↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many machine learning applications in chemistry and biology are hampered by the scarcity of labeled biochemical data. Existing methods, like self-supervised learning and active learning, have limitations in bridging the gap between pretraining and fine-tuning or require significant human effort, respectively. Domain knowledge-based approaches are also limited by bias and difficulty in universal integration.
In response, this paper introduces InstructMol, a novel semi-supervised learning approach. InstructMol uses an instructor model to evaluate the reliability of predicted labels from a target molecular model on unlabeled data. This instructor-based approach guides the target model, enhancing accuracy and avoiding knowledge transfer issues. The research demonstrates InstructMol’s superior performance on various real-world molecular datasets and OOD benchmarks, significantly advancing robust molecular property prediction.
Key Takeaways#
Why does it matter?#
This paper is highly important for researchers in chemistry, biology, and machine learning due to its novel approach to handling data scarcity in molecular property prediction. The InstructMol algorithm directly addresses the challenges of limited labeled data, a major bottleneck in applying ML to scientific discovery. Its success in predicting properties of novel drug molecules opens up new avenues for accelerating drug discovery and materials science. The flexible semi-supervised learning framework is broadly applicable, potentially impacting various other domains facing similar data limitations.
Visual Insights#

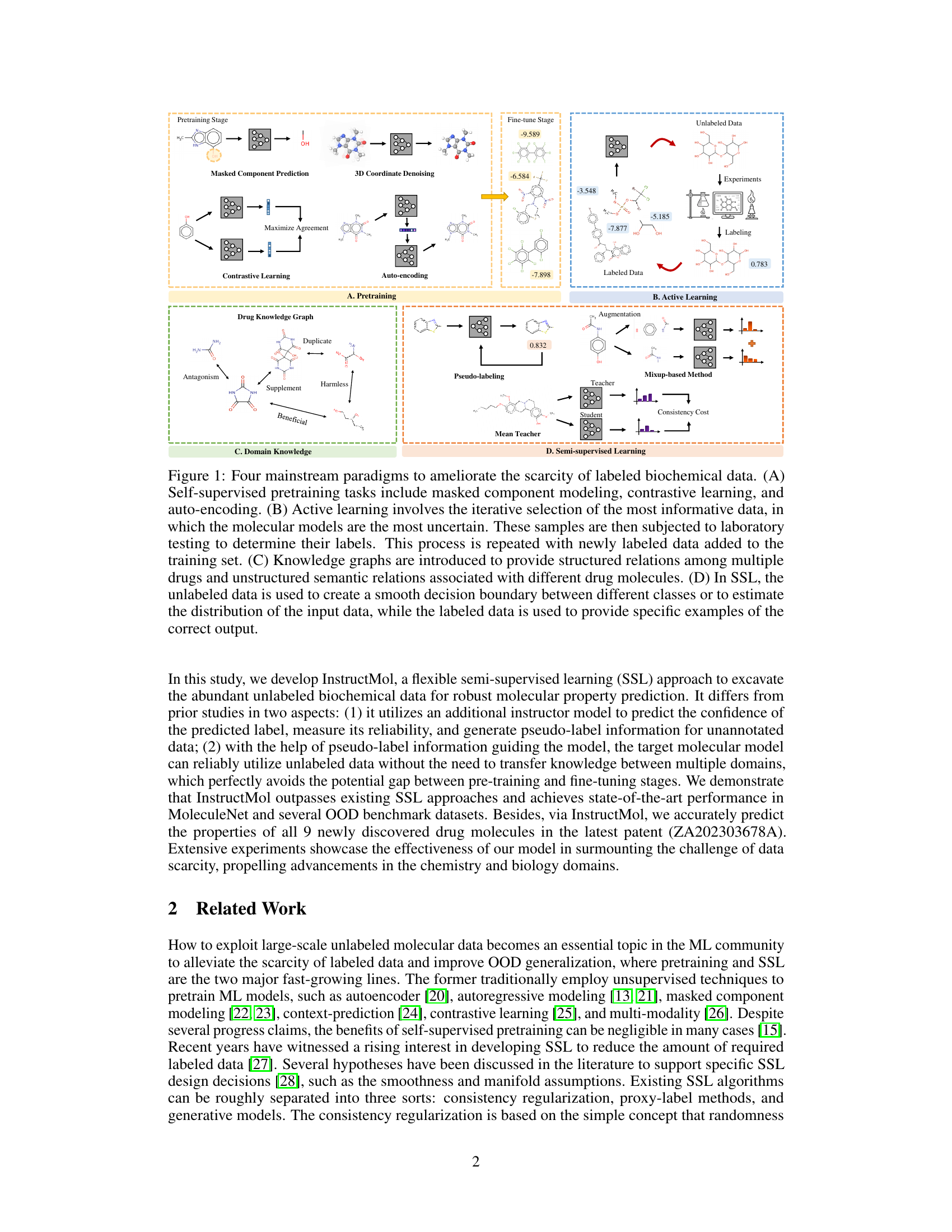
This figure illustrates four common approaches to address the challenge of limited labeled data in biochemical machine learning. (A) shows self-supervised pre-training, leveraging unlabeled data to learn representations. (B) depicts active learning, iteratively selecting data points for manual labeling based on model uncertainty. (C) highlights the use of domain knowledge, such as drug knowledge graphs, to improve model performance. (D) presents semi-supervised learning, combining labeled and unlabeled data to learn a more robust model.

