↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Current molecular generative models often lack adaptability, being tailored to specific downstream tasks. This limits their use in diverse drug design applications like structure-based, fragment-based, and ligand-based approaches. Furthermore, existing methods often require additional networks or retraining when adapting to new tasks, which adds complexity and computational cost.
UniGuide addresses these issues by providing a unified framework for controlled geometric guidance of unconditional diffusion models. It uses a ‘condition map’ to transform diverse geometric conditions into a format compatible with the model, enabling flexible conditioning during inference. The results show that UniGuide performs on-par or better than specialized models across various drug design scenarios, demonstrating its versatility and efficiency. UniGuide offers a streamlined approach to developing adaptable molecular generative models, eliminating the need for extra training or networks and enhancing their usability in diverse drug discovery tasks.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in drug discovery and generative modeling. It introduces a unified framework for geometry-conditioned molecular generation, addressing the limitations of task-specific models. This opens new avenues for developing adaptable generative models and streamlines the application of diffusion models in diverse scenarios, thus potentially accelerating drug discovery.
Visual Insights#

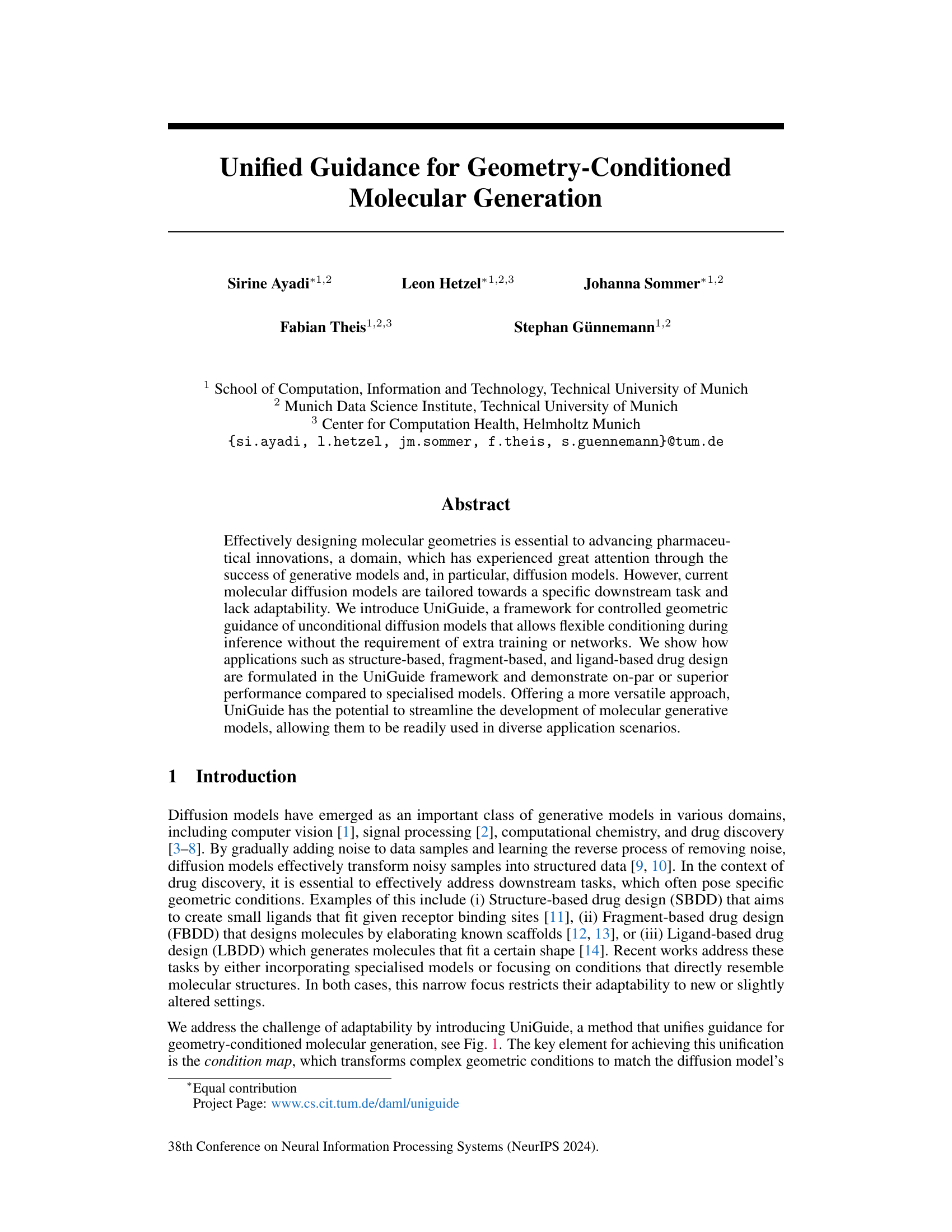
This figure illustrates the UniGuide framework’s ability to handle diverse conditioning modalities for geometry-conditioned molecular generation. It shows how UniGuide combines a source condition (representing various geometric constraints like target receptor shape in structure-based drug design, fragments in fragment-based drug design, or 3D shapes in ligand-based drug design) with an unconditional diffusion model. This combination, facilitated by a ‘condition map’, allows for flexible conditioning during inference without the need for extra training or networks. The figure highlights the framework’s versatility and adaptability to various drug discovery tasks, including its potential for handling new geometric tasks, exemplified by the example of conditioning on atomic densities.
