↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many existing methods for medical time series (MedTS) classification rely on handcrafted features and struggle with subject-independent classification. Transformer-based models have seen limited exploration in this area, and existing transformer models don’t fully capture the unique characteristics of MedTS data, such as inter-channel correlations and multi-scale temporal dynamics. This paper proposes Medformer, a novel multi-granularity patching transformer designed to address these issues.
Medformer incorporates three key mechanisms: cross-channel patching to leverage inter-channel correlations, multi-granularity embedding for capturing features at different scales, and two-stage multi-granularity self-attention. Extensive experiments on five public datasets show that Medformer surpasses 10 baselines, achieving the highest average ranking across various evaluation metrics, especially in the challenging subject-independent setup. This highlights the significant impact of Medformer on healthcare applications.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers working with medical time series data. It introduces a novel transformer-based model, Medformer, that significantly outperforms existing methods, especially in challenging subject-independent scenarios. This opens up new avenues for more accurate and reliable medical diagnoses using various physiological signals. The multi-granularity approach is particularly impactful, providing insights into both fine-grained and coarse-grained features, which is highly relevant to current trends in time series analysis.
Visual Insights#

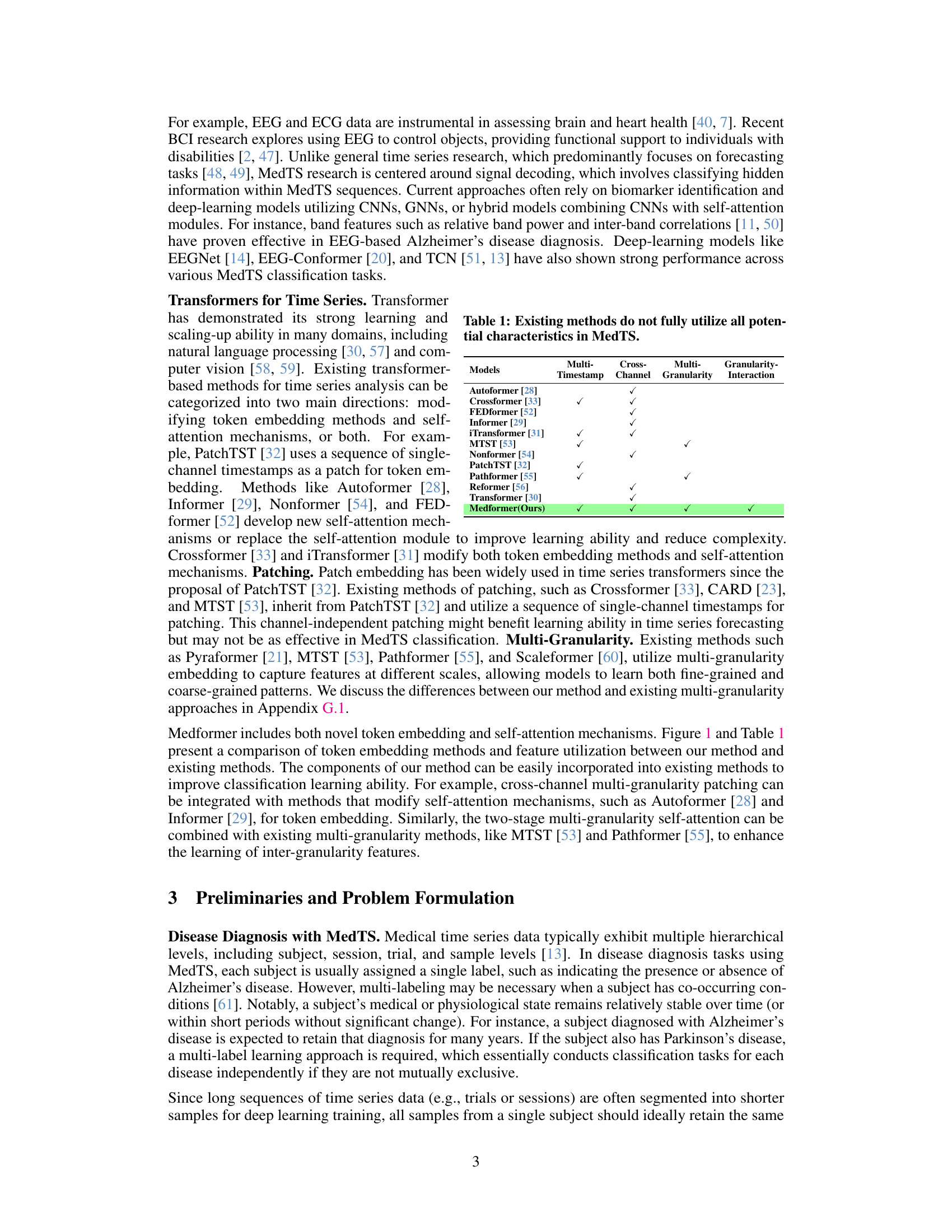
This figure compares different token embedding methods used in time series transformers. Vanilla Transformer, Autoformer, and Informer use a single timestamp from all channels. iTransformer uses all timestamps from a single channel. PatchTST and Crossformer use multiple timestamps from a single channel. The authors propose Medformer which uses multiple timestamps from all channels, incorporating cross-channel information and multiple temporal scales.
