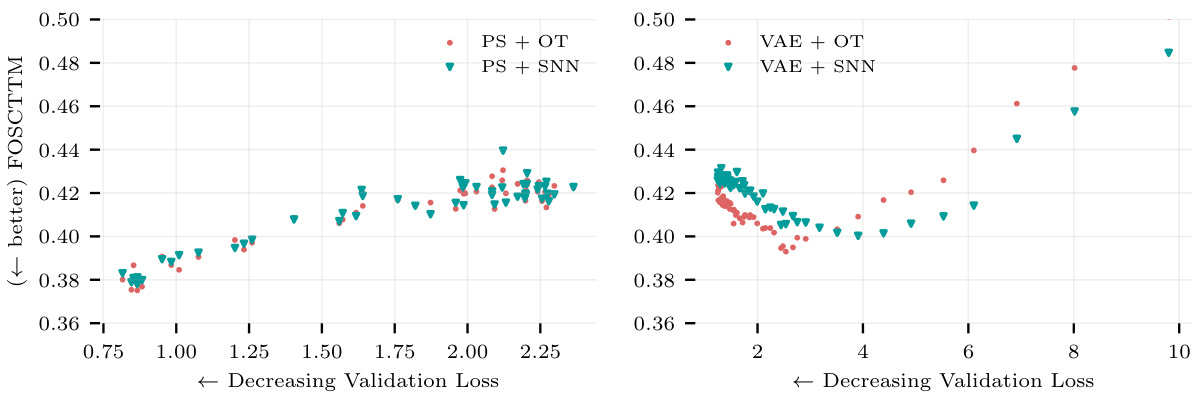
TL;DR#
Many multimodal representation learning techniques require paired samples, but collecting such data can be extremely challenging in many fields, particularly in biology where measurements may destroy samples. Existing approaches often struggle with unpaired data, which limits their applicability in diverse real-world scenarios. This paper tackles the problem by drawing an analogy between potential outcomes in causal inference and potential views in multimodal observations. It uses this analogy to estimate a common space for matching unpaired samples across modalities.
The core of the method is leveraging propensity scores, calculated from each modality, to identify and match samples with shared underlying latent states. The propensity score encapsulates all shared information, allowing definition of a distance between samples for alignment using two techniques: Shared Nearest Neighbors (SNN) and optimal transport (OT) matching. The paper demonstrates that OT matching with propensity score distances leads to significant improvements in alignment and prediction performance across various datasets (synthetic, CITE-seq, Perturb-Seq/single cell images). The improvements were observed in both in-distribution and out-of-distribution prediction tasks, showing the robustness and generalization ability of the approach.
Key Takeaways#
Why does it matter?#
This paper is important because it presents a novel approach to multimodal representation learning using propensity scores, addressing the challenge of aligning unpaired data which is prevalent in many fields like biology and medicine. The proposed methodology is simple, efficient and highly versatile, with the potential to significantly advance multimodal learning across various domains. It opens new avenues for research by leveraging causal inference and optimal transport techniques to tackle the challenge of unpaired data, and improving the performance of various downstream tasks. This is highly relevant given the increasing availability of large-scale unpaired multimodal data and the limitations of existing methods for handling such data.