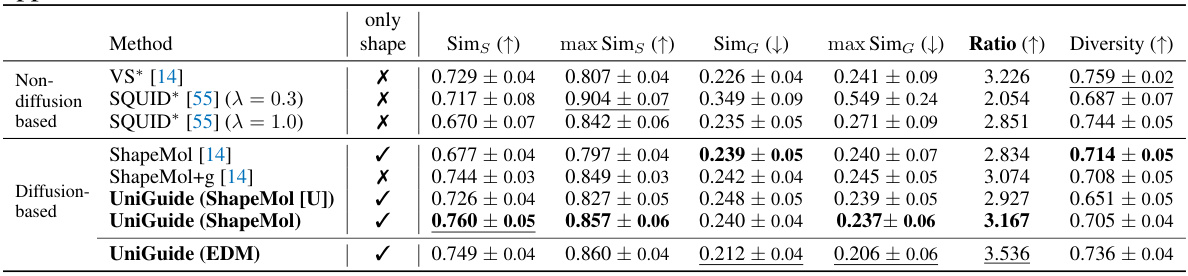
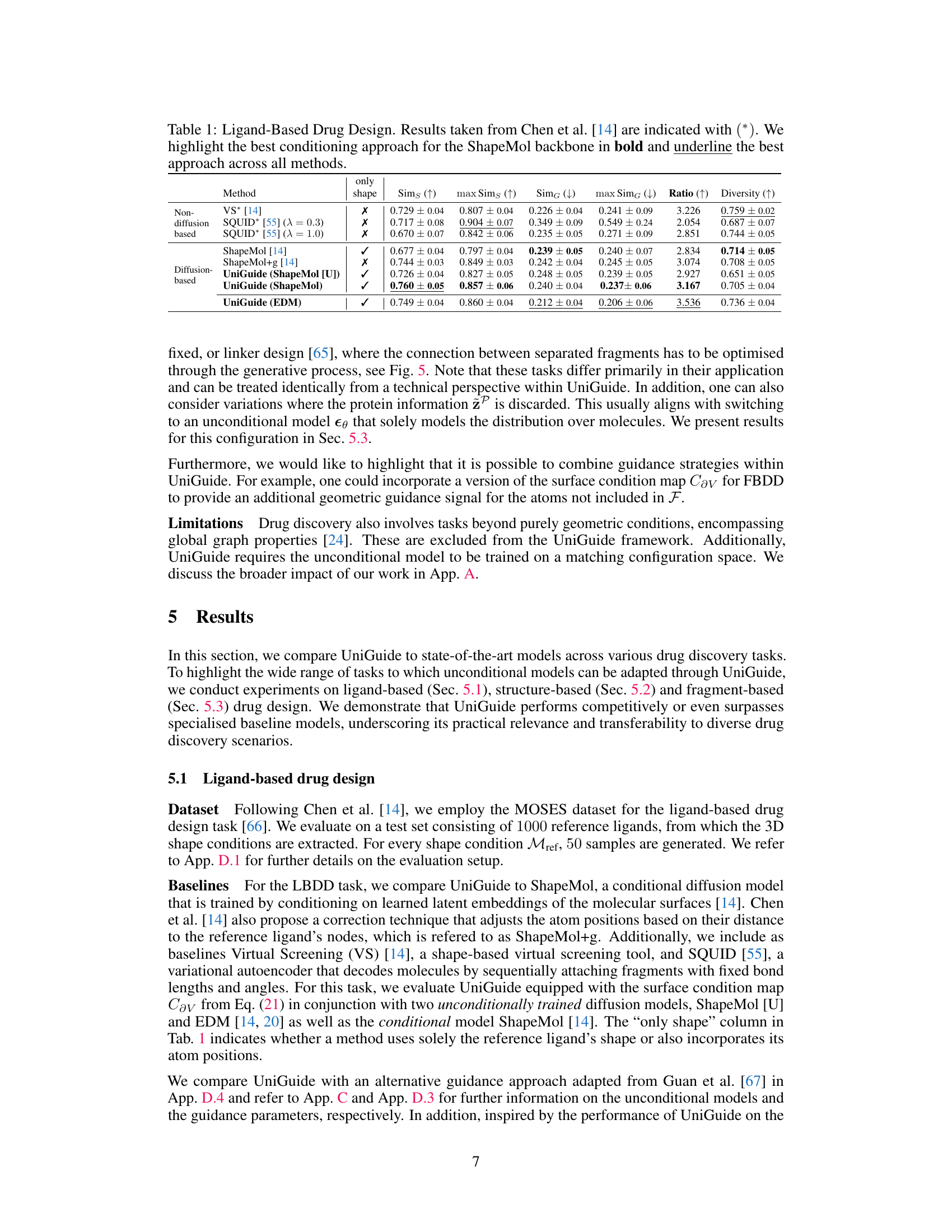
This table compares several methods for ligand-based drug design, focusing on their ability to generate molecules with similar 3D shapes to a reference ligand. The methods are categorized into non-diffusion-based and diffusion-based approaches. Metrics include shape similarity, graph similarity, the ratio between them, and diversity. The table highlights the superior performance of UniGuide compared to the specialized methods in generating ligands with similar shapes but diverse structures.
In-depth insights#
UniGuide Framework#
The UniGuide framework presents a novel, unified approach to geometrically-conditioned molecular generation. Its core innovation lies in decoupling geometric guidance from model training, eliminating the need for specialized models or additional training for different tasks. This versatility is achieved through a flexible condition map that transforms diverse geometric constraints (target receptors, fragments, 3D shapes) into a form compatible with an unconditional diffusion model. UniGuide demonstrates superior performance or on-par results compared to task-specific models, highlighting its adaptability and efficiency. The framework’s modularity simplifies development, allowing for straightforward application in structure-based, fragment-based, and ligand-based drug design, and potentially many more applications beyond.
Self-Guidance Method#
A self-guidance method in generative models cleverly sidesteps the need for extra training or networks by leveraging the model’s own predictions to guide the generation process. This is achieved through a condition map, which transforms complex geometric conditions into a format compatible with the diffusion model. The framework’s flexibility allows for diverse conditioning modalities. The core idea is to use the model’s inherent understanding of the data distribution to steer the generation, leading to improved performance and enhanced adaptability. This unification of guidance methods streamlines the development of geometry-conditioned molecular generation, enabling straightforward application across various downstream tasks. The framework’s self-guidance approach promotes versatility and potentially reduces reliance on extensive training data, crucial advantages in computationally expensive domains such as drug discovery. The elimination of extra networks simplifies implementation and potentially reduces computational costs, making the approach more efficient and accessible.
Drug Discovery Tasks#
Drug discovery is a complex process, and the paper’s focus on geometry-conditioned molecular generation highlights the importance of spatial considerations in designing effective drug candidates. The exploration of structure-based, fragment-based, and ligand-based drug design showcases the versatility of the proposed method, UniGuide, in tackling various geometric challenges inherent in different drug discovery approaches. UniGuide’s ability to unify these diverse tasks into a single framework is a major advantage, streamlining the development process and increasing the adaptability of generative models. The successful application to these tasks demonstrates the practicality and effectiveness of the method, highlighting its potential to improve the efficiency and success rate of drug discovery efforts. However, the paper should further discuss limitations and potential challenges associated with handling very complex molecules. The current method’s dependence on quality 3D shape information and its applicability to specific molecule types are potential areas for future investigation and improvement.
