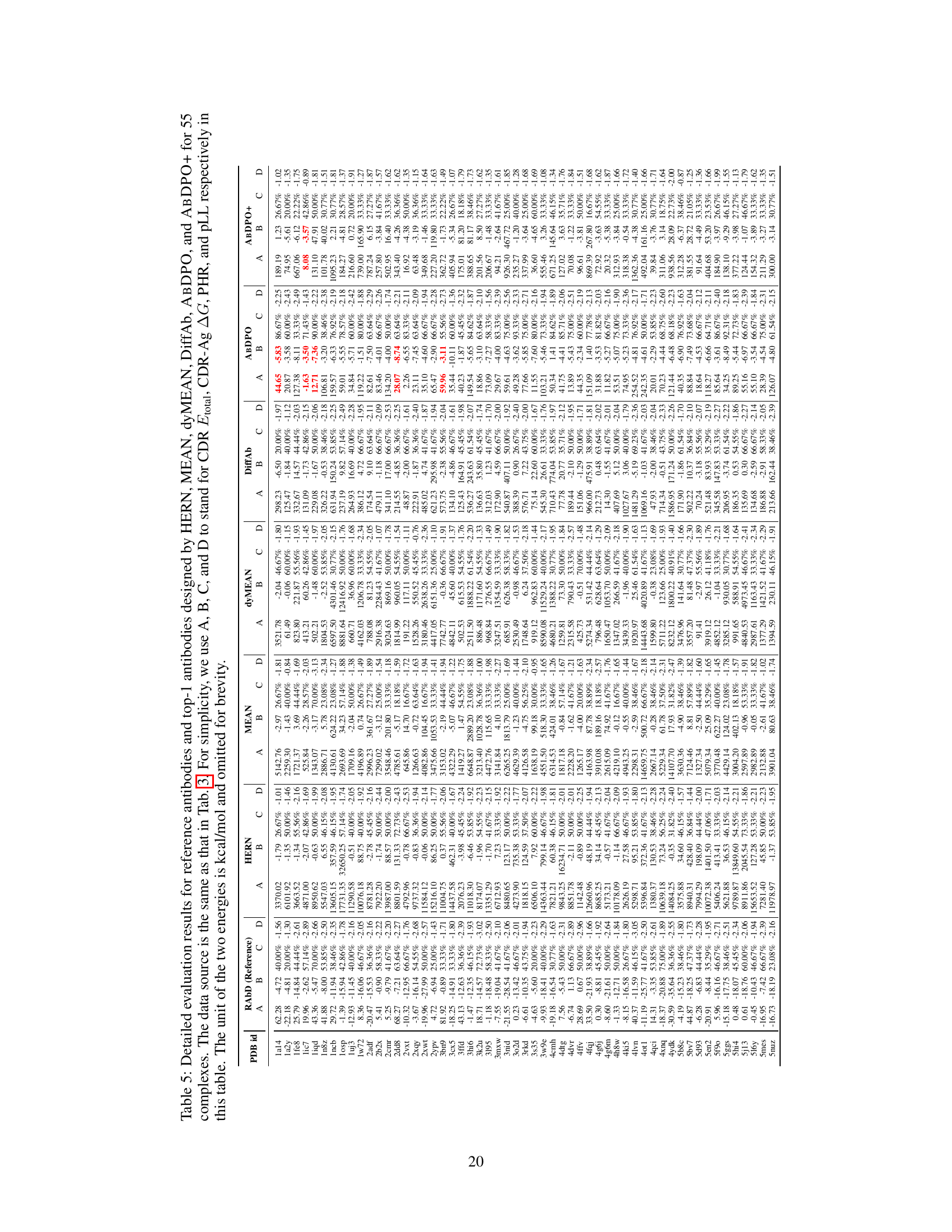
🔼 This figure visualizes three examples of antibodies generated by ABDPO and their corresponding reference antibodies from the RAbD dataset. Each example shows the structure of the reference antibody (left) and the synthetic antibody designed by ABDPO (right). The CDR-H3 regions are highlighted in yellow to emphasize the part of the antibody most relevant to antigen binding. The remaining parts of the antibodies are colored blue and the antigen is colored grey. The CDR total energy (CDR Etotal) and the change in Gibbs free energy upon binding (CDR-Ag AG) are provided for both the reference and ABDPO-generated antibodies, highlighting the difference in energy between the natural and synthetic antibodies. The figure illustrates ABDPO’s ability to generate antibodies with structures and energies resembling those of naturally occurring antibodies.
