↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Multimodal biomedical data analysis is crucial for better healthcare, but integrating diverse data sources (images, tabular data, etc.) is complex and current methods often fail to fully capture cross-modal information or struggle with missing data. This paper introduces HEALNet, a novel multimodal fusion architecture that tackles these challenges.
HEALNet uses a hybrid early-fusion approach, preserving each modality’s structural information while learning cross-modal interactions in a shared latent space. It’s designed to be robust to missing modalities during training and inference, improving the reliability of predictions and the model’s adaptability to real-world scenarios. Experiments on cancer datasets demonstrate that HEALNet achieves state-of-the-art performance, significantly outperforming existing methods.
Key Takeaways#
Why does it matter?#
This paper is important because it presents HEALNet, a novel and effective method for multimodal fusion in biomedical data analysis. This addresses a critical need in the field, where integrating heterogeneous data sources like images and tabular data is crucial but challenging. The results show state-of-the-art performance on survival prediction tasks, highlighting the model’s effectiveness and robustness. The availability of the code opens up new avenues for further research and development in multimodal deep learning, impacting various applications in healthcare.
Visual Insights#

This figure provides a detailed overview of the HEALNet architecture. It illustrates the hybrid early fusion approach, showcasing how modality-specific information is preserved through separate encoders while cross-modal interactions are captured in a shared latent space. The iterative attention mechanism and weight sharing between layers for efficiency are also depicted. The figure highlights the model’s ability to handle diverse data types (image and tabular) and its explainability due to the use of attention weights.