Performance Analysis#
A thorough performance analysis of a machine learning model should encompass several key aspects. It’s crucial to define appropriate metrics relevant to the specific task, such as accuracy, precision, recall, F1-score, AUC, etc., considering the model’s purpose. Beyond simple accuracy, a detailed error analysis is needed, identifying common error types and their potential causes. Computational efficiency, including training time and inference speed, are essential aspects, especially for real-world applications. The analysis must also consider resource utilization (GPU memory, CPU usage, etc.) Furthermore, robustness and generalizability should be tested, evaluating the model’s performance across diverse datasets and under various conditions (e.g., noisy data, different data distributions). Finally, a comparison to relevant baselines provides context and demonstrates the model’s relative strengths and weaknesses.
Future Directions#
Future research could explore extending UniGuide’s capabilities to handle diverse molecular properties beyond geometry, such as quantum properties or specific chemical functionalities. This would involve developing new condition maps that effectively capture and translate these properties into guidance signals compatible with diffusion models. Another promising avenue lies in improving UniGuide’s efficiency and scalability. Currently, generating large molecules or handling complex conditions might be computationally expensive. Optimizing algorithms, exploring more efficient data structures, and harnessing parallel computing could greatly enhance performance. Furthermore, investigating the application of UniGuide to other generative models, such as autoregressive or flow-based models, would broaden its applicability and potentially unlock new levels of control and versatility in molecular design.
More visual insights#
More on figures
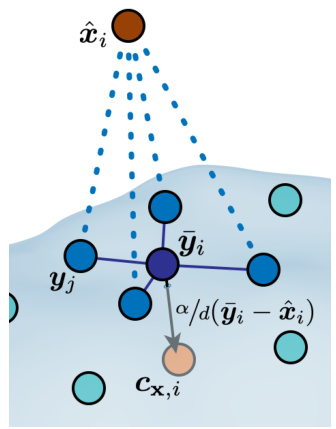
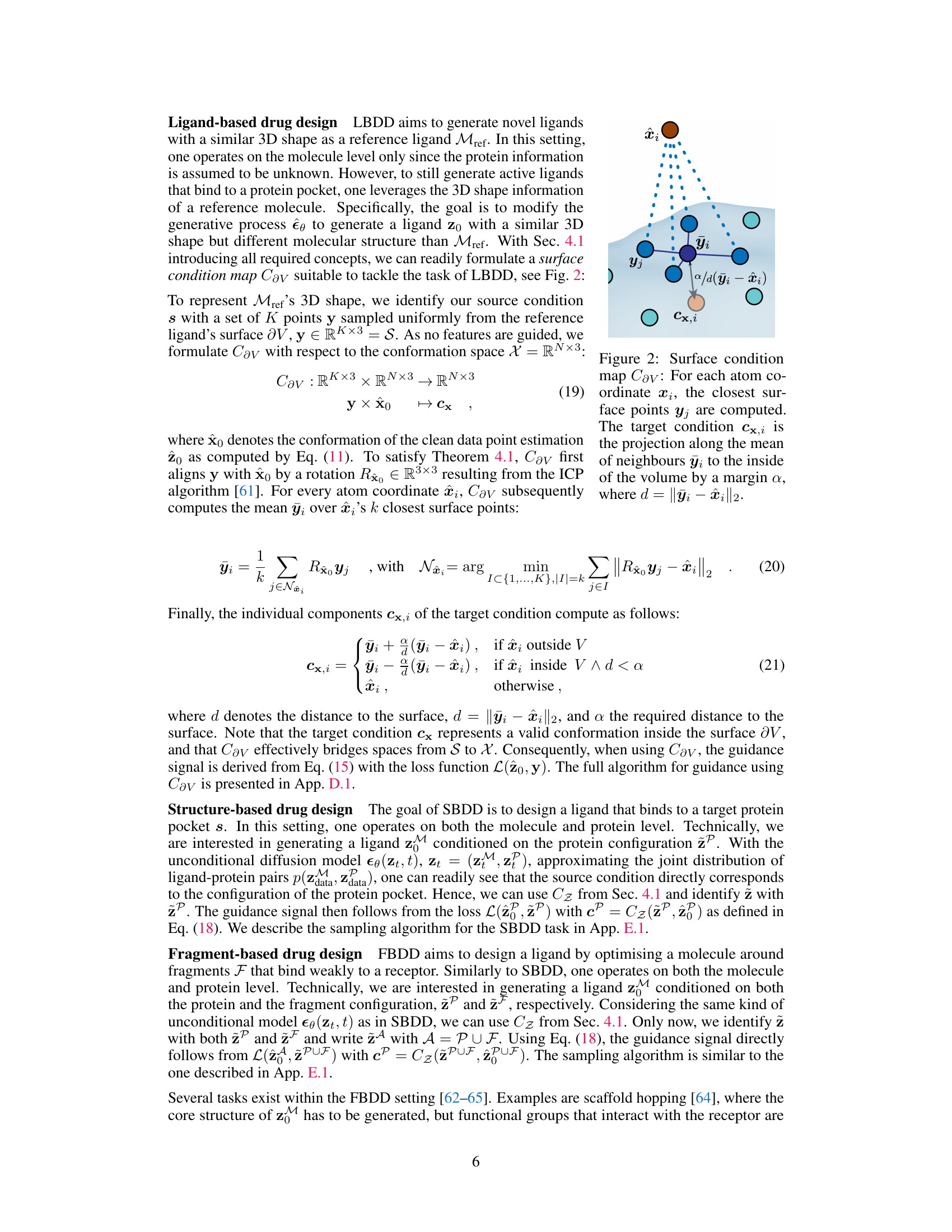
This figure shows how the surface condition map Cav works for ligand-based drug design (LBDD). The map takes as input a set of points (y) sampled from the surface of a reference ligand and the estimated clean conformation (x0) from an unconditional model. To align the reference ligand’s shape with the molecule being generated, a rotation (Rx) is applied using the Iterative Closest Point (ICP) algorithm. The algorithm then computes the mean of the k nearest neighbors (ỹi) for each atom (xi) and projects it inwards by a margin (a) to generate a target condition (cxi) for guiding the molecule generation process.