read the caption
Figure 3: Visualization of reference antibodies in RAbD and antibodies designed by ABDPO given specific antigens (PDB ID: 1iqd (left), 1ic7 (middle), and 2dd8 (right)). The unit of energy annotated is kcal/mol and omitted here for brevity.

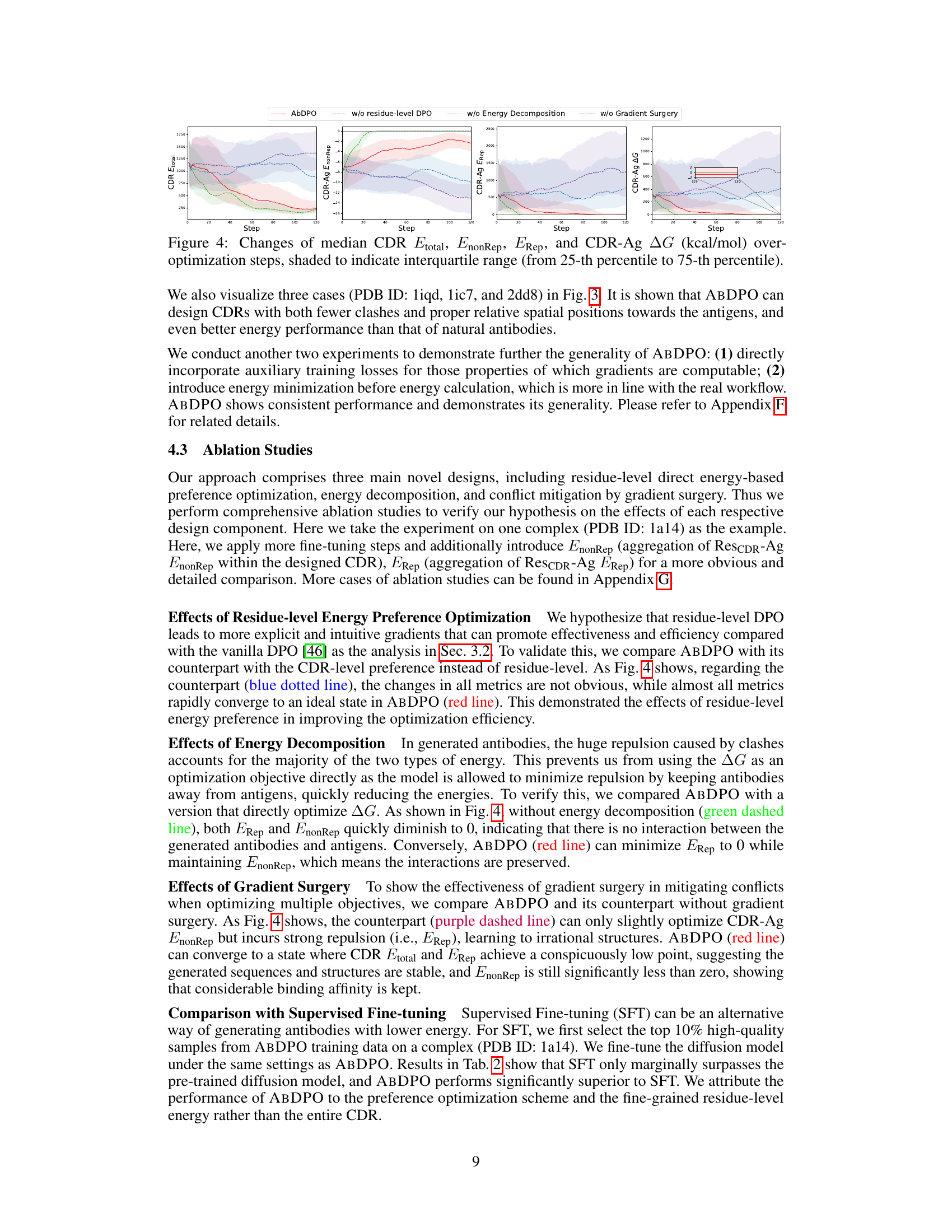
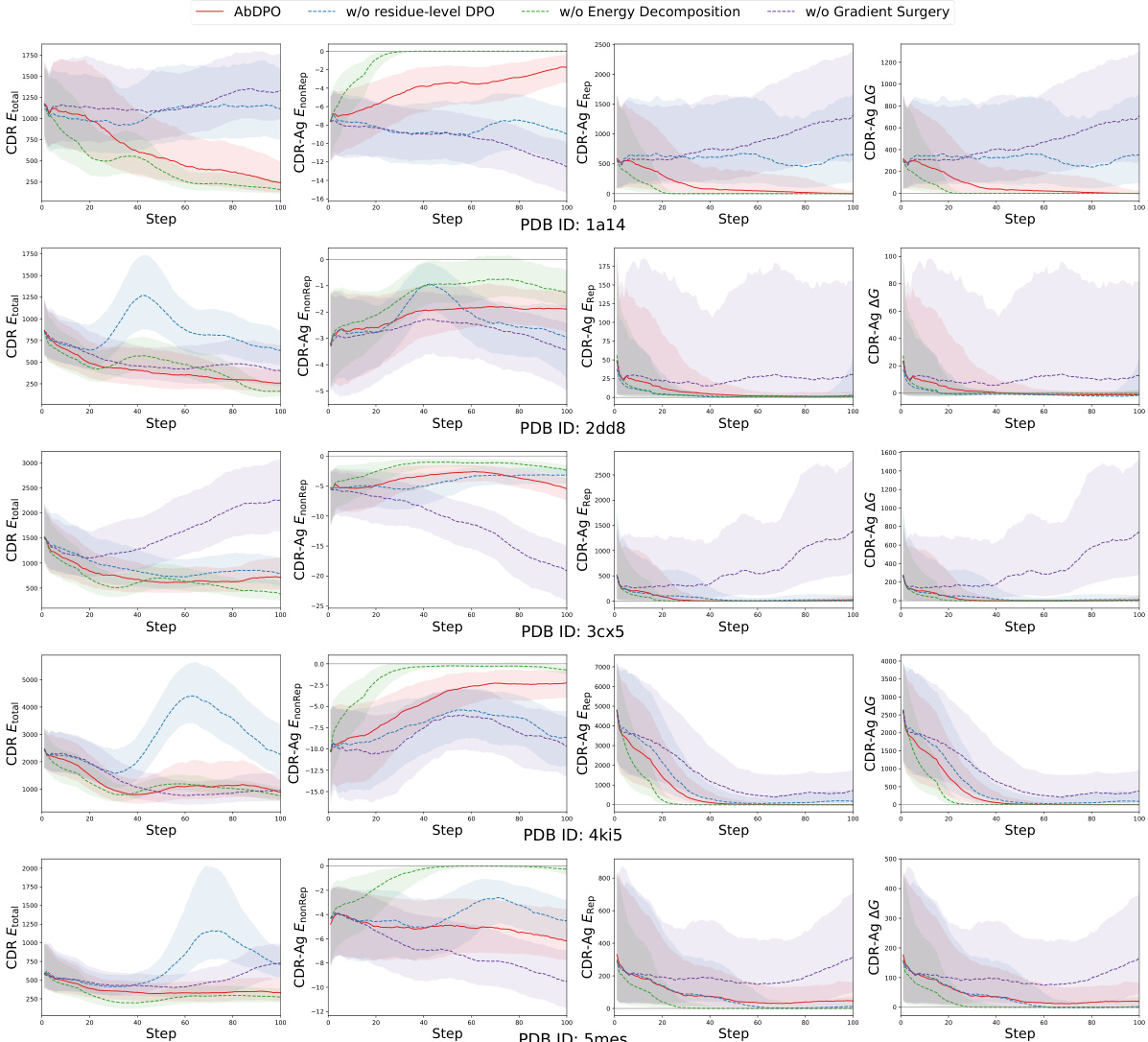
🔼 This figure displays the changes in median CDR Etotal, CDR-Ag EnonRep, CDR-Ag Erep, and CDR-Ag ∆G during optimization steps for five different PDB IDs (1a14, 2dd8, 3cx5, 4ki5, and 5mes). Shading indicates the interquartile range (25th to 75th percentile) showing variation across multiple runs. Each row represents one of these PDB IDs, illustrating the optimization process for each. The purpose is to demonstrate the effects of each element of the ABDPO method by comparing the results to the results of running the optimization without one of those elements.