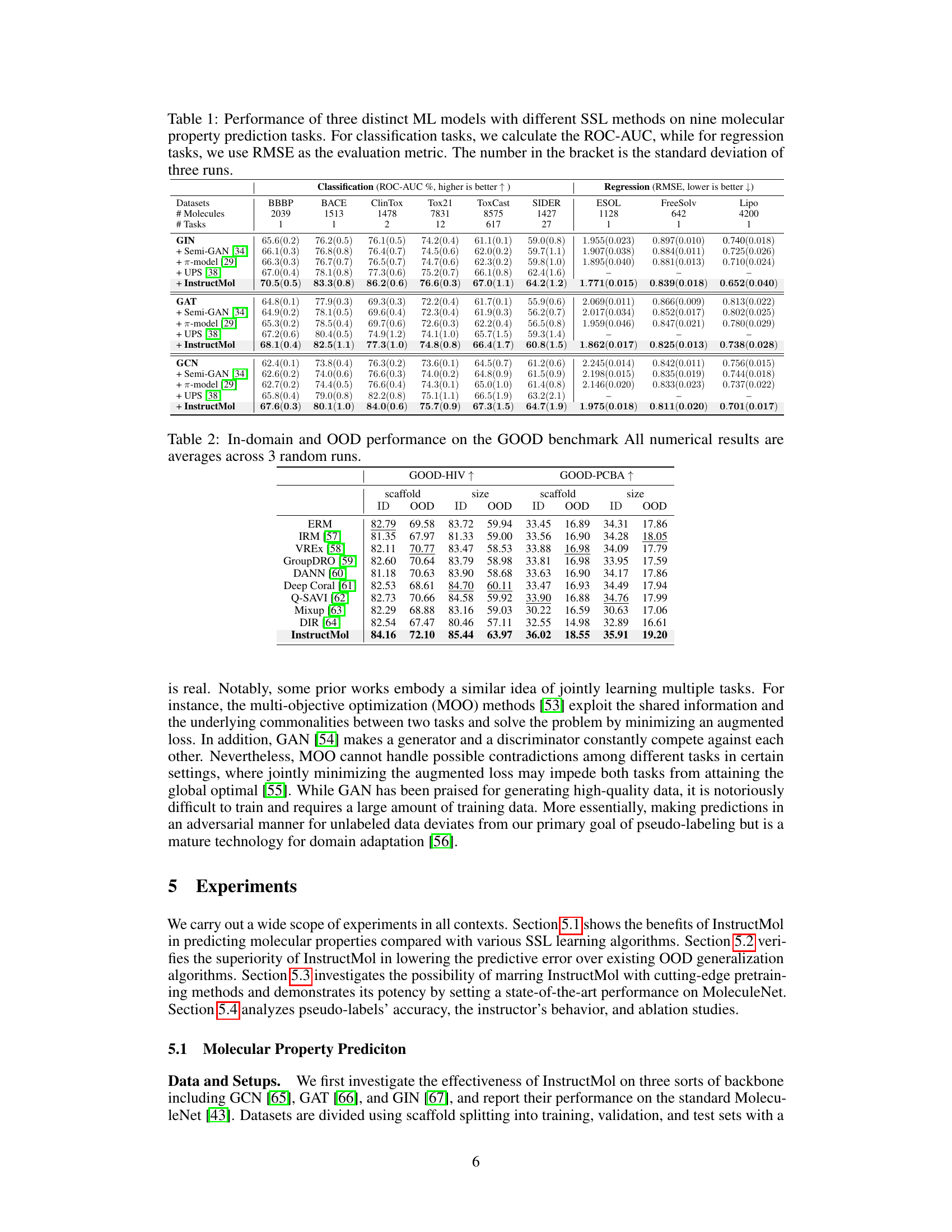
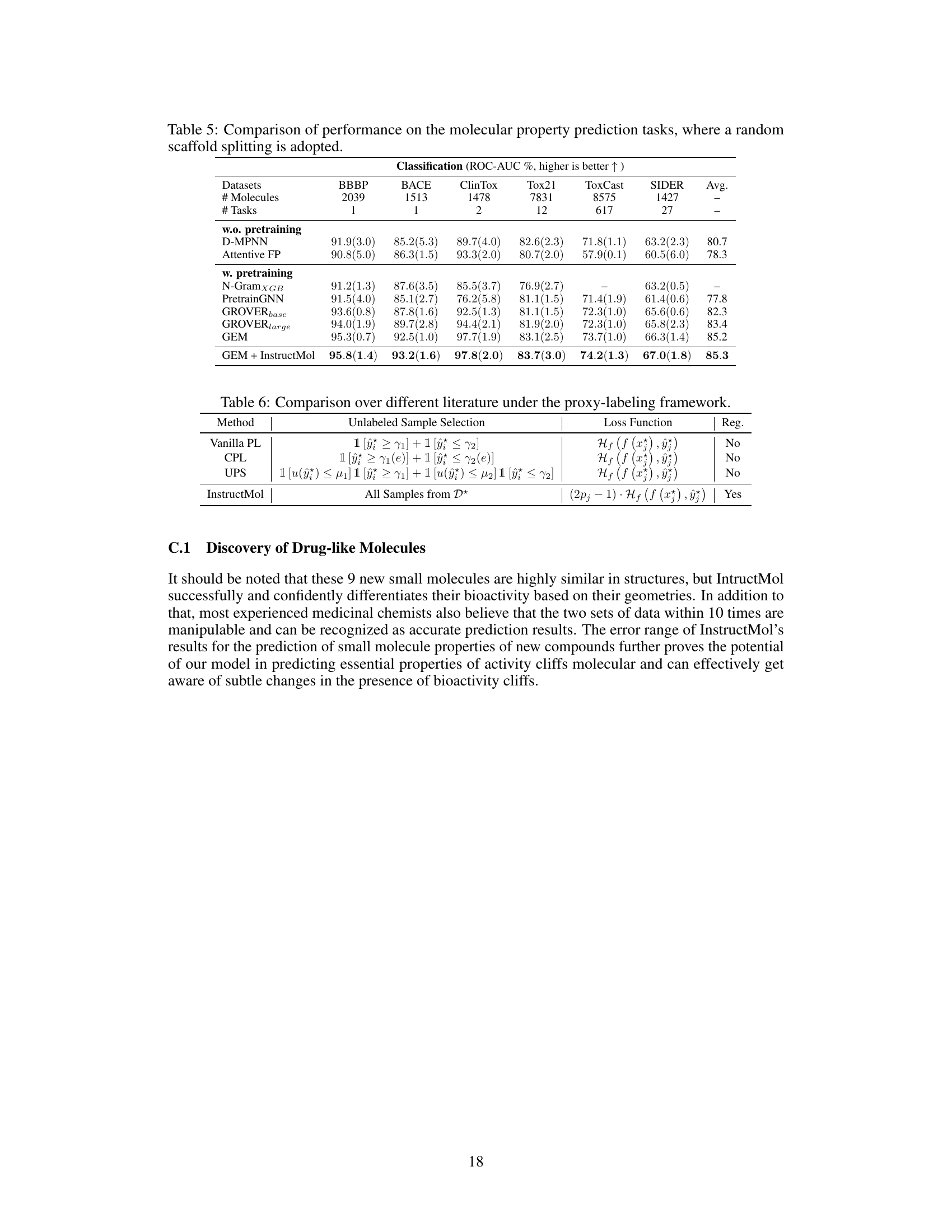
This table presents the performance comparison of three different machine learning models (GIN, GAT, GCN) using various semi-supervised learning (SSL) methods on nine different molecular property prediction tasks from the MoleculeNet dataset. The table shows the ROC-AUC scores (for classification tasks) and RMSE scores (for regression tasks), along with standard deviations across three runs. It aims to demonstrate the effectiveness of InstructMol compared to other SSL approaches.