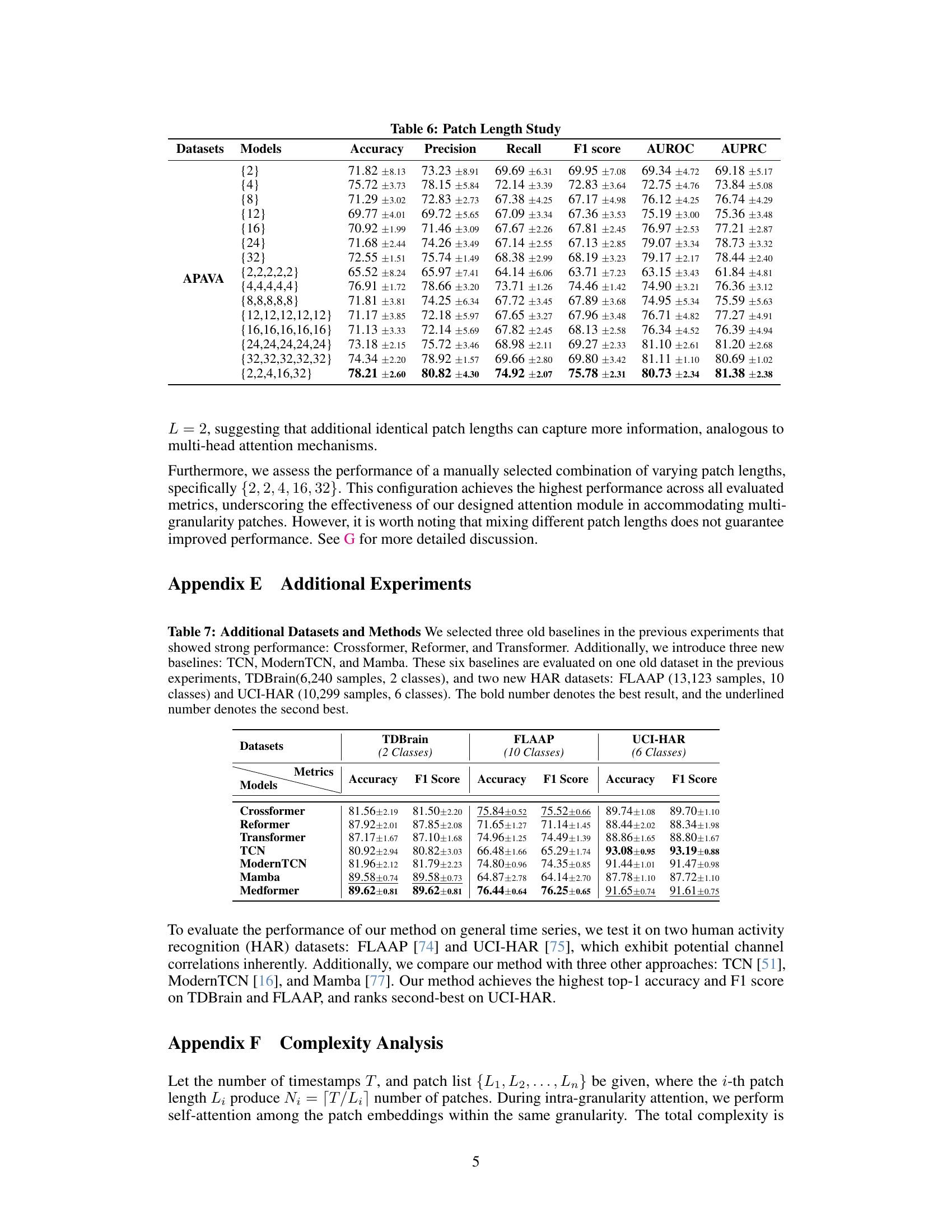
The table compares several existing transformer-based methods for time series analysis, highlighting their approaches to handling timestamps, channels, and granularities in the context of medical time series (MedTS) data. It shows that Medformer uniquely addresses all three aspects, unlike other methods which only partially utilize these characteristics. This is crucial for effective MedTS classification because the methods need to capture the relationships between multiple channels, the importance of temporal dynamics at various scales, and the inherent correlations between data points at different time resolutions.