read the caption
Figure 8: Changes of median CDR Etotal, CDR-Ag EnonRep, CDR-Ag Erep, and CDR-Ag ∆G (kcal/mol) over-optimization steps, shaded to indicate interquartile range (from 25-th percentile to 75-th percentile). The rows represent PDB 1a14, 2dd8, 3cx5, 4ki5, and 5mes respectively, in a top-down order.
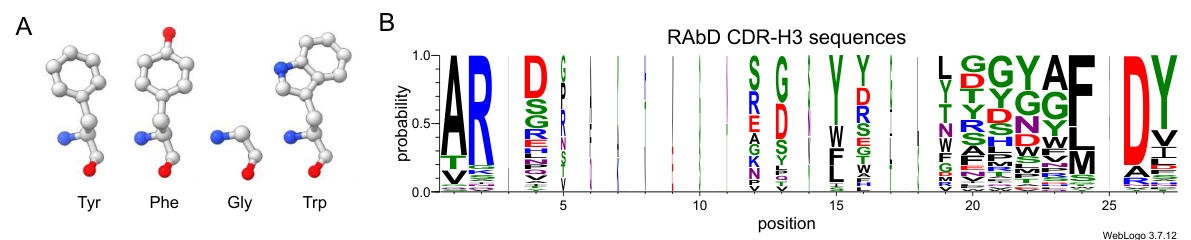
🔼 Figure 5 shows the differences in side chain size between amino acids, especially between Tyr/Phe and Gly/Trp. It also visualizes the frequency of amino acid occurrence at each position of CDR-H3 in the RAbD dataset using WebLogo. This highlights the limitations of AAR as an evaluation metric.
read the caption
Figure 5: A: Tyr (Y) and Phe (F) differ by only one oxygen atom. In contrast, there is a substantial difference between Gly (G) and Trp (W). Gly lacks a side chain, whereas Trp possesses the largest side chain of all amino acids. B: the visualization of the frequency of occurrence of each amino acid at various positions in RAbD CDR-H3 sequences. The sequences are initially aligned using MAFFT [26] and subsequently visualized with WebLogo [11]. The width of each column corresponds to the frequency of occurrence at that position.
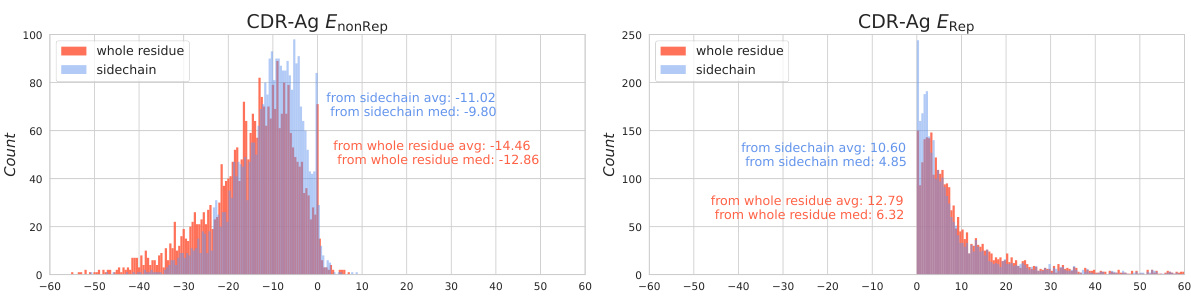
🔼 This figure shows the distribution of two types of energy: CDR-Ag EnonRep and CDR-Ag Erep. The energy calculations were performed using two different methods: using all atoms of the CDR region and using only the sidechain atoms. The distributions highlight the significant contribution of the side chain atoms to the interaction energy between CDR and antigen. The difference in the energy distribution between the whole residue and side chain methods visually demonstrates the importance of considering side chain contributions during antibody design.
read the caption
Figure 6: The distribution of CDR-Ag EnonRep (left) and CDR-Ag Erep (right) formed by the whole CDR atoms (colored in red) and solely by CDR side-chain atoms (colored in blue) among SAbDab dataset.