This figure shows two examples of ligand molecules generated using the UniGuide method. The goal of the ligand-based drug design (LBDD) task is to create new molecules that have a similar 3D shape to a reference molecule, but different molecular structures. The figure demonstrates that UniGuide successfully generated two ligands (in blue) that are very similar in 3D shape to their respective reference molecules (in grey). Importantly, the figure emphasizes that the generated ligands have low molecular graph similarity to their reference molecules, indicating that UniGuide can generate molecules with the same 3D shape but substantially different structures. This is a key aspect of the LBDD task, which aims to find novel molecules with similar shapes but potentially different interactions with a target protein.

This figure shows two examples of ligands generated using UniGuide with shape conditioning. The goal of the method is to create molecules with similar 3D shapes to a reference ligand, but different molecular structures (low graph similarity, high shape similarity). The figure visually demonstrates that UniGuide can successfully generate molecules fulfilling this objective.
This figure illustrates the versatility of UniGuide in handling diverse conditioning modalities for geometry-conditioned molecular generation. It shows how UniGuide incorporates different types of geometric constraints (target receptor for structure-based drug design, fragments for fragment-based drug design, and 3D shape for ligand-based drug design) into an unconditional diffusion model using a condition map. The figure highlights the flexibility of UniGuide, emphasizing its adaptability to various geometric tasks beyond the three examples shown, such as conditioning on atomic densities.
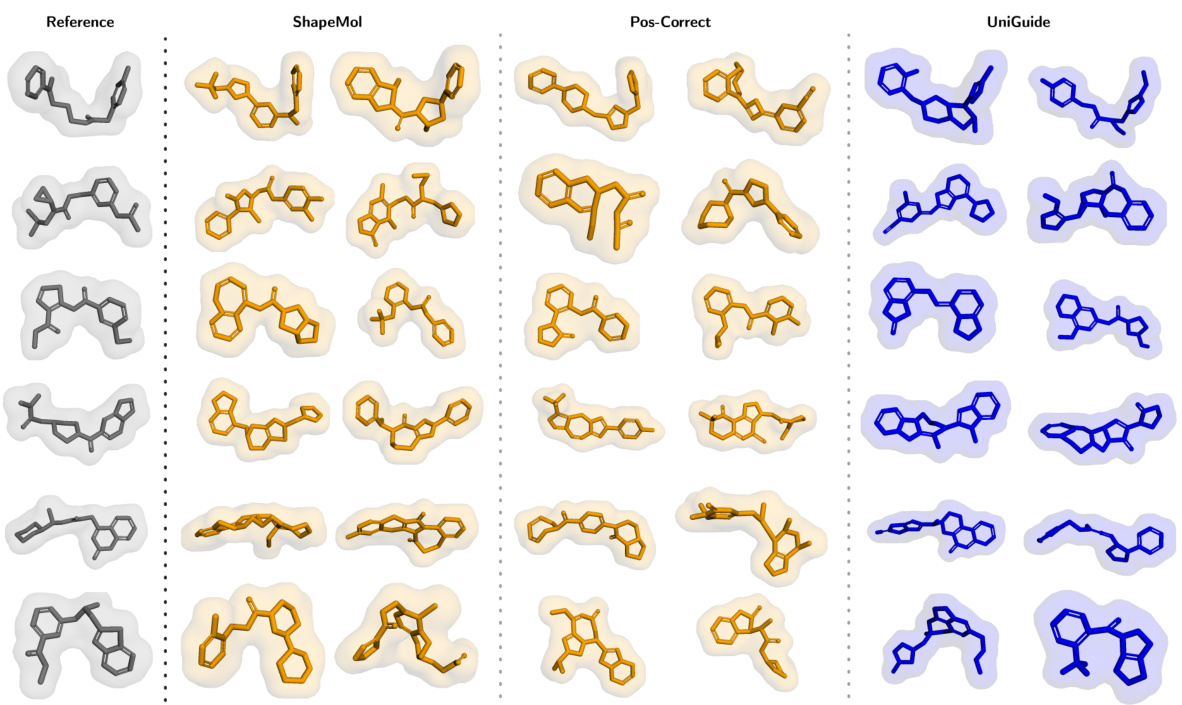
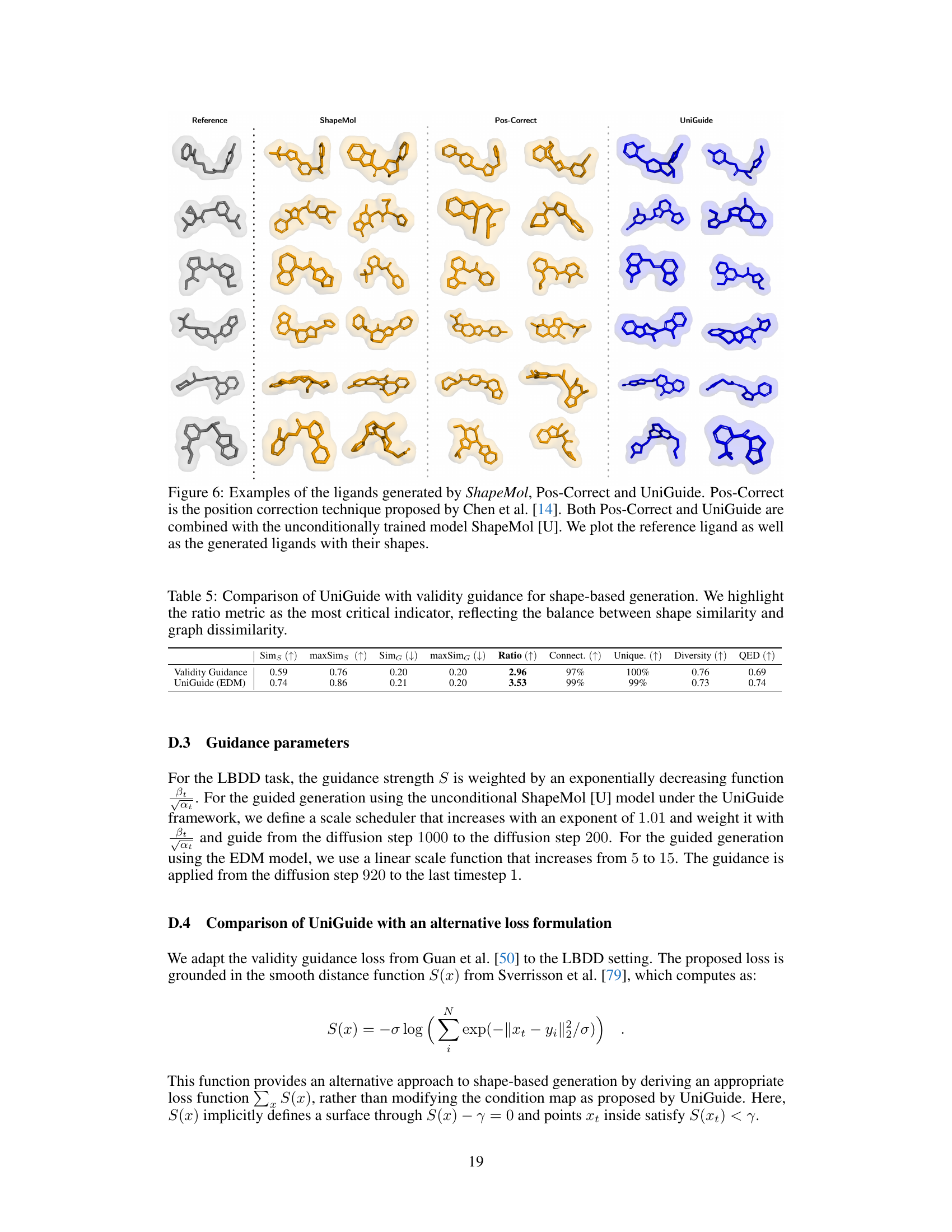
This figure compares the 3D shapes of ligands generated by three different methods: ShapeMol, Pos-Correct, and UniGuide. All three methods used an unconditionally trained ShapeMol model. The goal was to generate ligands that have similar 3D shapes to a reference ligand, but different molecular structures. The figure shows that UniGuide is able to generate ligands with similar shapes to the reference ligand, but with a wider variety of structures compared to ShapeMol and Pos-Correct.