Visual Insights#

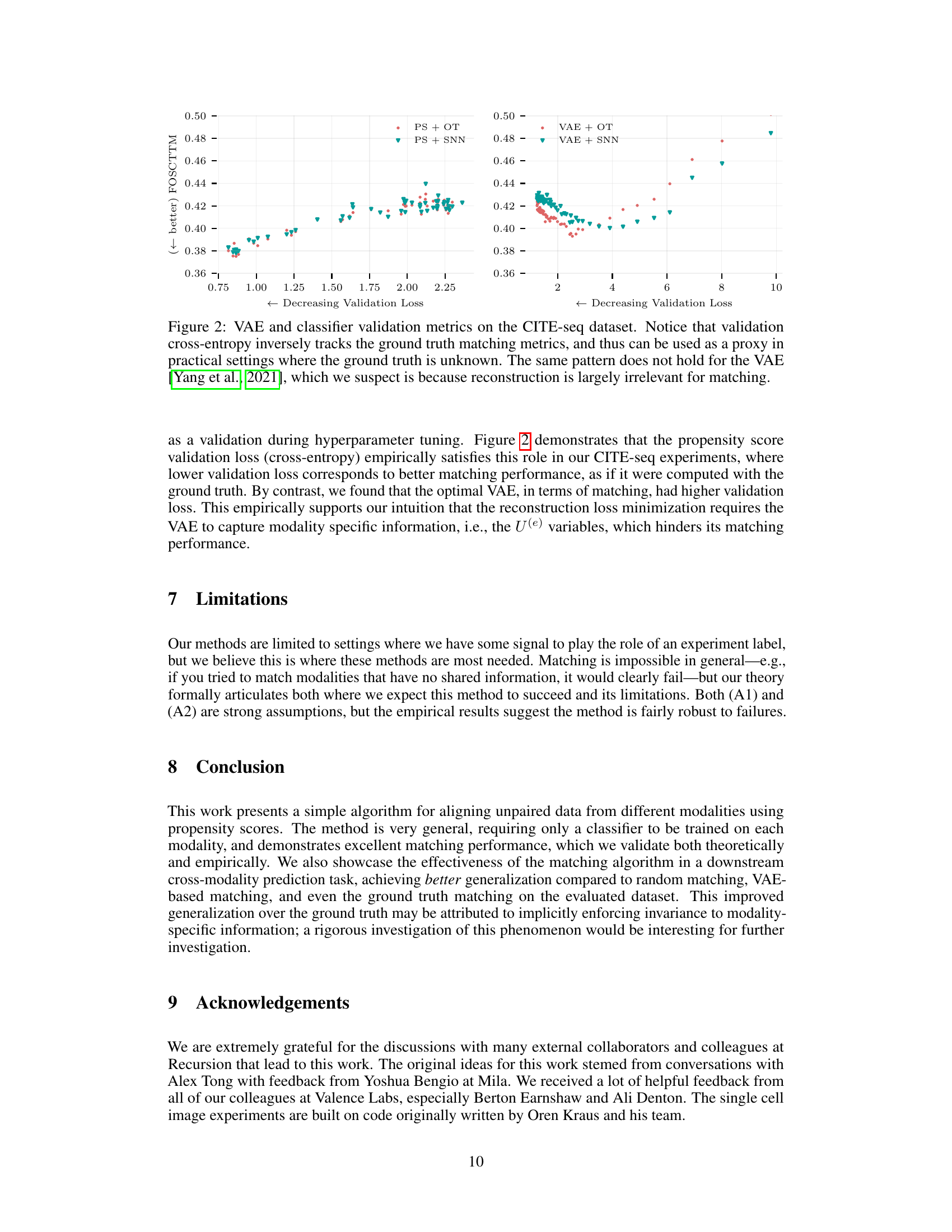
🔼 This figure illustrates the process of propensity score matching for aligning unpaired multimodal data. Two modalities (e.g., microscopy images and gene expression data) are shown. Separate classifiers estimate propensity scores for each modality, reflecting the shared information between the modalities and a treatment variable (t). These scores are then used to match samples across modalities within each treatment group, effectively creating pseudo-paired data by finding samples with similar propensity scores despite coming from different data sources.
read the caption
Figure 1: Visualization of propensity score matching for two modalities (e.g., Microscopy images and RNA expression data). We first train classifiers to estimate the propensity score for samples from each modalities; the propensity score reveals the shared information p(t|z_i), which allows us to re-pair the observed disconnected modalities. The matching procedure is then performed within each perturbation class based on the similarity bewteen the propensity scores.
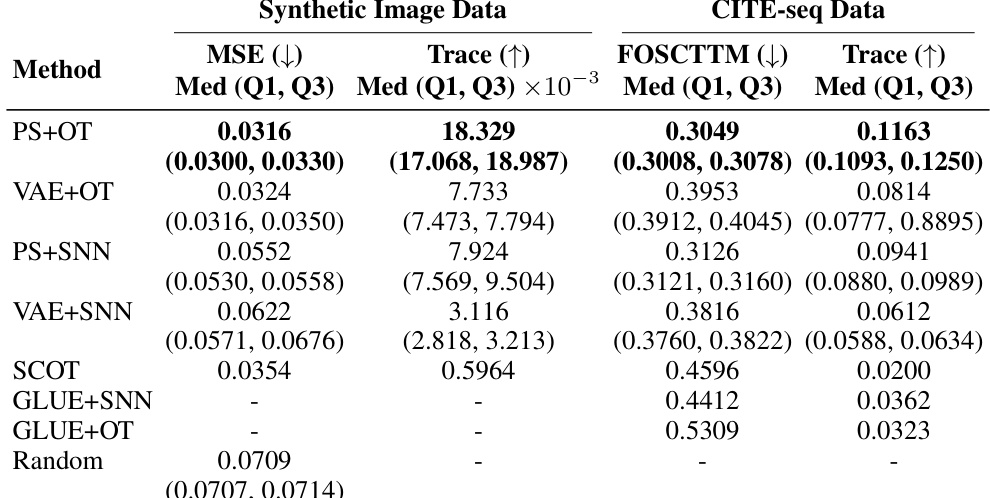
🔼 This table presents the results of two different alignment methods (OT and SNN) applied to two datasets: synthetic interventional images and CITE-seq data. The methods are compared using four metrics: Mean Squared Error (MSE), Trace, Fraction Of Samples Closer Than the True Match (FOSCTTM), and Trace. Lower MSE and FOSCTTM values are preferred, while higher Trace values are better. The results show the performance of propensity score-based matching (PS) compared to the performance of a Variational Autoencoder (VAE)-based approach for each alignment method and dataset.