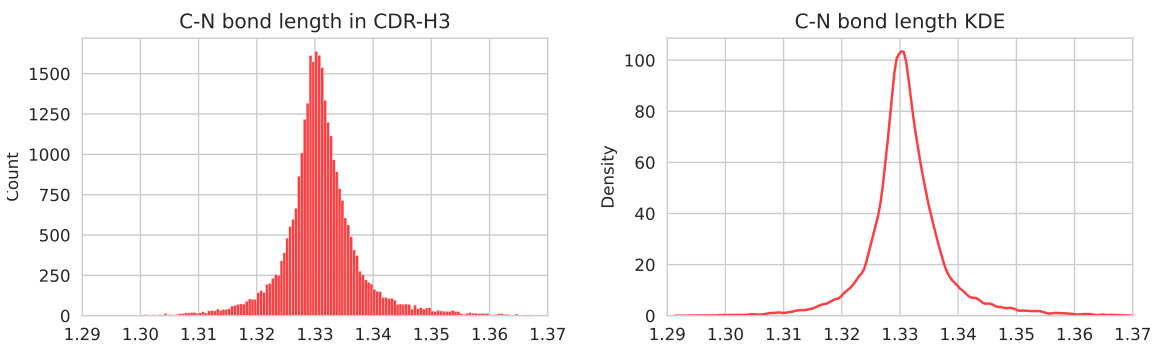
🔼 This figure shows the distribution of peptide bond lengths in CDR-H3 from the SAbDab dataset (left panel) and the kernel density estimation (KDE) of the distribution (right panel). The KDE provides a smoothed representation of the distribution, highlighting the concentration around the average peptide bond length. This visualization is used to assess the consistency of generated antibodies’ peptide bond lengths compared to naturally occurring antibodies, a key aspect of structural fidelity and rationality in antibody design. The figure appears in section F of the paper, in the subsection discussing the introduction of an auxiliary loss for peptide bond length in the ABDPO++ model.
read the caption
Figure 7: Left: the distribution of peptide bond length within CDR-H3 in the SAbDab dataset; Right: the kernel density estimation (KDE) function fit on the natural peptide bond length distribution.
🔼 The figure visualizes the changes in median CDR Etotal, EnonRep, ERep, and CDR-Ag ∆G over optimization steps for one specific antibody-antigen complex. Shading represents the interquartile range (25th to 75th percentile), showing the distribution of values. It illustrates how these energy components change during the optimization process, providing insight into the effectiveness of the ABDPO method in optimizing multiple aspects of antibody design. The graph’s overall trend indicates that ABDPO is successful in reducing the total energy and improving the binding affinity of the antibodies.
read the caption
Figure 4: Changes of median CDR Etotal, EnonRep, ERep, and CDR-Ag ∆G (kcal/mol) over optimization steps, shaded to indicate interquartile range (from 25-th percentile to 75-th percentile).
More on tables
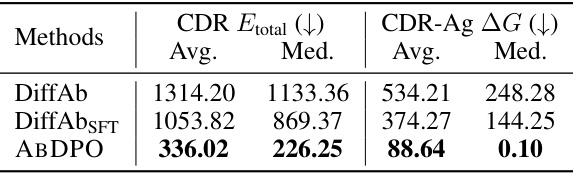
🔼 This table compares the performance of ABDPO and supervised fine-tuning (SFT) on a single antibody-antigen complex (PDB ID: 1a14). It shows the average and median values of CDR Etotal and CDR-Ag AG (in kcal/mol) for both methods. ABDPO demonstrates significantly lower values for both metrics than SFT.
read the caption
Table 2: Comparison of ABDPO and supervised fine-tuning (SFT) on 1a14.