In-depth insights#
InstructMol: Method#
InstructMol’s methodology centers on a two-phase semi-supervised learning approach. Phase one involves using a pre-trained molecular model to generate pseudo-labels for unlabeled data. Crucially, phase two introduces an instructor model which assesses the reliability of these pseudo-labels, assigning confidence scores. This confidence measure is then used to intelligently re-weight the contribution of each data point (labeled or pseudo-labeled) during the training of the primary molecular model. This innovative weighting mechanism avoids the pitfalls of standard pseudo-labeling techniques, which often overemphasize unreliable predictions. The use of an instructor model is a key innovation, enabling InstructMol to effectively leverage large unlabeled datasets while mitigating the risk of error propagation from less reliable pseudo-labels. The combined approach promotes robustness and improved performance, particularly valuable in the context of limited labeled data, a common constraint in chemical and biological domains.
OOD Generalization#
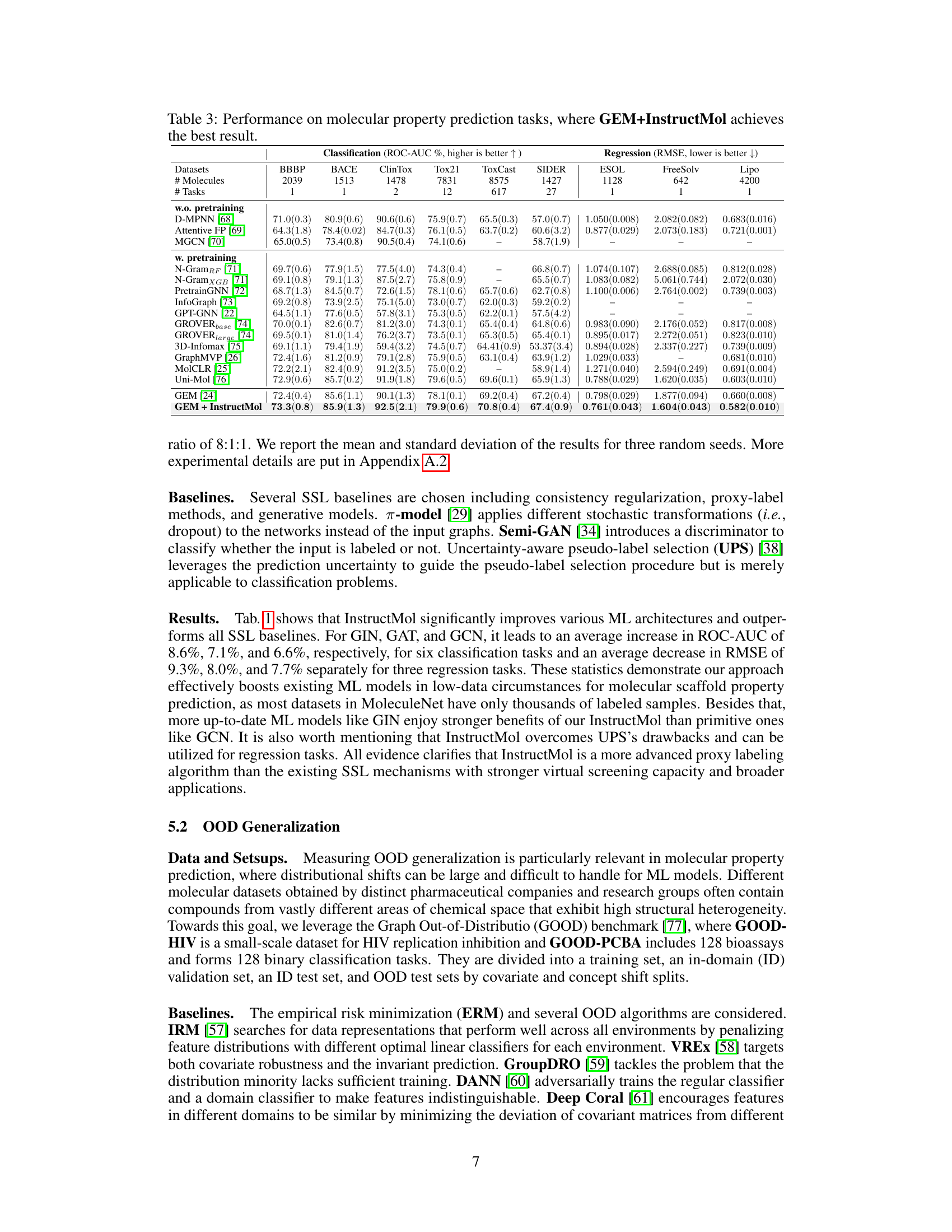
The section on “OOD Generalization” in the research paper is crucial for evaluating the robustness and reliability of the proposed InstructMol model. It directly addresses the model’s ability to generalize to unseen data, a critical aspect in real-world applications where encountering out-of-distribution (OOD) samples is inevitable. The use of the GOOD benchmark is a strong methodological choice, as it specifically assesses OOD generalization performance through different types of distribution shifts. The results demonstrating that InstructMol significantly outperforms other methods, including ERM and various OOD algorithms, highlight its ability to handle data that differs from the training set. This is particularly valuable in the context of molecular property prediction, where substantial heterogeneity across datasets is common. The consistent improvement across various splits, including in-domain and out-of-distribution tests, further strengthens the claim of InstructMol’s superior robustness. This section effectively showcases the practical advantages of the proposed method in scenarios beyond perfectly matched training and testing data, underscoring its potential for broader impact and real-world applicability.