read the caption
Table 1: Alignment metrics results using synthetic interventional image dataset and CITE-seq data.
In-depth insights#
Unpaired Multimodal#
The concept of “Unpaired Multimodal” learning tackles the challenge of integrating information from multiple sources (modalities) where data isn’t paired. This is a significant hurdle because traditional multimodal techniques rely on aligned data, such as image-caption pairs. The lack of pairing necessitates novel approaches to establish correspondences between different modalities. Propensity score matching, for example, emerges as a powerful strategy to tackle this. By leveraging the shared information between modalities, even in the absence of explicit pairings, models can learn relationships and create a common representation space. This approach is especially valuable in domains where obtaining paired data is difficult or impossible, such as in biology or medical imaging. However, it introduces new complexities, including the need for methods to handle uncertainty and missing data, and careful consideration must be given to biases that could be introduced by unpaired data and the effectiveness of the matching techniques employed.
Propensity Score#
Propensity score methods are crucial for causal inference, particularly when dealing with observational data. They aim to mitigate selection bias by creating balanced groups for comparison. In the context of multimodal data, propensity scores offer a powerful way to align unpaired observations across different modalities. By modeling the probability of a sample belonging to a certain treatment group based on observed features, propensity scores can help to estimate a common space where samples can be meaningfully matched, even when direct pairings are not available. This matching procedure is particularly valuable in situations where collecting paired samples is costly or impossible, as in biological experiments. Optimal transport (OT) techniques can further enhance propensity score-based alignment by leveraging the resulting propensity score distances to construct effective matching matrices. The overall approach relies on the identifiability of the propensity score, which ensures that this quantity can be estimated from individual modalities. Limitations lie in the assumptions about the data generating process, specifically regarding the independence of treatment and modality-specific noise. However, the empirical results demonstrate robustness to violations of these assumptions, suggesting that the method offers a powerful approach to a difficult problem in multimodal data analysis.
OT Matching#
Optimal Transport (OT) matching, a core component of the research, offers a powerful technique for aligning unpaired multimodal data. Unlike simpler methods relying on direct feature comparisons, OT leverages a cost function derived from propensity scores to find the optimal alignment between disparate modalities. This approach is particularly robust to differences in feature spaces and noise levels inherent in real-world data, making it highly relevant for applications in biology and other domains with noisy and heterogeneous data. The use of OT enables soft matching, meaning multiple samples can contribute to the alignment process unlike traditional methods. This is crucial for handling the inherent uncertainty and sparsity in unpaired data. The researchers found that OT consistently outperformed alternative alignment methods in experiments, demonstrating its capacity to integrate information across modalities effectively, even under noisy conditions. The choice of OT underscores the importance of mathematically grounded approaches when dealing with complex multimodal problems.
Downstream Tasks#
The section on “Downstream Tasks” in this research paper is crucial as it demonstrates the practical utility and effectiveness of the proposed propensity score alignment method. Instead of merely focusing on the alignment quality, the authors cleverly employ this aligned data to tackle a challenging cross-modality prediction problem. This approach is commendable as it directly addresses a real-world scenario where paired data is scarce. The use of both barycentric projection and stochastic gradients is a noteworthy technical detail, showcasing a sophisticated understanding of how to leverage the soft matching matrix for downstream tasks. The evaluation metrics are carefully chosen to reflect both the accuracy of the predictions and the generalizability of the model. Experiments on real-world datasets, such as the CITE-seq data, further strengthen the claims by demonstrating significant improvements over existing approaches. The focus on a practical application, rather than purely theoretical metrics, makes this section highly valuable, highlighting the practical impact of the research.