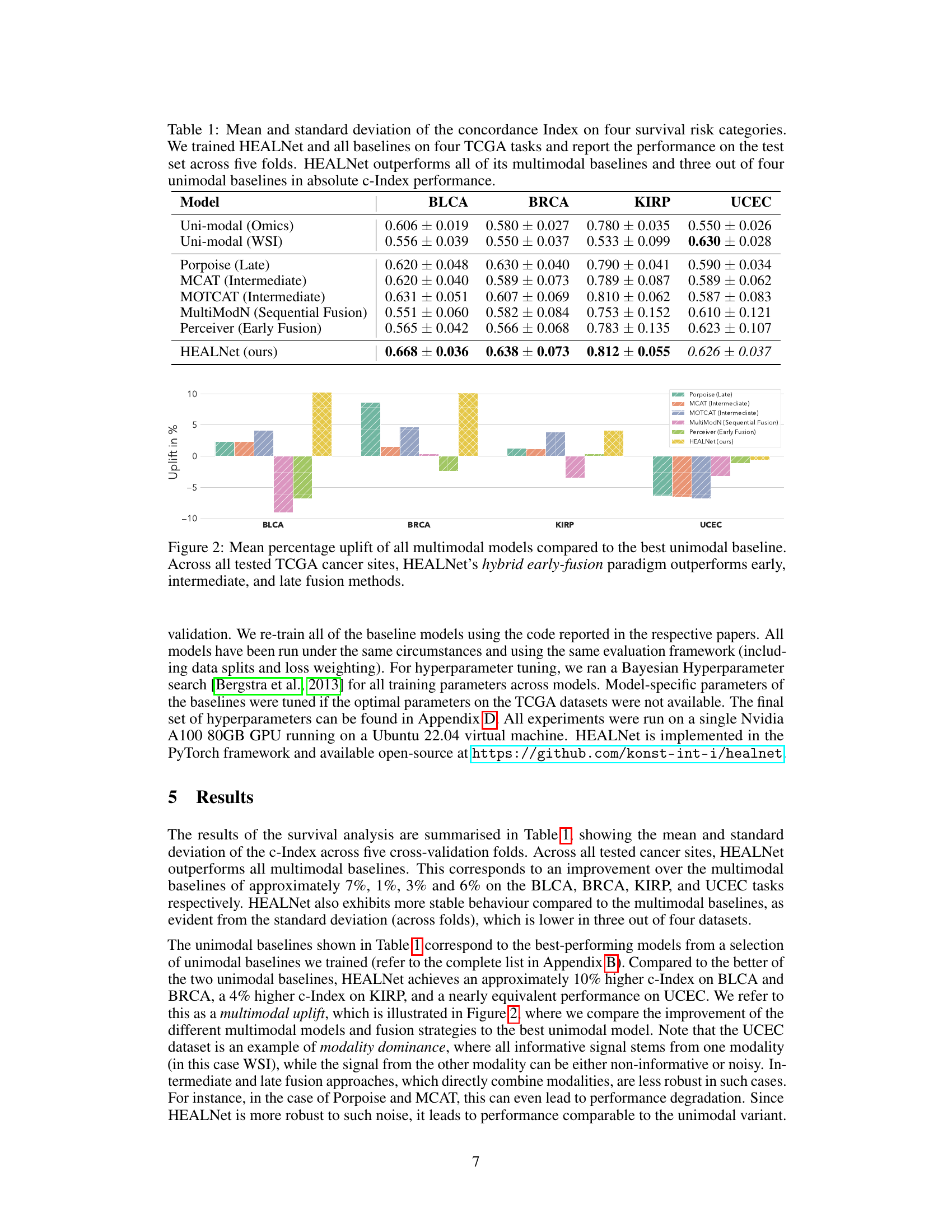
This table presents the mean and standard deviation of the concordance index (c-index) achieved by HEALNet and various baseline models across four different cancer datasets from The Cancer Genome Atlas (TCGA). The c-index is a measure of the predictive accuracy of survival models. The table shows that HEALNet outperforms most other models in terms of both mean c-index and consistency (lower standard deviation).
In-depth insights#
Hybrid Fusion’s Edge#
The concept of “Hybrid Fusion’s Edge” in multimodal learning suggests a significant advantage by combining the strengths of early and intermediate fusion methods. Early fusion, while simple, often loses crucial modality-specific information. Intermediate fusion, although preserving individual structures, can struggle with interpretability and handling missing data. A hybrid approach, therefore, seeks to leverage the benefits of both: the efficiency of early fusion with the preservation of detailed information from intermediate fusion. This likely involves a carefully designed architecture that integrates modalities at an early stage, yet incorporates mechanisms (like attention) to learn and maintain individual modality nuances within a shared representation. The “edge” arises from improved performance, better interpretability, and robustness to missing data. A successful hybrid fusion model should outperform both unimodal and purely early or intermediate fusion approaches, offering a compelling solution for complex multimodal tasks.
Missing Modality Robustness#
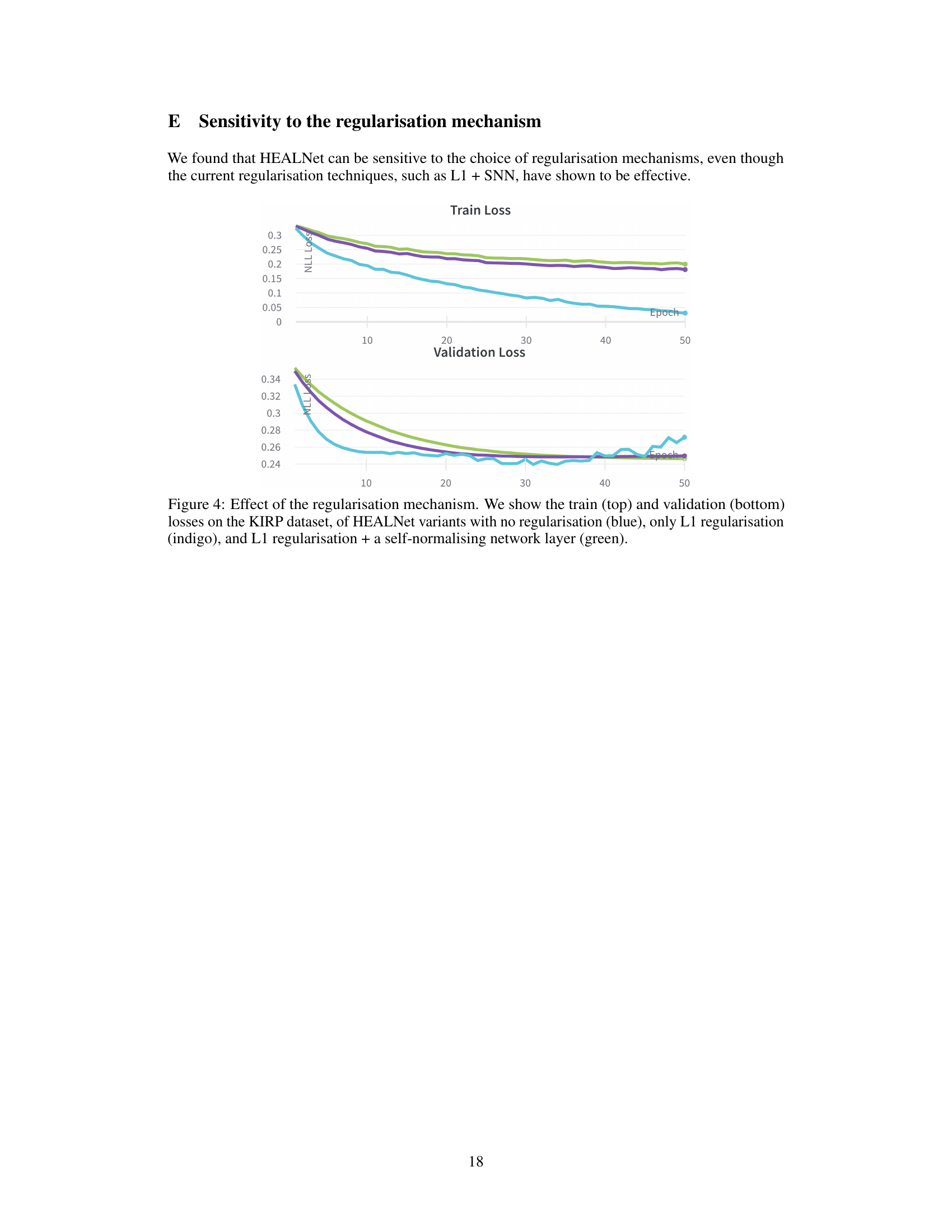
Missing modality robustness is a crucial aspect of multimodal learning, especially in real-world applications where data is often incomplete. A robust model should gracefully handle the absence of one or more modalities during inference without significant performance degradation. The core challenge lies in designing models that can effectively leverage the available information while mitigating the negative impact of missing data. This might involve techniques such as attention mechanisms to selectively focus on available features, or sophisticated imputation methods to estimate missing data. A key consideration is the balance between performance on complete data and resilience to missing modalities, as overly complex imputation methods might introduce noise and hinder performance. A successful approach often demands a deep understanding of the data’s inherent structure and relationships between different modalities to effectively capture the underlying patterns even with missing pieces. Evaluation of missing modality robustness requires careful consideration of different missing data scenarios, including random and systematic missingness, to fully understand the model’s limitations and strengths.
Attention Mechanism Insights#