In-depth insights#
MedTS Transformer#
A MedTS Transformer is a deep learning model specifically designed for medical time-series data classification. It leverages the power of transformer architectures, known for their success in natural language processing and other sequential data tasks, to effectively handle the unique characteristics of physiological time series like ECG and EEG. Key improvements over traditional methods like CNNs often involve the incorporation of mechanisms to capture both temporal dynamics and inter-channel relationships. Multi-granularity approaches are particularly valuable, enabling the model to learn from different temporal scales and frequencies within the data. Attention mechanisms play a crucial role, allowing the model to focus on relevant features and relationships among different channels and time points. The subject-independent evaluation is a critical aspect of MedTS transformers, as it better reflects real-world clinical scenarios where models need to generalize across patients. While promising, challenges remain in effectively addressing noise and high variability in MedTS data, as well as computational cost associated with the complexity of transformer models. Further research is needed to refine model architectures and training strategies for even more robust and efficient performance.
Patching Mechanisms#
Patching mechanisms in time-series analysis, particularly within transformer architectures, represent a crucial innovation for efficiently processing long sequences. Medformer’s approach distinguishes itself through its multi-granularity and cross-channel design. Instead of treating each time-step or channel independently, it leverages inter-channel correlations and incorporates multiple patch lengths. This multi-granularity strategy allows for the simultaneous capture of both fine-grained and coarse-grained features, effectively mimicking the ability to analyze data across various frequency bands. The cross-channel patching further enhances performance by considering the interconnected nature of various channels in medical time-series data. By combining these strategies, Medformer creates a richer representation, improving the model’s capacity to learn complex relationships and leading to superior performance in classification tasks. The method avoids the limitations of single-granularity and single-channel methods, resulting in a more robust and effective model.
Multi-Granularity#
The concept of “Multi-Granularity” in the context of time series analysis, particularly for medical data, signifies the simultaneous consideration of multiple temporal scales within the data. This approach acknowledges that relevant information may exist at different levels of granularity, such as fine-grained details (individual heartbeats) and coarse-grained patterns (overall heart rhythm). Effectively capturing these diverse scales is crucial for accurate diagnostics and comprehensive understanding, as various medical conditions manifest themselves differently at these various granularities. The advantages of such a method include the potential to discover nuanced patterns that may be missed by focusing only on single scales, resulting in more robust