This figure illustrates the UniGuide framework’s ability to handle various geometric conditions for molecular generation. It shows how UniGuide combines a source condition (e.g., target receptor, molecular fragments, 3D shape) with an unconditional diffusion model to guide the generation process. The key innovation is the condition map, which transforms complex geometric conditions into a format compatible with the diffusion model, enabling flexible conditioning during inference without additional training or networks. The framework can adapt to new geometric tasks beyond the examples shown (e.g., conditioning on atomic densities).
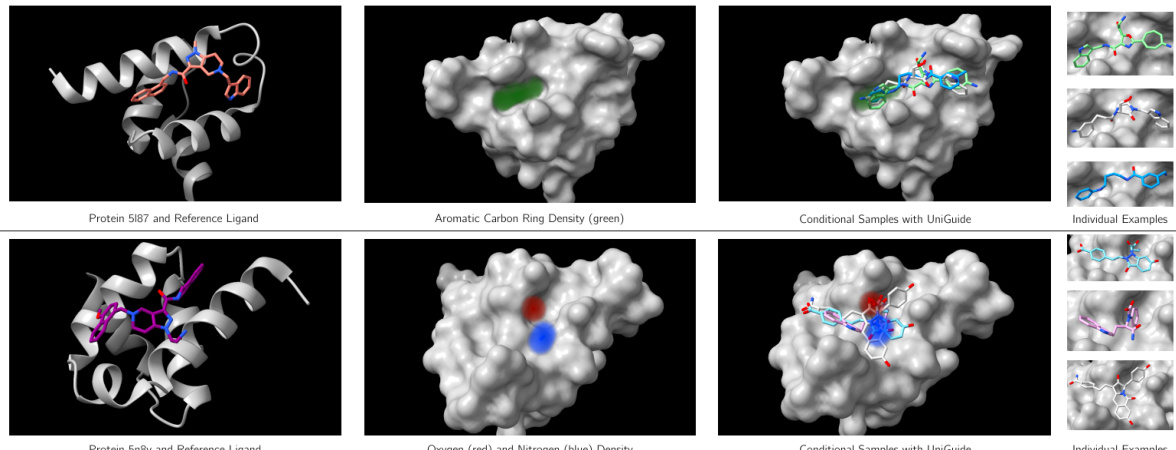
This figure shows two examples of how UniGuide can be extended to generate molecules with desired atom densities. The top row shows a protein (5I87) and its reference ligand, along with a density map highlighting aromatic carbon rings (in green). UniGuide is then used to generate new ligands, shown in the ‘Conditional Samples with UniGuide’ section and also individually in the ‘Individual Examples’ section. These generated ligands incorporate the desired aromatic carbon rings. The bottom row follows the same procedure, but focuses on generating ligands with specific oxygen (red) and nitrogen (blue) atom densities for a different protein (5n8v). The generated ligands successfully include these desired atoms.