An attention mechanism, in the context of a multimodal deep learning model processing biomedical data, offers a powerful way to weigh the importance of different input modalities. Early fusion approaches often suffer from loss of structural information. However, a well-designed attention mechanism can capture cross-modal interactions and learn the relationships between different data types (e.g., images, tabular data, graphs) even when the modalities have different structures. This selective weighting allows the model to focus on the most relevant features and learn more effective representations. Modality-specific attention further enables the model to retain crucial structural information from each modality, avoiding complete homogenization of the input. Hybrid early-fusion techniques which leverage both shared and modality-specific attention offer an effective balance between leveraging cross-modal interactions while maintaining individual modality structures. The interpretability of attention weights is a critical advantage, providing insights into which modalities are most influential in the model’s predictions. This can be instrumental in understanding the model’s decision-making process and potentially for clinical applications. Careful consideration of the attention mechanism’s design (e.g., type of attention, number of layers) is vital for optimal model performance, balancing the expressiveness of the model with computational efficiency. Missing data handling can also benefit greatly from well-designed attention as the model can dynamically adapt to missing inputs, providing robustness in real-world scenarios.
High-Dimensional Data Handling#
Handling high-dimensional biomedical data, such as gigapixel whole slide images (WSIs) and multi-omic datasets, presents significant challenges. HEALNet addresses this by employing a hybrid early-fusion approach, combining raw data early in the process to avoid issues with dimensionality explosion. The use of iterative attention mechanisms allows for efficient learning and reduces computational burden associated with large input sizes. The model’s design naturally handles missing data modalities at inference time, a critical advantage in clinical settings where complete data is often unavailable. Furthermore, the architecture’s focus on learning from raw data, rather than opaque embeddings, promotes model interpretability and explainability. By capturing cross-modal interactions and structural information effectively, HEALNet demonstrates superior performance in handling the complexity of high-dimensional biomedical data while retaining both accuracy and robustness.
Future Research Scope#
Future research could explore extending HEALNet’s capabilities to a wider array of biomedical data modalities, such as incorporating other omics data (e.g., metabolomics, proteomics) or different imaging modalities (e.g., MRI, CT scans). Investigating the impact of different attention mechanisms and exploring alternative fusion strategies within the hybrid early-fusion framework could lead to further performance improvements. A crucial area for future work is robustness testing with significantly larger and more diverse datasets, ideally spanning multiple institutions and patient populations. This would help to validate the generalizability of HEALNet and its ability to handle the complexities inherent in real-world clinical data. Additionally, research should focus on enhancing model interpretability by developing visualization techniques that offer clearer insights into the learned cross-modal relationships and the model’s decision-making processes. Finally, exploring applications of HEALNet to other clinical prediction tasks, including diagnosis and treatment response prediction, would broaden its impact.
More visual insights#
More on figures