SSL & Pretraining#
The combination of self-supervised learning (SSL) and pretraining techniques offers a powerful approach to address the challenge of limited labeled data in molecular property prediction. SSL leverages unlabeled data to improve model generalization and robustness, while pretraining helps establish strong foundational representations. The study explores how pretraining on large-scale unlabeled molecular datasets can create robust initial models which are then fine-tuned using the instructive learning approach. This combined strategy mitigates the potential domain gap between pretraining and fine-tuning stages, a common issue in transfer learning. The results demonstrate that the synergy between pretraining and instructive learning significantly enhances the accuracy and generalizability of molecular property prediction, especially in out-of-distribution scenarios. The effectiveness of this combined approach showcases the potential to advance scientific discovery by leveraging readily available unlabeled data to build more powerful models for complex tasks like predicting drug properties and toxicity. The research suggests a future direction of exploring more advanced SSL methods in combination with pretraining strategies.
Ablation Studies#
Ablation studies systematically remove components of a model or process to assess their individual contributions. In machine learning, this often involves removing layers, features, or hyperparameters to understand their impact on performance. Well-designed ablation studies are crucial for establishing causality and avoiding spurious correlations; they show whether observed improvements are due to the proposed changes or other factors. A good ablation study should present a baseline result, and then systematically remove features to isolate the effect of the component of interest. This provides strong evidence for the importance and effectiveness of specific components, separating their benefits from other variables such as increased model complexity. A thorough ablation study would consider multiple variants of the modifications to examine the robustness of the findings. It is critical that ablation studies are clearly presented and easy to interpret to reveal valuable insights into the relative contributions of different aspects of the model’s design or the experimental procedure. This ensures that the conclusions drawn from the research are robust and generalizable.
Future Work#
Future research directions stemming from this InstructMol model could explore several promising avenues. Improving the instructor model’s calibration is crucial; while it effectively distinguishes between reliable and unreliable pseudo-labels, further refinement in accuracy would significantly enhance performance. Investigating alternative architectures and training strategies for the instructor model, perhaps incorporating Bayesian methods or advanced calibration techniques, is warranted. Extending InstructMol to handle diverse molecular data formats (beyond the 2D graphs used here) and tasks (such as reaction prediction or property optimization) is another key area. Exploring the synergy between InstructMol and other self-supervised learning techniques (such as contrastive learning) offers significant potential for further performance gains. Finally, a thorough investigation into the model’s generalization ability across a wider range of chemical spaces and out-of-distribution benchmarks would be beneficial, along with a deeper analysis of the model’s bias and potential limitations.
More visual insights#
More on figures
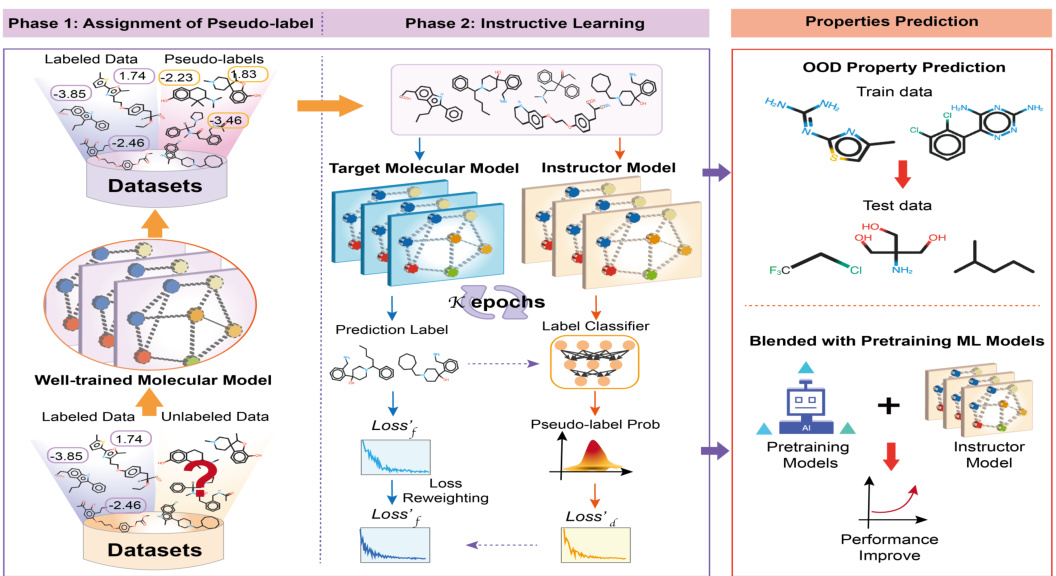
This figure illustrates the two phases of the InstructMol algorithm. In Phase 1, a pre-trained molecular model assigns pseudo-labels to unlabeled data. Then, in Phase 2, an instructor model evaluates the reliability of these pseudo-labels. This information guides the target model in its learning process, weighting the importance of different data points (labeled and pseudo-labeled) to improve performance in predicting molecular properties and out-of-distribution (OOD) benchmarks. The instructor model helps to mitigate the impact of potentially noisy pseudo-labels.