More on tables
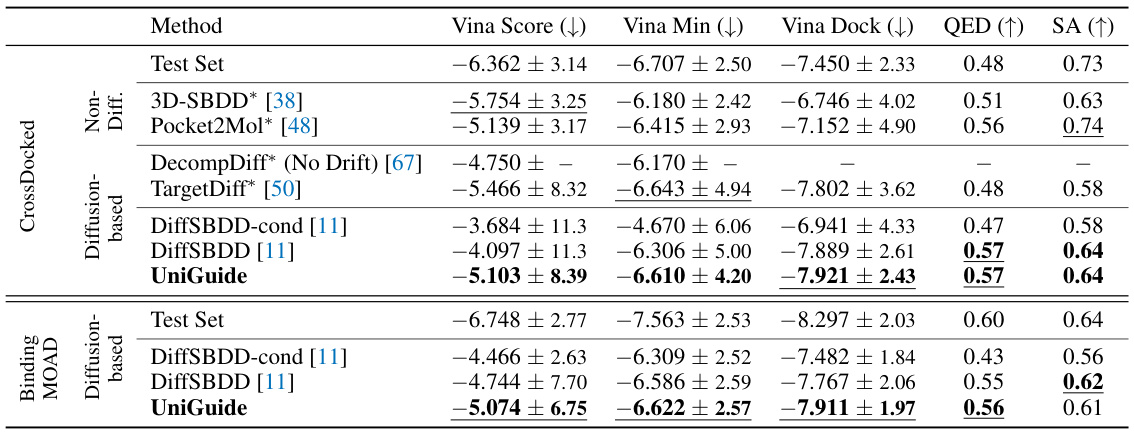
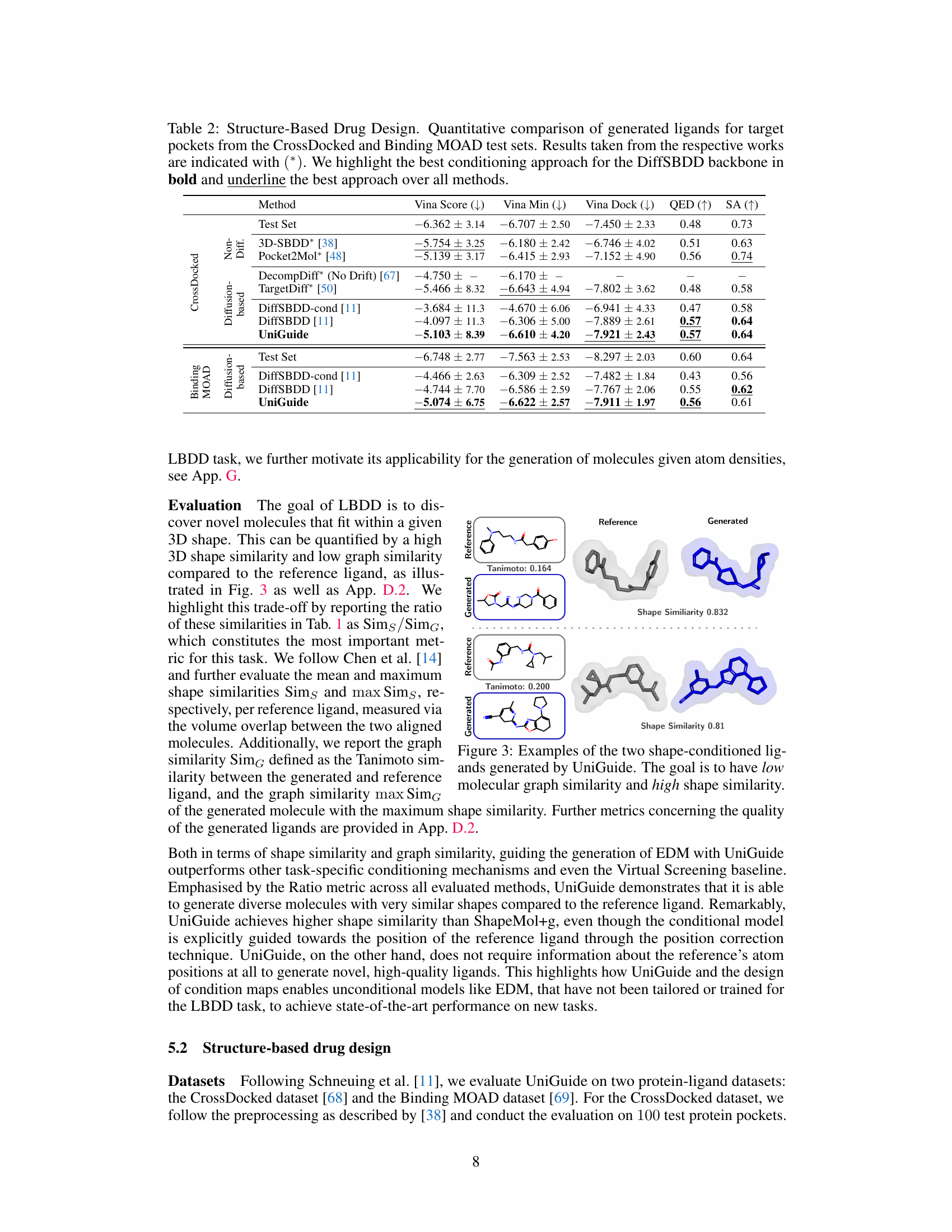
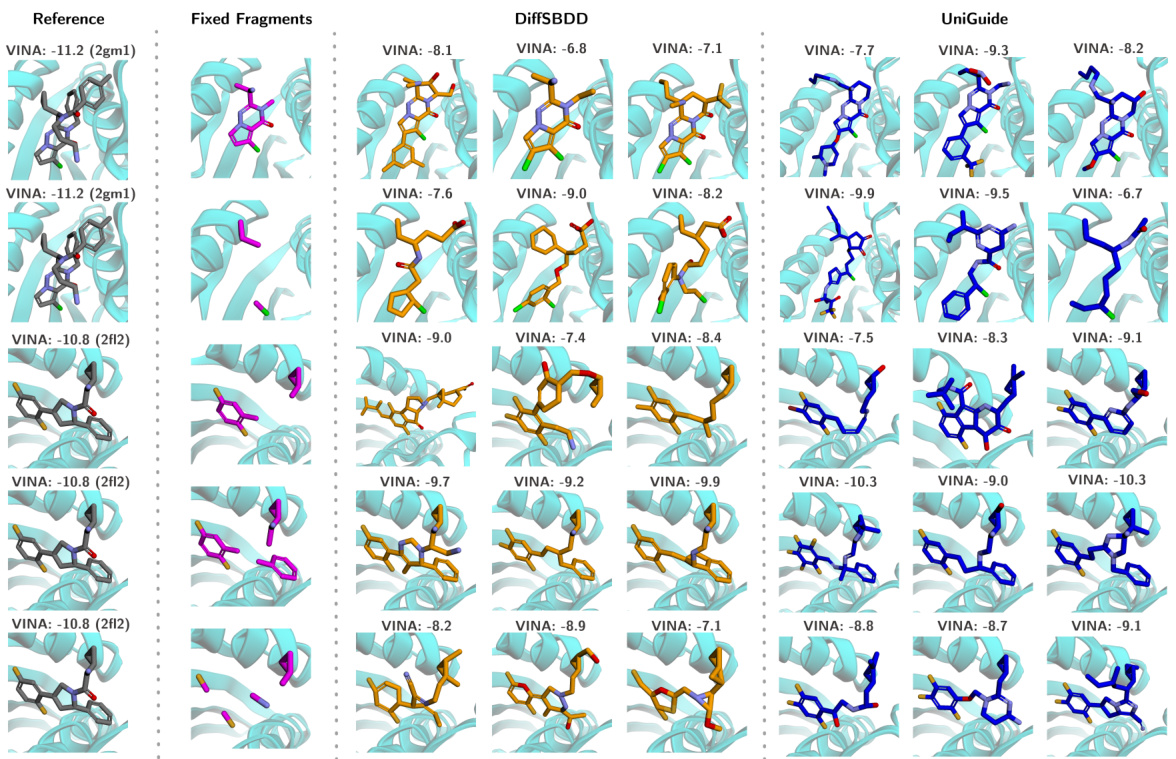
This table quantitatively compares the performance of UniGuide against several state-of-the-art baselines on two structure-based drug design datasets (CrossDocked and Binding MOAD). The metrics used are Vina Score (lower is better, indicating higher binding affinity), Vina Min (lower is better, indicating better binding in the best docking pose), Vina Dock (lower is better), QED (Quantitative Estimate of Drug-likeness, higher is better), and SA (Synthetic Accessibility, higher is better). The table highlights UniGuide’s competitive performance, often outperforming specialized methods.