and informative models. A multi-granularity approach addresses the challenge of capturing both local temporal features and global temporal trends, which are equally important for a thorough and accurate analysis of complex medical time-series data. By integrating information from multiple granularities, the model can achieve a more comprehensive and robust understanding of the data. However, the implementation of multi-granularity can significantly increase computational complexity, and careful design of algorithms is needed to balance the advantages of improved accuracy against this increase in complexity.
Subject-Independent#
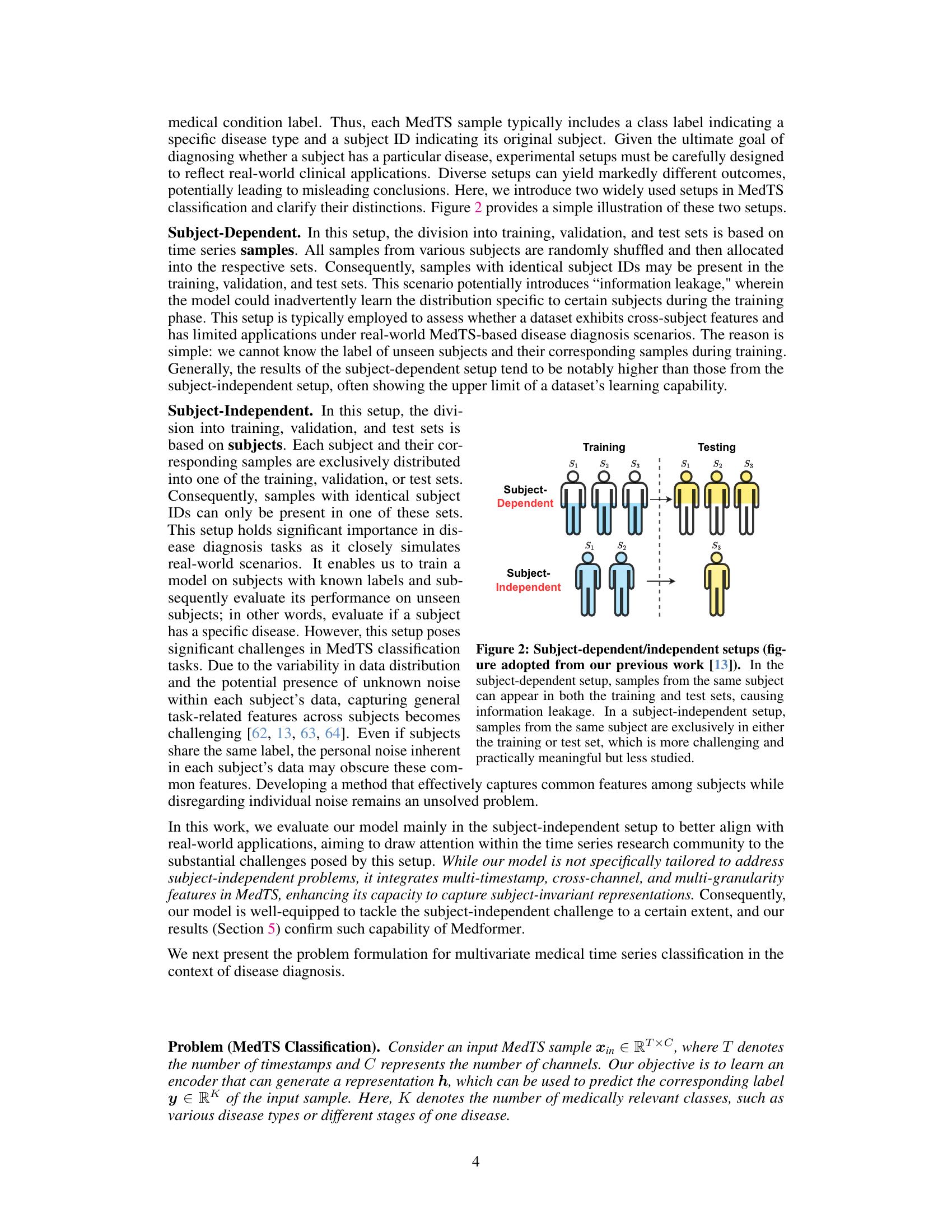
The subject-independent setup in medical time series (MedTS) classification presents a more realistic and challenging scenario compared to its subject-dependent counterpart. Unlike subject-dependent setups where the model might inadvertently learn subject-specific patterns, leading to overly optimistic performance, subject-independent evaluations assess the model’s ability to generalize across unseen subjects. This directly mirrors real-world applications, where the goal is to accurately diagnose patients not previously encountered during training. The subject-independent setup highlights the model’s robustness and its capacity to capture common disease features while mitigating patient-specific noise. Successfully navigating this challenge underscores a model’s practical utility and reliability in healthcare applications. Medformer’s superior performance under this rigorous evaluation, as highlighted in the paper, attests to its potential for reliable and widespread clinical application.
Future Directions#
Future research directions stemming from this work could explore several promising avenues. Improving the model’s robustness to noisy data is crucial for real-world applications, where physiological signals often contain artifacts and inaccuracies. Investigating more sophisticated data augmentation techniques could enhance this. Further, exploring alternative token embedding strategies beyond cross-channel patching, perhaps incorporating techniques sensitive to temporal dependencies or using hybrid approaches, warrants further investigation. The model’s capacity for handling diverse MedTS modalities beyond EEG and ECG should be evaluated, and the architecture’s adaptability to various temporal resolutions tested. Lastly, developing methods for effective multi-label classification is a significant challenge given the frequent co-occurrence of conditions in clinical practice. Addressing this will considerably enhance the model’s usefulness in diagnostic settings.
More visual insights#
More on figures
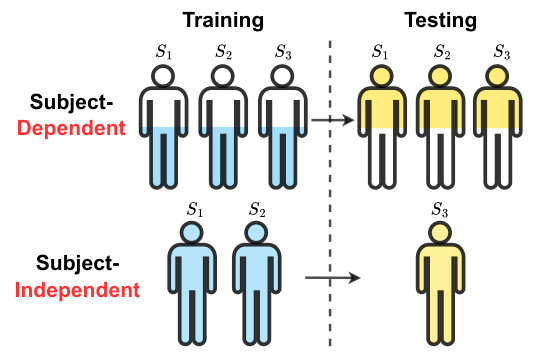
This figure illustrates the difference between subject-dependent and subject-independent experimental setups for medical time series classification. In subject-dependent setup, data from the same subject may appear in both training and testing sets, leading to potential information leakage. The subject-independent setup is more realistic as it simulates real-world scenarios where the model is tested on unseen subjects, making it more challenging but also more meaningful.