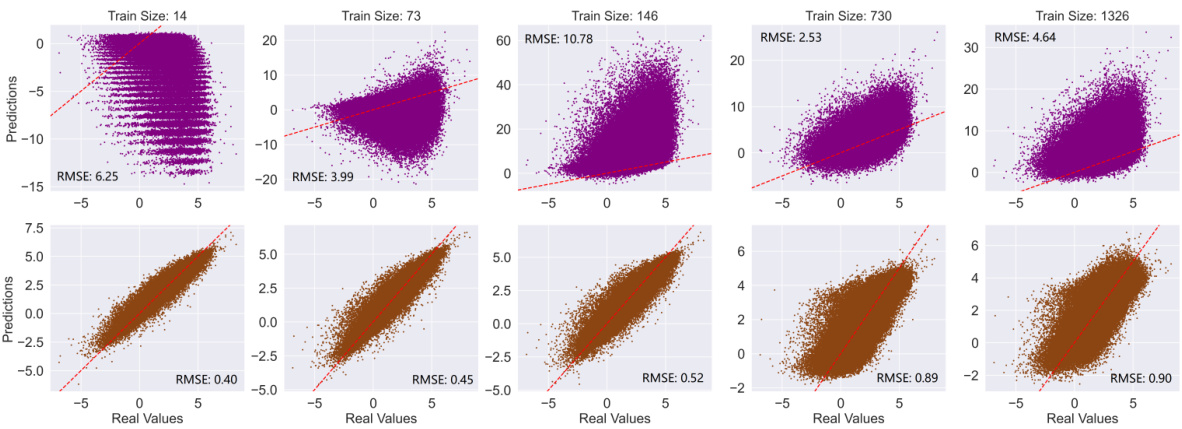
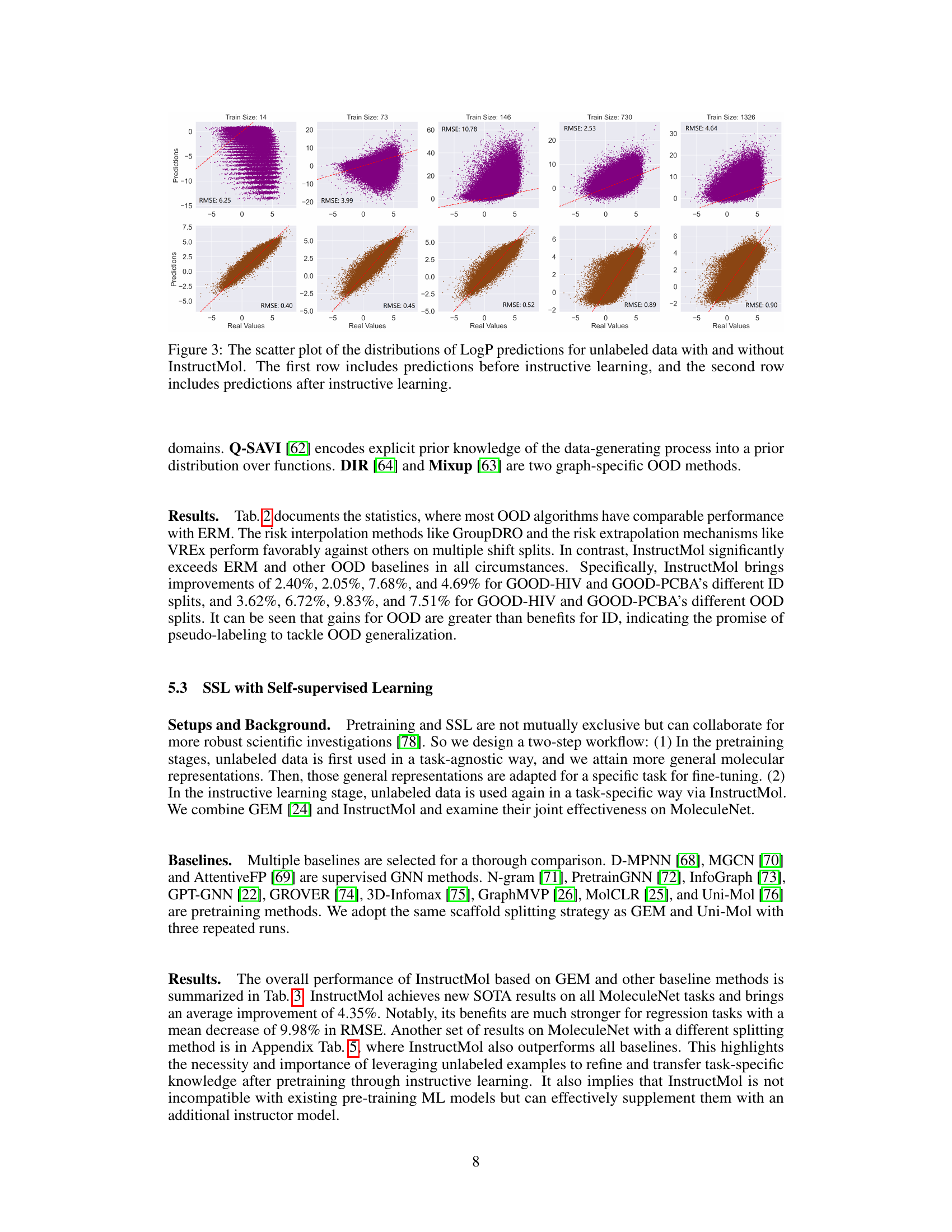
This figure shows the scatter plots of the predicted LogP values against the actual LogP values for unlabeled data, comparing the performance of InstructMol with and without instructive learning. The top row displays the predictions before InstructMol’s instructive learning phase, while the bottom row shows the predictions after this phase. Each subplot represents a different training data size (14, 73, 146, 730, and 1326 molecules), demonstrating how the model’s accuracy improves with instructive learning, even with limited training data. The RMSE (Root Mean Squared Error) value is provided for each subplot, quantifying the prediction error. The plots visually illustrate the improved accuracy and reduced error of InstructMol’s LogP predictions after instructive learning.