Biological Data#
In the realm of biological research, the integration of diverse data modalities presents a significant challenge. Multimodal learning techniques, aiming to unify and analyze data from various sources like genomics, imaging, and proteomics, require careful consideration. A key challenge is the scarcity of paired datasets, meaning that corresponding measurements across modalities are often unavailable, hindering the development and validation of effective multimodal models. This limitation becomes critical in scenarios like single-cell analysis where measurements may be destructive, making paired observations impossible. Therefore, methods which can effectively handle unpaired multimodal data through approaches like propensity score alignment are crucial for extracting valuable insights from biological experiments. Propensity score alignment helps to overcome this limitation by establishing a common space for comparison of samples from different sources, thereby enabling the development of robust and comprehensive models capable of leveraging rich and complex biological data.
More visual insights#
More on figures
🔼 This figure illustrates the propensity score matching method for aligning unpaired multimodal data. It shows how classifiers are trained on each modality to estimate a propensity score, which captures shared information between modalities. These scores then allow for matching samples across modalities based on similarity, particularly within the same experimental perturbation group. The example modalities shown are microscopy images and RNA expression data.
read the caption
Figure 1: Visualization of propensity score matching for two modalities (e.g., Microscopy images and RNA expression data). We first train classifiers to estimate the propensity score for samples from each modalities; the propensity score reveals the shared information p(tzi), which allows us to re-pair the observed disconnected modalities. The matching procedure is then performed within each perturbation class based on the similarity bewteen the propensity scores.

🔼 This figure illustrates the propensity score matching method for aligning unpaired multimodal data. It shows how classifiers are trained on each modality to estimate propensity scores, which capture shared information between modalities. These scores are then used to find the best matches between samples across modalities within the same treatment group, effectively creating paired data from unpaired data.
read the caption
Figure 1: Visualization of propensity score matching for two modalities (e.g., Microscopy images and RNA expression data). We first train classifiers to estimate the propensity score for samples from each modalities; the propensity score reveals the shared information p(t|zi), which allows us to re-pair the observed disconnected modalities. The matching procedure is then performed within each perturbation class based on the similarity bewteen the propensity scores.