This figure illustrates the architecture of Medformer, a multi-granularity patching transformer for medical time-series classification. It shows the workflow of the model, the cross-channel multi-granularity patch embedding mechanism, and the multi-granularity self-attention mechanism. The workflow diagram details the processing steps from input time series to output. The patch embedding section highlights how Medformer handles multiple granularities of temporal information simultaneously, across different channels. Finally, the self-attention section depicts how the model learns intra-granularity and inter-granularity relationships via a two-stage attention mechanism.

This figure compares different token embedding methods used in various transformer-based time series models. It highlights the limitations of existing methods in capturing the unique characteristics of medical time series data, such as cross-channel correlations and multi-scale temporal patterns. The figure shows how Medformer addresses these limitations by incorporating cross-channel patching, multi-timestamp embedding, and multi-granularity to learn features more effectively for medical time series classification.
More on tables

This table presents the characteristics of the five datasets used in the experiments. For each dataset, it provides the number of subjects, samples, classes (disease types), channels (signals recorded), number of timestamps per sample, sampling rate, modality (EEG or ECG), and file size. The table helps to understand the scale and nature of the data used in evaluating the performance of Medformer and the baseline methods.

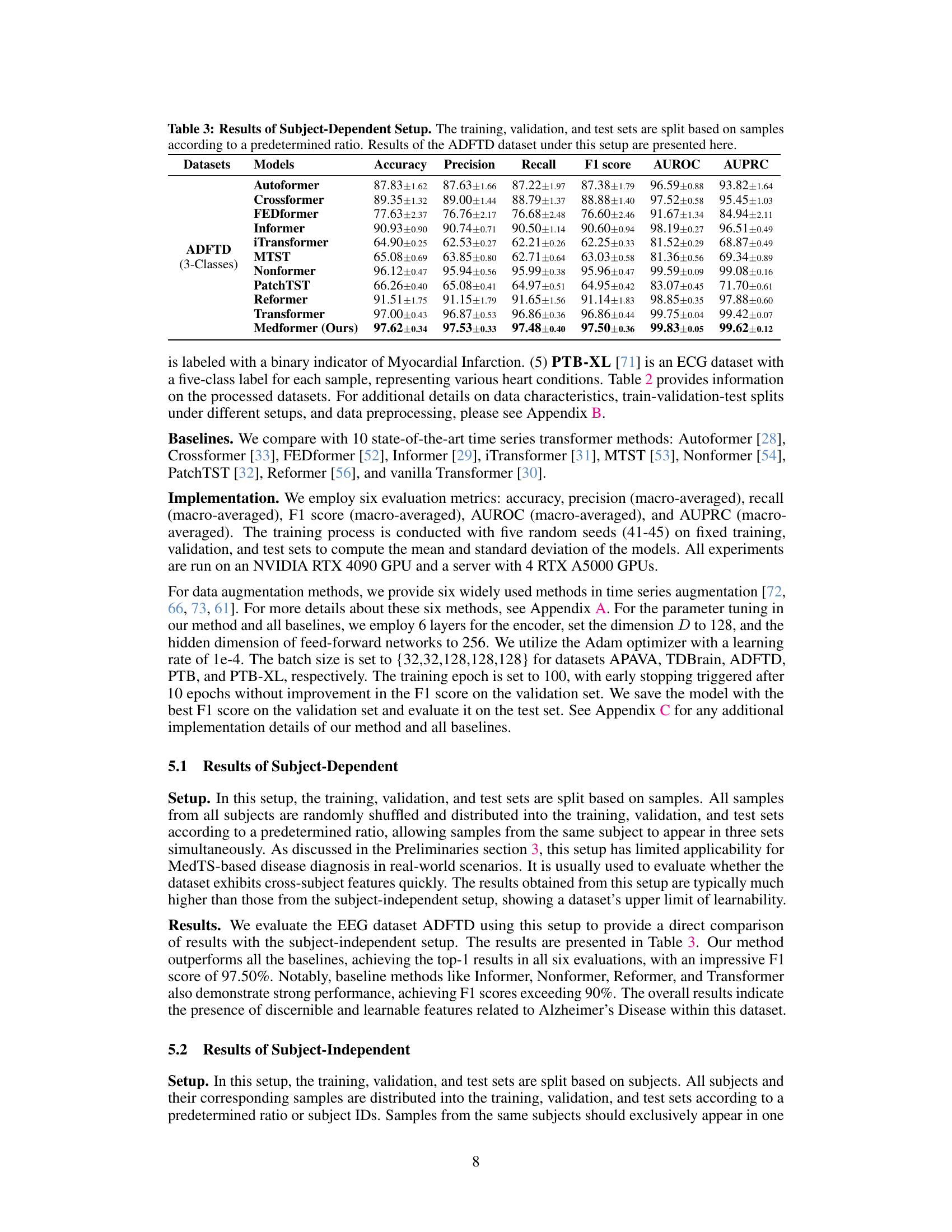
This table presents the results of a subject-dependent experimental setup where training, validation, and test data are randomly sampled without considering subject IDs. The results for the ADFTD dataset (3 classes) are shown, comparing Medformer’s performance (accuracy, precision, recall, F1 score, AUROC, AUPRC) against ten other baseline models. This setup is less realistic than subject-independent evaluations, but provides a performance upper bound for the dataset.