This figure shows the distributions of confidence scores produced by the instructor model for both real and fake labels throughout the training process. The x-axis represents the confidence scores, ranging from 0.0 to 1.0, with 1.0 indicating high confidence. The y-axis shows the probability density. Separate distributions are shown for real labels (those assigned manually) and fake labels (those generated by the target model). The figure displays how the instructor model’s ability to distinguish between real and fake labels improves over training iterations (0K to 10K). Initially, the distributions overlap significantly, indicating low confidence and poor discrimination. Over time, the distributions separate, with real labels exhibiting higher confidence scores and the fake labels having lower scores, demonstrating improved reliability.
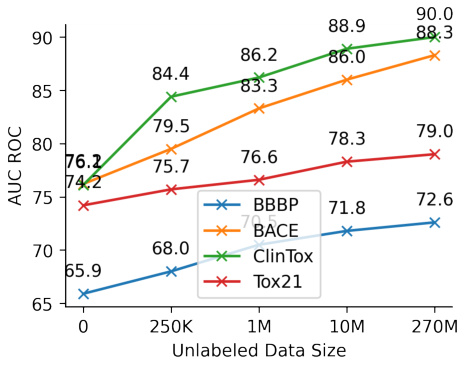
This figure displays the impact of varying the size of the unlabeled dataset on the performance of the model across four different molecular property prediction tasks (BBBP, BACE, ClinTox, and Tox21). Each line represents a different task, showcasing the AUC ROC score achieved as the number of unlabeled data points increases. The graph illustrates that