🔼 This figure illustrates the propensity score matching method for two modalities. It begins by training separate classifiers on each modality to estimate a propensity score for each sample. This score reflects the shared information between the modalities and the treatment applied (the experimental perturbation). Then, within each treatment group, a matching procedure (such as shared nearest neighbours or optimal transport) is used to match samples based on the similarity of their propensity scores, effectively creating aligned pairs from the originally unpaired data.
read the caption
Figure 1: Visualization of propensity score matching for two modalities (e.g., Microscopy images and RNA expression data). We first train classifiers to estimate the propensity score for samples from each modalities; the propensity score reveals the shared information p(tzi), which allows us to re-pair the observed disconnected modalities. The matching procedure is then performed within each perturbation class based on the similarity bewteen the propensity scores.
More on tables

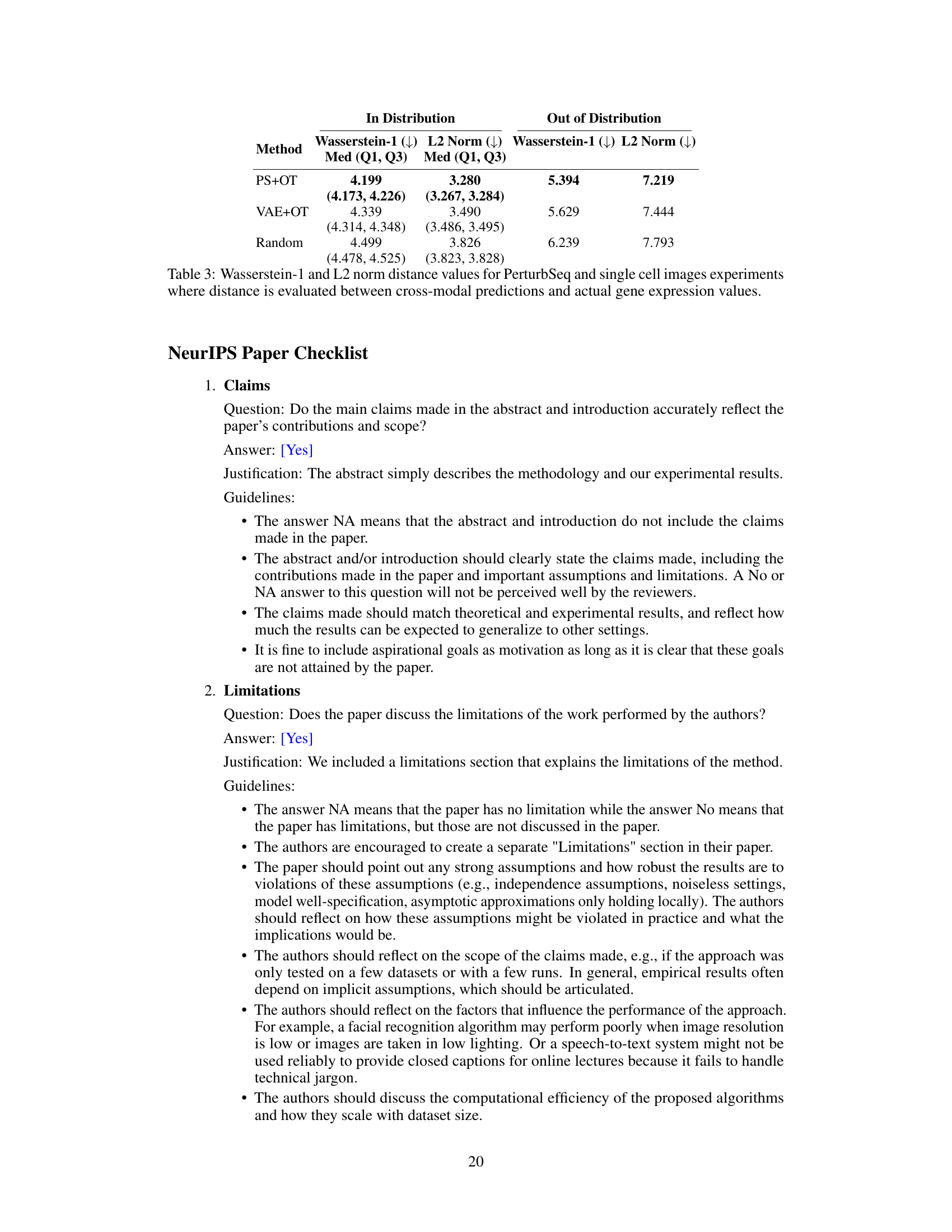
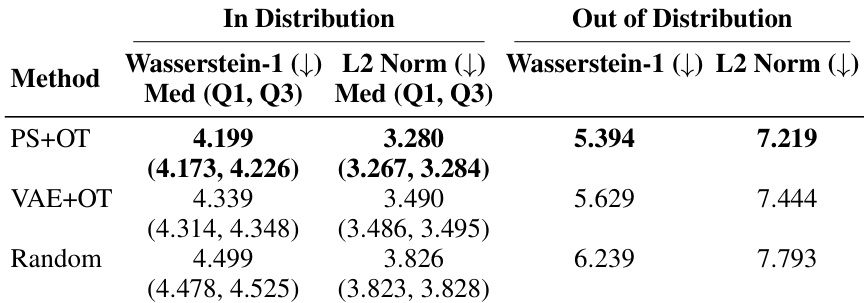
🔼 This table presents the results of cross-modality prediction experiments using two datasets: CITE-seq and PerturbSeq/single cell image data. It evaluates the performance of different matching methods (Random, VAE+OT, PS+OT) using two different loss functions (MSE and Unbiased Loss) for the CITE-seq data. For the PerturbSeq/single cell image data, it assesses the performance using Kullback-Leibler (KL) divergence as a metric, both for in-distribution and out-of-distribution data. The results are expressed as medians with interquartile ranges (Q1, Q3). The table also includes the performance of using true pairs for comparison.
read the caption
Table 2: Cross-modal prediction results using CITE-seq data and PerturbSeq/single cell image data including an out of distribution distance evaluation for PerturbSeq/single cell images.
🔼 This table presents the results of applying different matching methods on two datasets: synthetic interventional images and CITE-seq data. The methods compared include propensity score matching with optimal transport (PS+OT), propensity score matching with shared nearest neighbors (PS+SNN), VAE-based matching with optimal transport (VAE+OT), VAE-based matching with shared nearest neighbors (VAE+SNN), SCOT, GLUE+SNN, GLUE+OT, and a random matching baseline. For each method and dataset, the median, first quartile (Q1), and third quartile (Q3) of two evaluation metrics are reported: Mean Squared Error (MSE) and Trace. Lower MSE values and higher Trace values indicate better performance. The results show that propensity score matching with optimal transport consistently outperforms other methods across both datasets.
read the caption
Table 1: Alignment metrics results using synthetic interventional image dataset and CITE-seq data.
Full paper#