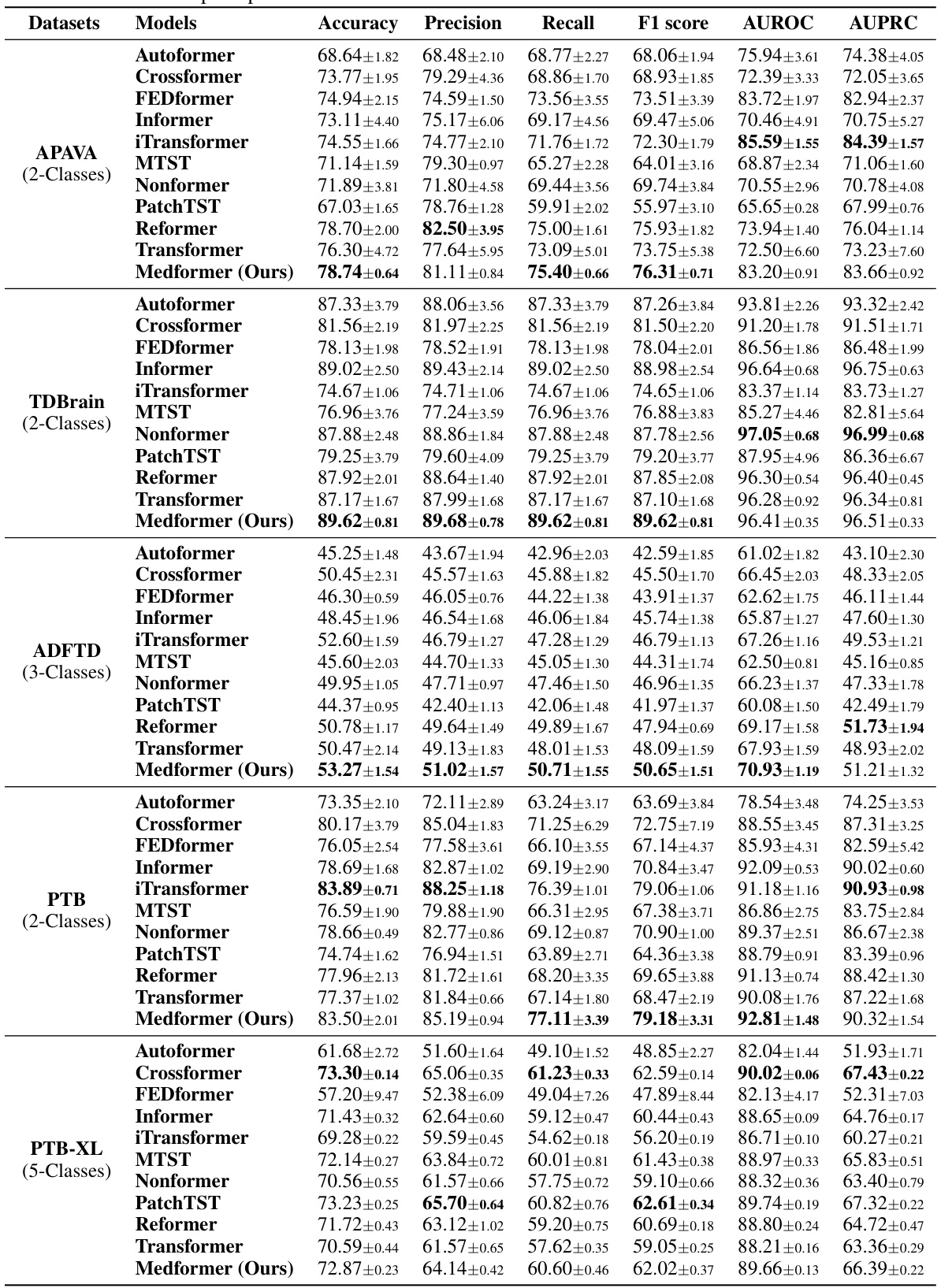
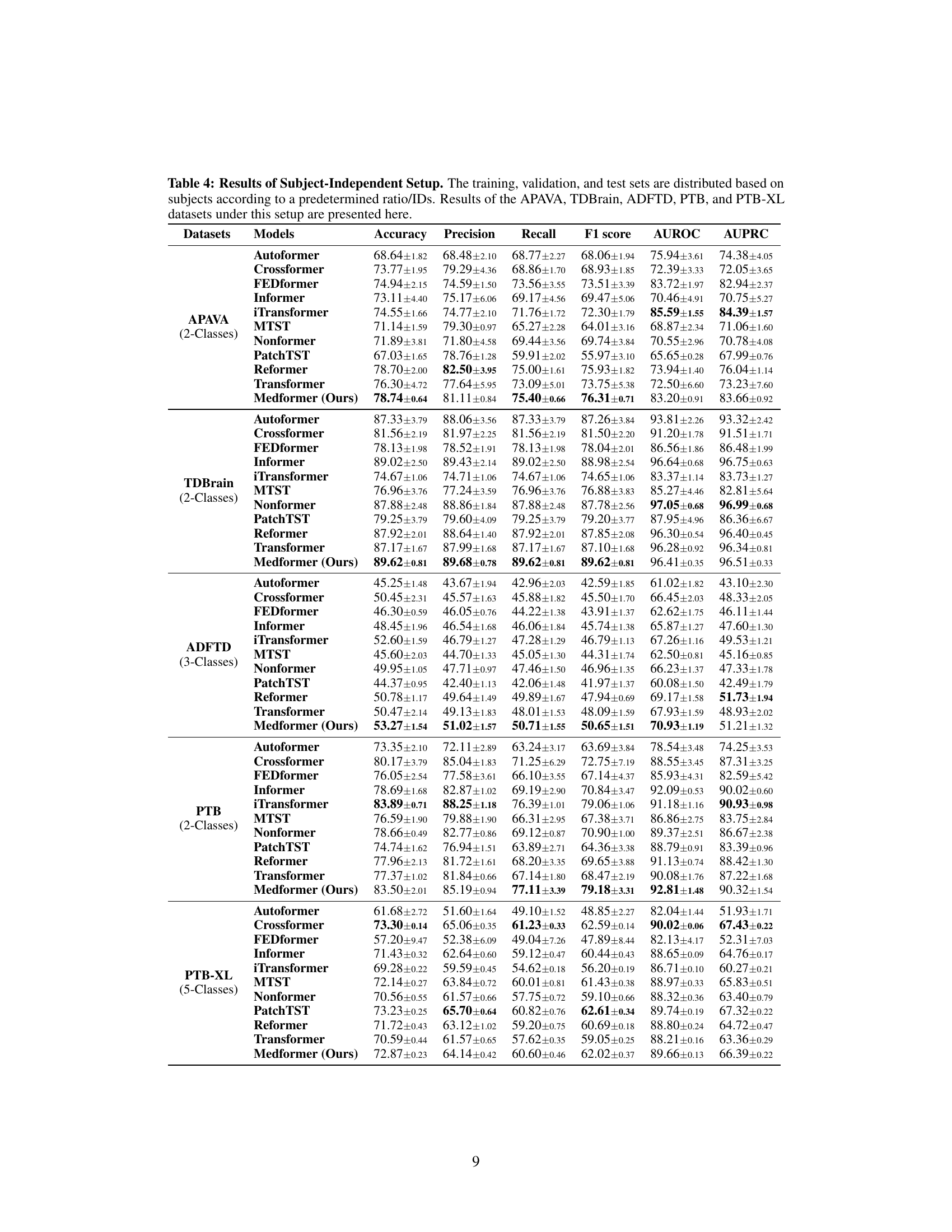
This table presents the results of the subject-independent setup experiments. It compares the performance of Medformer against 10 baseline models across five different datasets using six evaluation metrics (Accuracy, Precision, Recall, F1 score, AUROC, and AUPRC). The subject-independent setup is considered more realistic for real-world applications because it evaluates the model’s ability to generalize to unseen subjects.