🔼 This table presents a comparison of the performance of different antibody design methods, including the proposed ABDPO method and several baselines, across various metrics. The metrics assess different aspects of the generated antibodies, including amino acid recovery (AAR), root mean square deviation (RMSD), total energy of the CDR (CDR Etotal), change in Gibbs free energy upon binding to the antigen (CDR-Ag AG), pseudo log-likelihood (pLL), percentage of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). Lower values are better for RMSD, CDR Etotal, CDR-Ag AG, and PHR, while higher values are better for AAR, pLL, and Nsuccess.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.

🔼 This table summarizes the performance of different antibody design methods, including the proposed ABDPO and several baselines (HERN, MEAN, dyMEAN, DiffAb). The metrics used to evaluate performance are amino acid recovery (AAR), root mean square deviation (RMSD), total energy of the CDR (CDR Etotal), change in Gibbs free energy between the bound and unbound states of the CDR-antigen complex (CDR-Ag AG), pseudo log-likelihood (pLL), percent of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). Lower is better for RMSD, CDR Etotal, and CDR-Ag AG; higher is better for AAR, pLL, and Nsuccess; and lower is better for PHR. The table highlights the superior performance of ABDPO compared to the baselines across multiple evaluation metrics.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.
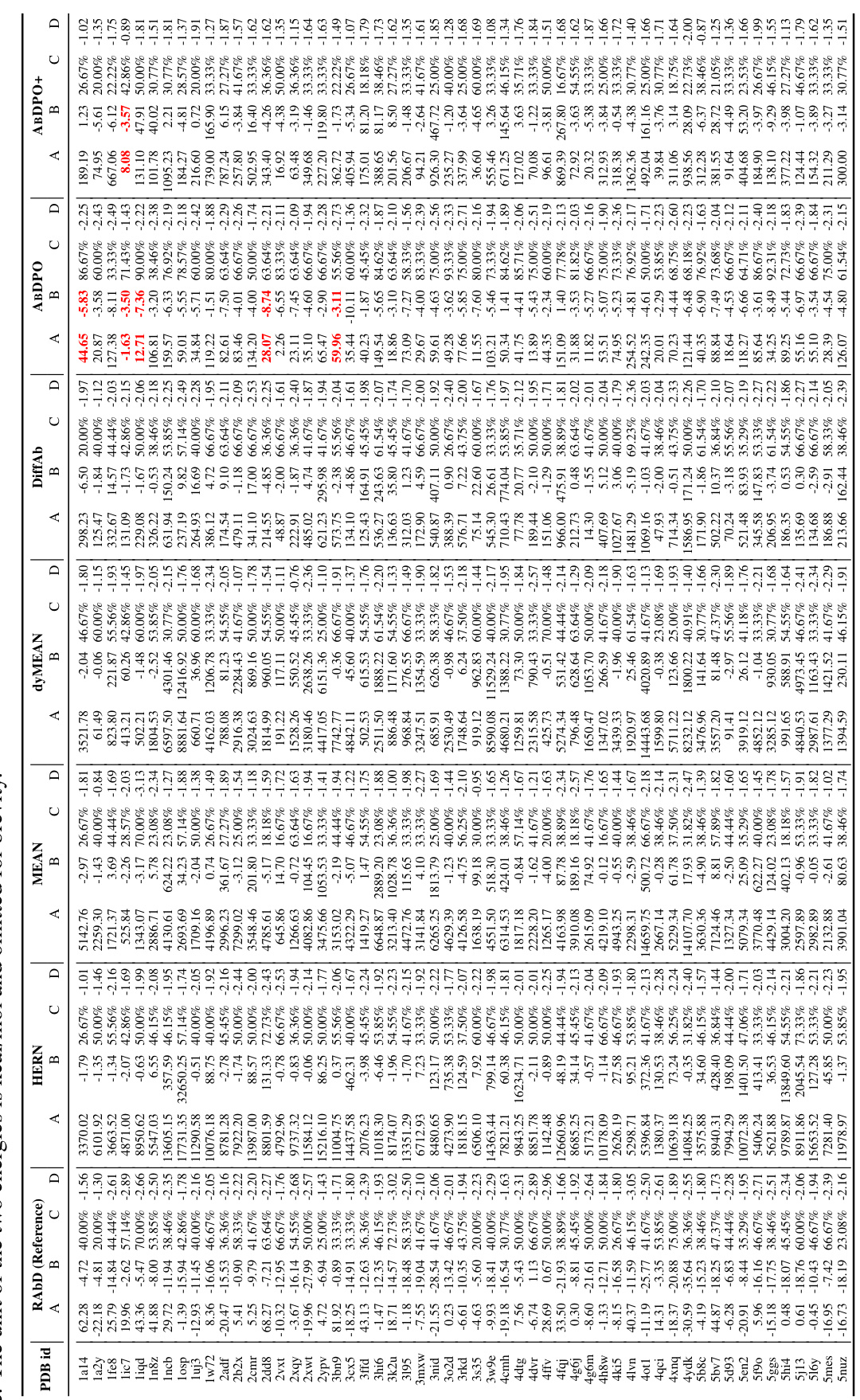
🔼 This table presents a comparison of the performance of different antibody design methods, including the proposed ABDPO method and several baselines (HERN, MEAN, dyMEAN, DiffAb). The performance is evaluated using several metrics, including amino acid recovery (AAR), root mean square deviation (RMSD), total CDR energy (CDR Etotal), antigen-binding energy (CDR-Ag AG), pseudo log-likelihood (pLL), percent of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). Lower values are generally preferred for RMSD, CDR Etotal, and CDR-Ag AG, while higher values are better for AAR, pLL, and Nsuccess.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.