augmenting the model with more unlabeled data consistently improves its predictive performance on all tasks, especially for the classification tasks. The increase is more prominent at lower data sizes and flattens out as the quantity of unlabeled data becomes very large.
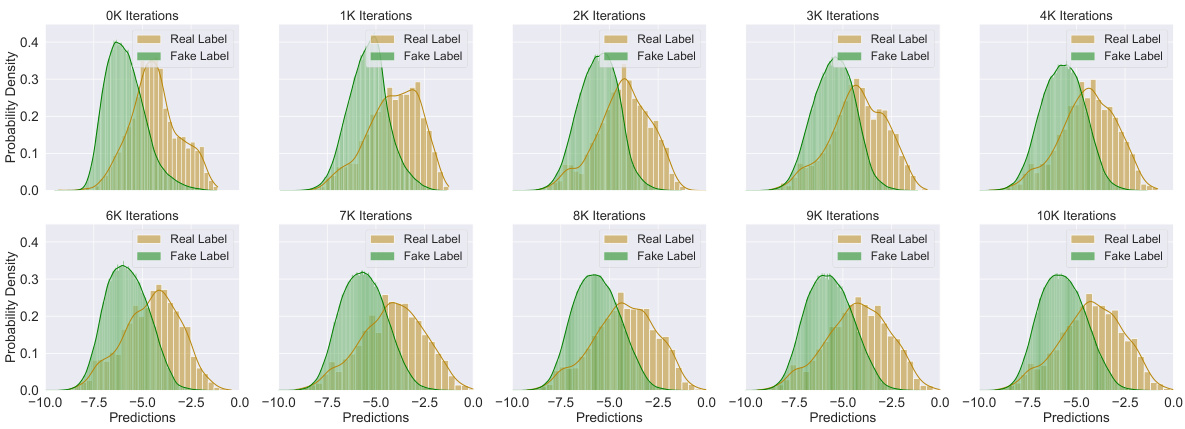
This figure shows the distribution of predictions from the target model for both labeled and unlabeled data at various training stages. The distributions are plotted as histograms to visualize the model’s uncertainty and how it changes as the training progresses. This helps to understand how the model learns to differentiate between real and pseudo-labels over time.
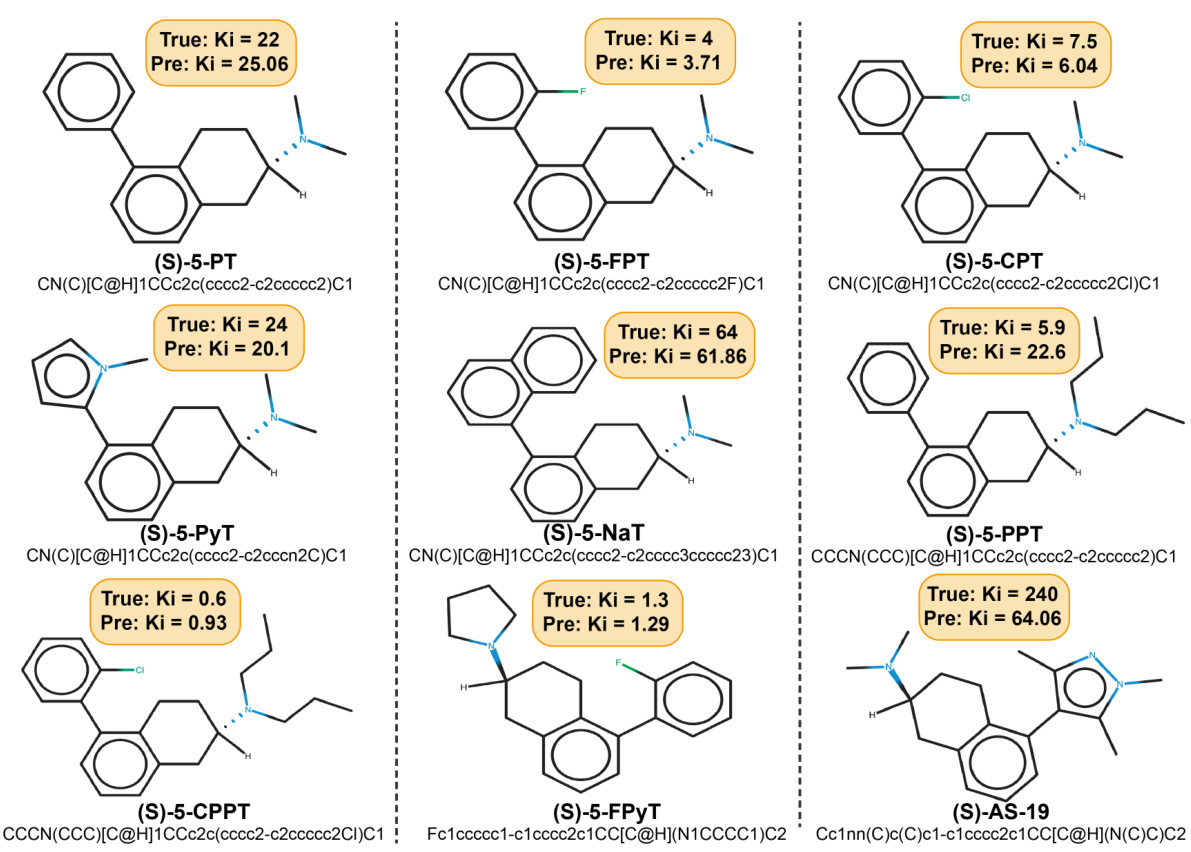
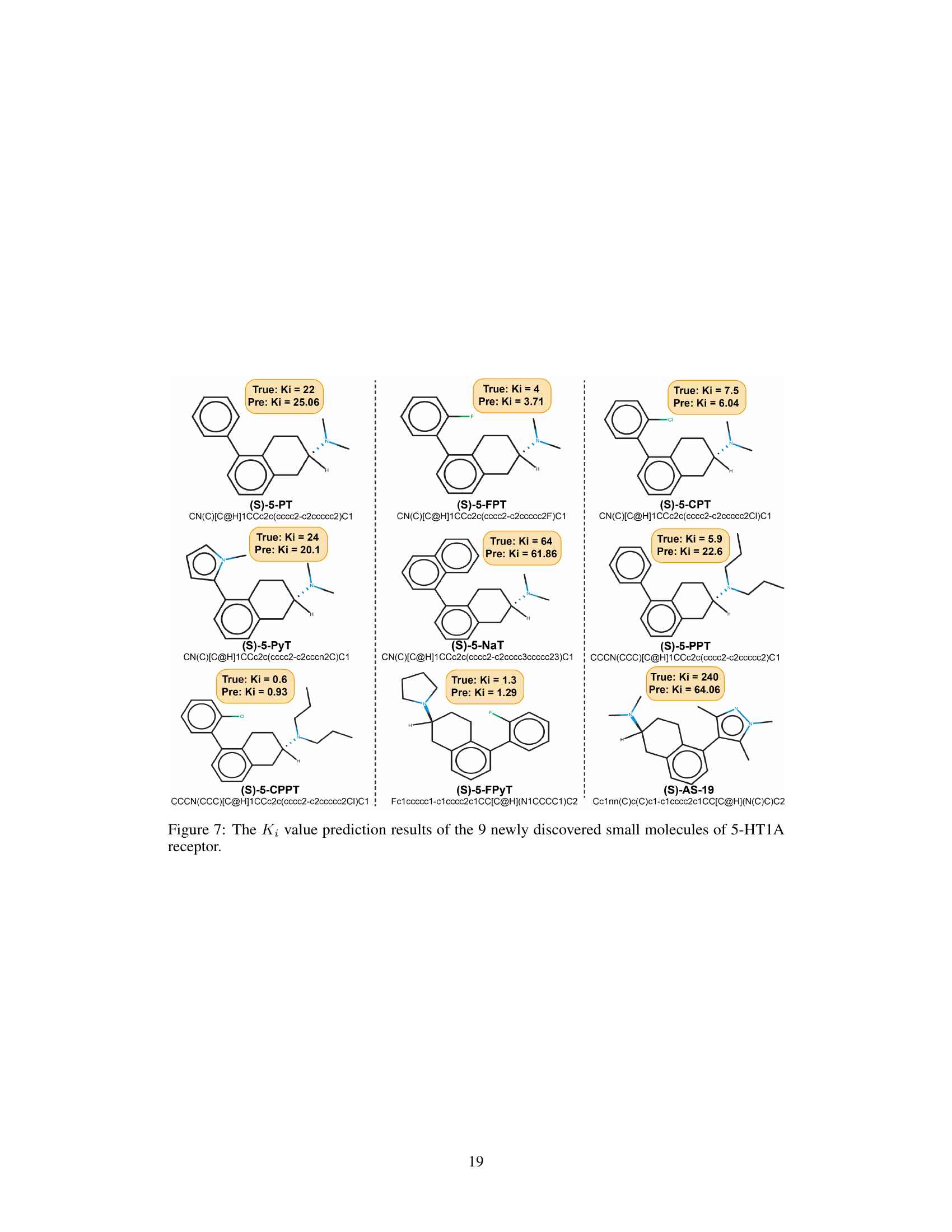
This figure shows the predicted and actual Ki values for nine newly discovered small molecules that target the 5-HT1A receptor. The predictions were made using the InstructMol model. Each molecule is depicted with its structure, SMILES notation, actual Ki value from experimental testing, and the Ki value predicted by InstructMol. The close correspondence between predicted and actual Ki values demonstrates the model’s accuracy in predicting the properties of novel drug molecules.