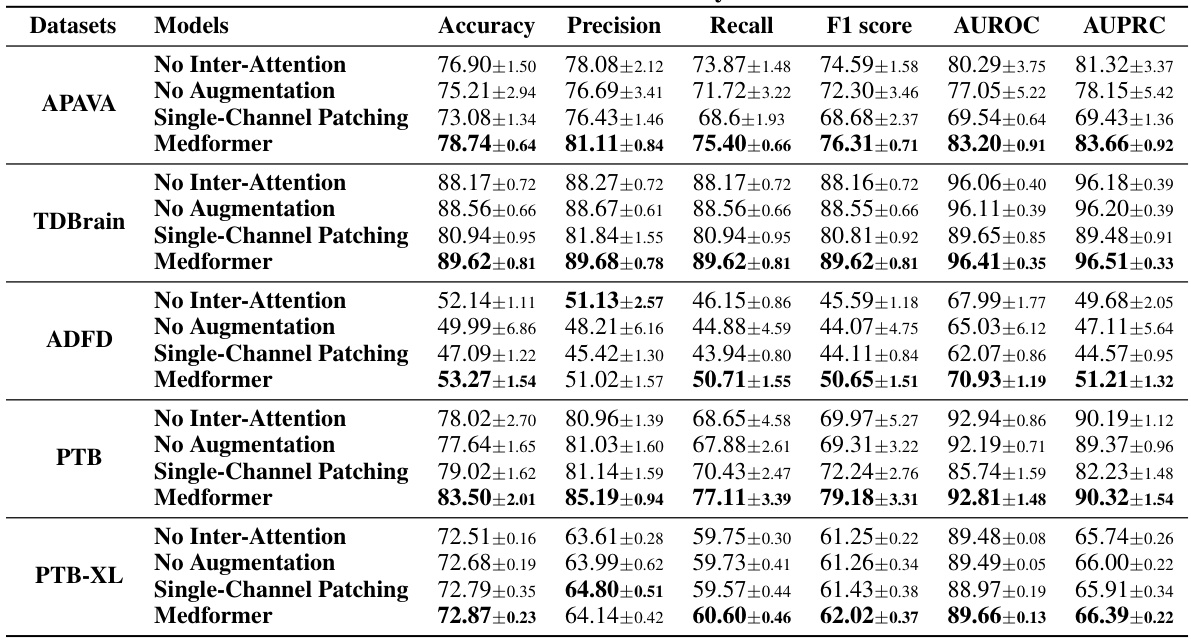
This table presents the results of the subject-dependent setup for the ADFTD dataset. The subject-dependent setup randomly assigns samples from all subjects into training, validation and test sets without considering subject ID. This means a subject’s samples could appear in multiple sets. The table shows various evaluation metrics (Accuracy, Precision, Recall, F1 score, AUROC, AUPRC) for several models, including the proposed Medformer model and several baselines. The results demonstrate Medformer’s superior performance in this setting.
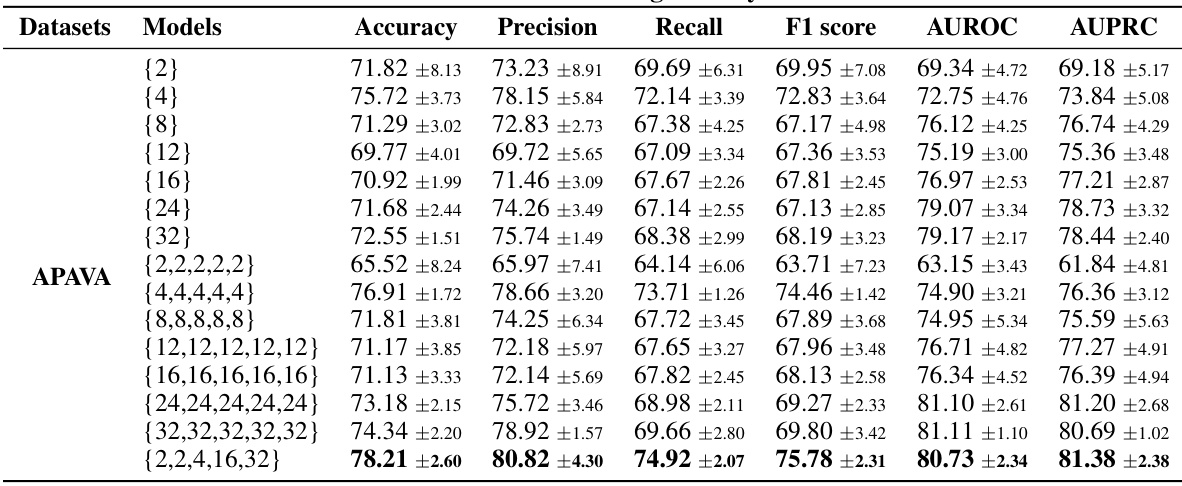
This table presents the results of an ablation study on the APAVA dataset, investigating the impact of different patch lengths on model performance. It compares the performance of using a single patch length versus using multiple identical patch lengths and a mix of patch lengths. The results show that using multiple identical patch lengths generally enhances performance compared to a single patch length. A mixed set of patch lengths also demonstrates effectiveness.

This table presents the performance of Medformer and ten baseline models on five medical time series datasets under a subject-independent setup. The table shows the results for six evaluation metrics (Accuracy, Precision, Recall, F1 score, AUROC, and AUPRC) across datasets with different characteristics (number of classes, number of channels, etc.). This setup is more realistic for real-world applications as it assesses the generalizability of the models to unseen subjects.
Full paper#