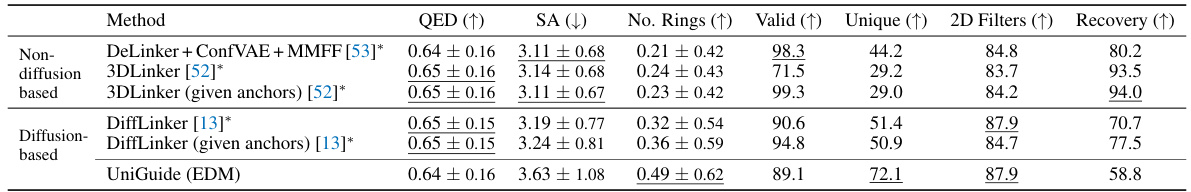
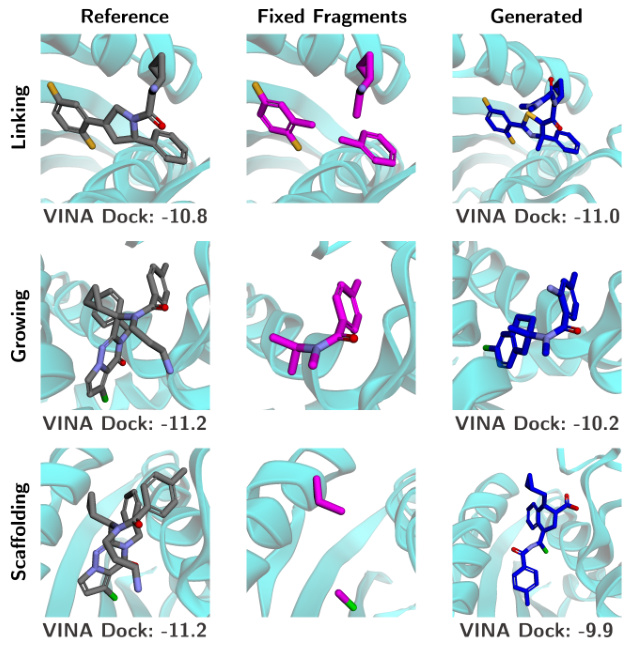
This table compares the performance of different methods for linker design, a subtask of fragment-based drug design. It evaluates several methods including DeLinker, 3DLinker, DiffLinker and UniGuide across various metrics: QED (quantitative estimate of drug-likeness), SA (synthetic accessibility), Number of Rings, Valid (percentage of valid molecules), Unique (percentage of unique molecules), 2D Filters (number of 2D filters), and Recovery (percentage of recovered ligands). The results show that UniGuide (using EDM), while not specifically designed for this task, achieves comparable or better results than specialized methods like DiffLinker, particularly in terms of generating unique and valid linkers. The results highlight the versatility of the UniGuide framework.
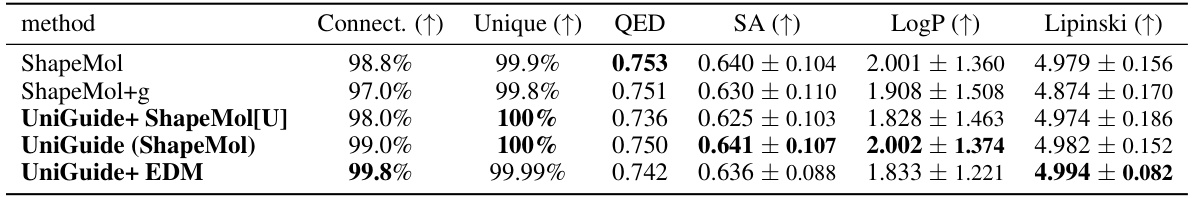
This table compares various methods for ligand-based drug design, focusing on shape similarity and diversity. It presents quantitative results from different approaches, including non-diffusion-based methods (VS, SQUID, ShapeMol) and diffusion-based methods (ShapeMol+g, UniGuide with ShapeMol and EDM). The metrics used for evaluation are shape similarity, maximum shape similarity, graph similarity, maximum graph similarity, ratio of shape to graph similarity, diversity, and the number of valid molecules generated. The results demonstrate that UniGuide, even when using simpler unconditional models, can achieve performance comparable to or better than specialized models.

This table presents a comparison of different methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference ligand. The table includes several non-diffusion and diffusion-based methods, evaluating metrics such as shape similarity, graph similarity, ratio (shape similarity to graph similarity), diversity, and drug-likeness (QED). The results highlight UniGuide’s performance against specialized models for LBDD.
This table presents the quantitative comparison of different methods for ligand-based drug design. The results from Chen et al. [14] are marked with an asterisk (*). The table highlights the best conditioning approach for the ShapeMol backbone (a specific method) in bold font and underlines the overall best performing method across all compared approaches. Metrics used for comparison include shape similarity, maximum shape similarity, graph similarity, maximum graph similarity, ratio of shape and graph similarity, diversity of generated molecules, and quantitative estimation of drug-likeness.

This table presents the quantitative comparison of different methods for ligand-based drug design, specifically focusing on the ability to generate molecules with similar 3D shapes to a reference ligand while maintaining diversity and other desirable properties. The results are organized by model type (non-diffusion-based vs. diffusion-based), and key metrics include shape similarity, maximum shape similarity, graph similarity, maximum graph similarity, ratio of shape similarity to graph similarity, diversity, and QED. The table shows that UniGuide (with ShapeMol and EDM as base models) performs either comparably to or better than task-specific baselines like ShapeMol and SQUID.

This table compares different methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference molecule while maintaining diversity. It shows the performance of various methods using several metrics, including shape similarity, graph similarity (to measure dissimilarity in molecular structure), diversity, and drug-likeness properties (QED, Ratio, and Lipinski). The results indicate that UniGuide with EDM achieves competitive or superior results to other specialized methods.