🔼 This table presents a comparison of the performance of several antibody design methods, including the authors’ proposed method (ABDPO), and several baselines (HERN, MEAN, dyMEAN, DiffAb). The metrics used for comparison include amino acid recovery (AAR), root mean square deviation (RMSD), total energy of the CDR (CDR Etotal), change in Gibbs free energy between the bound and unbound states of the CDR and antigen (CDR-Ag AG), pseudo log-likelihood (pLL), percent of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). Lower values are better for RMSD, CDR Etotal, CDR-Ag AG, and PHR, while higher values are preferred for AAR, pLL, and Nsuccess.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.
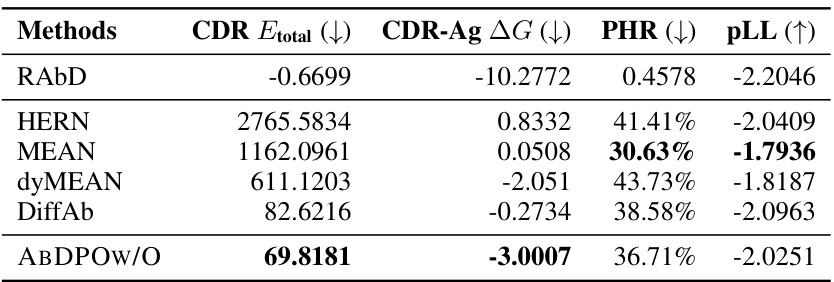
🔼 This table summarizes the performance of various antibody design methods, including the proposed ABDPO method and several baselines (HERN, MEAN, dyMEAN, DiffAb), across multiple evaluation metrics. These metrics assess different aspects of antibody quality, such as sequence similarity to natural antibodies (AAR), structural similarity (RMSD), total energy of the CDR region (CDR Etotal), binding affinity (CDR-Ag AG), pseudo log-likelihood (pLL), percent of hydrophobic residues (PHR), and the number of successfully designed antibodies (Nsuccess). The table allows for a comparison of the methods’ performance in terms of both rationality and functionality of the generated antibodies.
read the caption
Table 1: Summary of AAR, RMSD, CDR Etotal, CDR-Ag AG (kcal/mol), pLL, PHR, and Nsuccess of antibodies designed by our model and baselines. (↓) / (↑) denotes a smaller / larger number is better.
Full paper#