This figure demonstrates HEALNet’s explainability by visualizing attention weights. It shows a whole slide image (WSI) from a high-risk UCEC patient, an attention map highlighting regions the model focused on, a zoomed-in view of those regions with cell types identified, and a bar chart showing the attention weights given to different omics features. The high-risk regions are shown to have high concentrations of epithelial cells, a known indicator of various cancer types.

This figure shows the architecture of HEALNet, a hybrid early-fusion attention learning network for multimodal fusion. It highlights the use of both shared and modality-specific parameter spaces, iterative attention layers to capture cross-modal information, and a self-normalizing network to improve robustness and efficiency. The figure displays the iterative update of a shared latent embedding through modality-specific attention mechanisms, leading to a final multimodal representation.
More on tables

This table shows the performance of HEALNet when some modalities are missing during the testing phase. It compares HEALNet’s performance to unimodal baselines in four different scenarios: (1) only omic data, (2) only WSI data, (3) a mix of both, and (4) using only the available modality. HEALNet consistently achieves better results, demonstrating its robustness to missing data and ability to leverage cross-modal information learned during training.

This table presents the performance of HEALNet and various baseline models on four different cancer datasets from The Cancer Genome Atlas (TCGA). The performance metric is the concordance index (c-index), a measure of how well the model predicts survival times. The table shows the mean and standard deviation of the c-index across five-fold cross-validation for each model. The models include unimodal models (using only one data type), and multimodal models using various fusion methods (late, intermediate, early). The results demonstrate that HEALNet generally outperforms the other models, showcasing its effectiveness in multimodal survival analysis.

This table presents the results of a classification experiment using the MIMIC-III dataset. The experiment compares the performance of HEALNet against several other unimodal and multimodal baselines on two tasks: predicting patient mortality (MORT) and disease classification (ICD9). The performance metric used is AUC (Area Under the Curve) for ICD9 and Macro-AUC for MORT. The results are averaged across five folds of cross-validation.

This table presents a summary of the four cancer datasets from The Cancer Genome Atlas (TCGA) used in the HEALNet experiments. For each dataset (BLCA, BRCA, KIRP, UCEC), it lists the number of slide samples, the number of omic samples, the overlap between slide and omic samples used in the analysis, the number of omic features used, the resolution of the whole slide images (WSI), the percentage of censored data (where the outcome was not observed), the survival bin sizes used for discretizing survival time, and the amount of disk space used for storing the data.

This table presents the performance of HEALNet and various baseline models (unimodal and multimodal) on four different cancer datasets from The Cancer Genome Atlas (TCGA). The performance is measured using the concordance index (c-index), a common metric for evaluating survival prediction models. The results show that HEALNet achieves state-of-the-art performance, outperforming other multimodal methods and several unimodal baselines.
Full paper#