This table compares different methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference molecule. It presents results from various methods, including those based on diffusion models and other techniques. The metrics used to assess performance include shape similarity, maximum shape similarity, graph similarity, maximum graph similarity, the ratio of shape to graph similarity, diversity, and QED. The table highlights the best performance achieved using each method and across all methods.
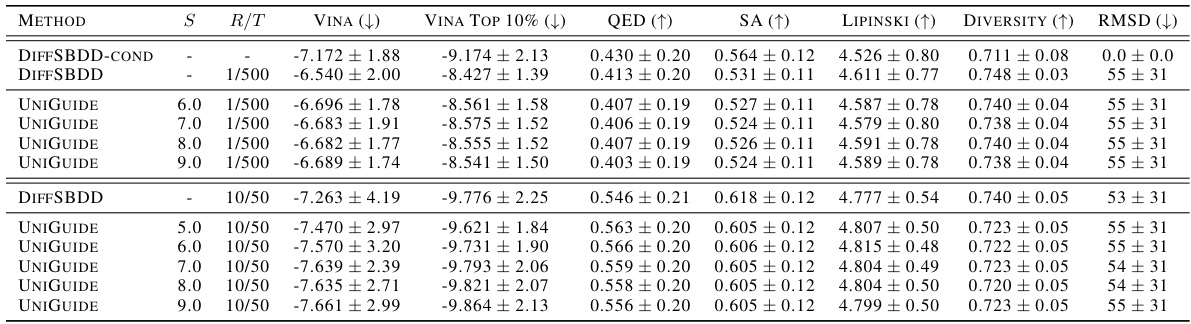
This table compares various methods for ligand-based drug design, focusing on shape similarity and diversity. It shows the performance of different models, including several diffusion-based approaches and a non-diffusion-based virtual screening method, across multiple metrics. The table highlights UniGuide’s competitive performance and superior diversity.
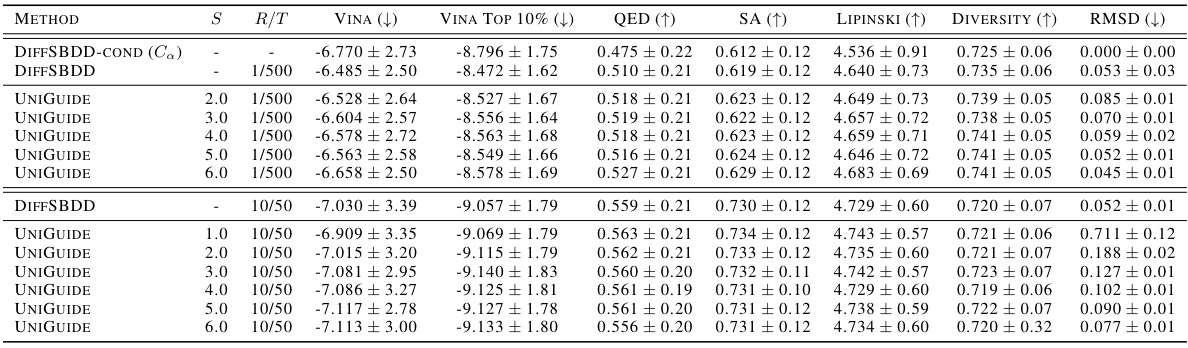
This table presents a comparison of various methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference ligand. The metrics used to evaluate performance include shape similarity, maximum shape similarity, graph similarity, maximum graph similarity, the ratio of shape similarity to graph similarity (a key metric reflecting the tradeoff between similar shape and dissimilar structure), diversity, and quantitative estimated drug-likeness (QED). Several baselines are compared against UniGuide, showcasing UniGuide’s performance with different unconditional models.
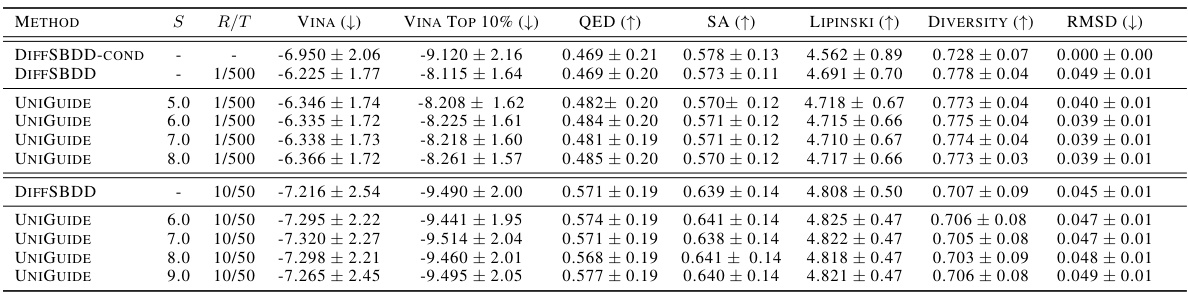
This table compares the performance of UniGuide against other methods for ligand-based drug design. Several metrics are used to evaluate the generated molecules, including shape similarity (Sims, maxSims), graph similarity (Simg, maxSimg), diversity, and the ratio of shape to graph similarity. The results demonstrate that UniGuide, even when using an unconditional model, can outperform specialized, conditional models.

This table presents a comparison of different methods for ligand-based drug design. The methods are categorized into non-diffusion-based and diffusion-based approaches. For each method, the table shows the shape similarity (Sims and maxSims), graph similarity (Simg and maxSimg), the ratio of shape similarity to graph similarity, and the diversity of generated molecules. The best performing method in each category is highlighted in bold, and the overall best-performing method is underlined. Results from a prior study by Chen et al. are marked with an asterisk.

This table presents a comparison of different methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference ligand while maintaining diversity and other desirable properties. The results are evaluated using metrics such as shape similarity, graph similarity, ratio of shape to graph similarity, diversity, and drug-likeness.

This table presents a comparison of different methods for ligand-based drug design, focusing on the ability to generate molecules with similar 3D shapes to a reference ligand. The table includes both non-diffusion and diffusion-based methods, and various metrics are reported to evaluate the performance, including shape similarity, graph similarity, ratio of shape to graph similarity (to capture the tradeoff between generating similar shapes while maintaining diversity), diversity of generated molecules, and the overall quality of the generated molecules (QED). The results highlight UniGuide’s competitive or superior performance compared to other specialized models.
Full paper#