More on tables
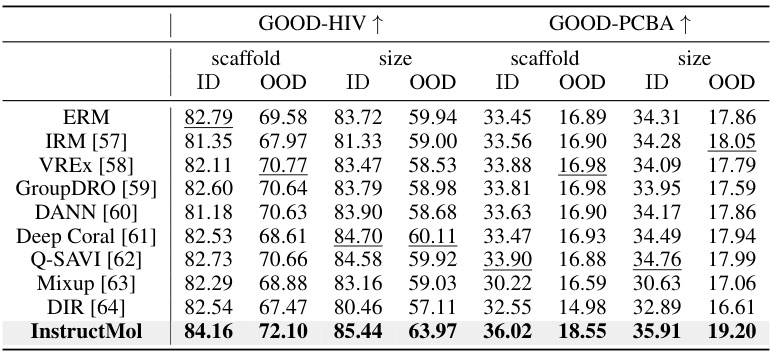
This table presents the in-domain and out-of-distribution (OOD) generalization performance of various methods on the GOOD benchmark. The results are averaged over three random runs, showing performance metrics for both in-domain (ID) and OOD data for two different datasets, GOOD-HIV and GOOD-PCBA, and with two different ways of splitting the data (scaffold size). It demonstrates the ability of the different methods to generalize to unseen data.
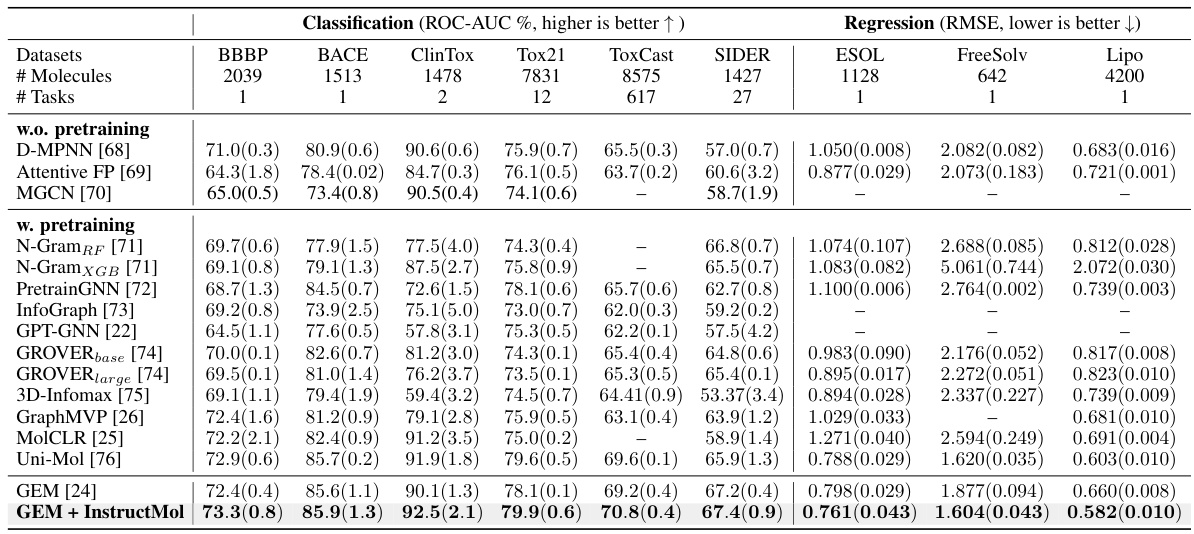
This table presents the performance comparison of three different machine learning models (GIN, GAT, and GCN) using various semi-supervised learning (SSL) methods on nine molecular property prediction datasets. It shows the ROC-AUC scores (for classification tasks) and RMSE values (for regression tasks), along with their standard deviations across three independent runs. The table highlights the improvement achieved by the InstructMol method compared to other SSL techniques.
This table presents the performance comparison of three machine learning models (GIN, GAT, and GCN) using different semi-supervised learning (SSL) methods on nine molecular property prediction tasks from the MoleculeNet dataset. The performance is measured using ROC-AUC for classification tasks and RMSE for regression tasks. The table shows the effectiveness of InstructMol compared to other SSL techniques and how it affects the performance of each model.
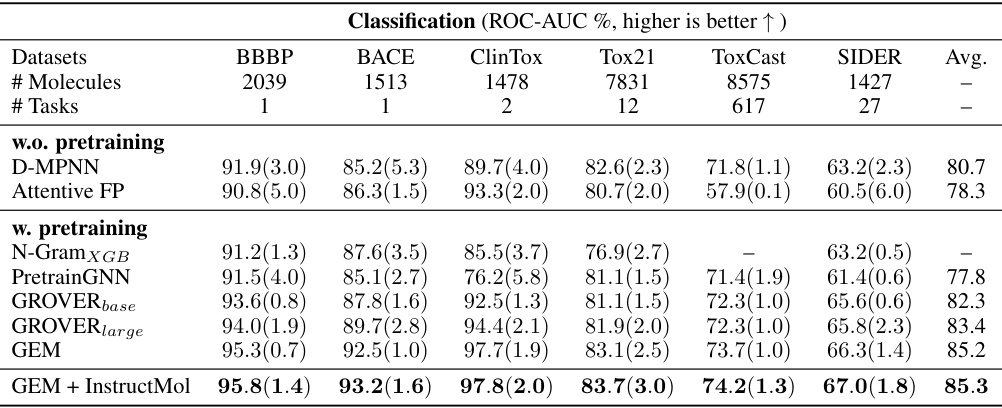
This table presents the performance comparison of three different machine learning models (GIN, GAT, GCN) combined with several semi-supervised learning (SSL) methods on nine molecular property prediction tasks from the MoleculeNet dataset. The results are shown in terms of ROC-AUC (for classification tasks) and RMSE (for regression tasks). The table highlights the improvement achieved by using the InstructMol method compared to other SSL techniques.

This table presents the performance comparison of three different machine learning models (GIN, GAT, and GCN) using various semi-supervised learning (SSL) methods on nine molecular property prediction tasks. The performance is measured using ROC-AUC for classification tasks and RMSE for regression tasks. The results show the mean performance across three independent runs, with standard deviations included to show variability.
Full paper#