↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many scientific fields rely on accurate dynamical systems models for understanding complex processes. Current methods for building these models often rely on human experts and are expensive, time-consuming, and limited by existing knowledge. This creates a significant bottleneck for research in areas like pharmacology, where accurate drug models are essential for safety and efficacy.
The paper introduces the Data-Driven Discovery (D3) framework, which uses Large Language Models (LLMs) to automate the process of model discovery and refinement. D3 allows the LLM to propose new models, acquire relevant data, and evaluate the models’ performance, iteratively improving the model’s accuracy. The authors demonstrate D3’s effectiveness using a pharmacokinetic dataset for Warfarin, where it discovers a new model that is both accurate and interpretable.
Key Takeaways#
Why does it matter?#
This paper is important because it presents a novel framework for discovering and refining interpretable dynamical systems models using Large Language Models (LLMs). This addresses the limitations of traditional methods which are often costly, lack scalability, and rely heavily on human expertise. The proposed Data-Driven Discovery (D3) framework offers a significant advancement in the field, opening avenues for more efficient and insightful investigations into complex systems across various domains. The application to pharmacokinetics, demonstrating the discovery of a new, well-fitting model for Warfarin, highlights the potential for precision medicine and personalized treatment.
Visual Insights#
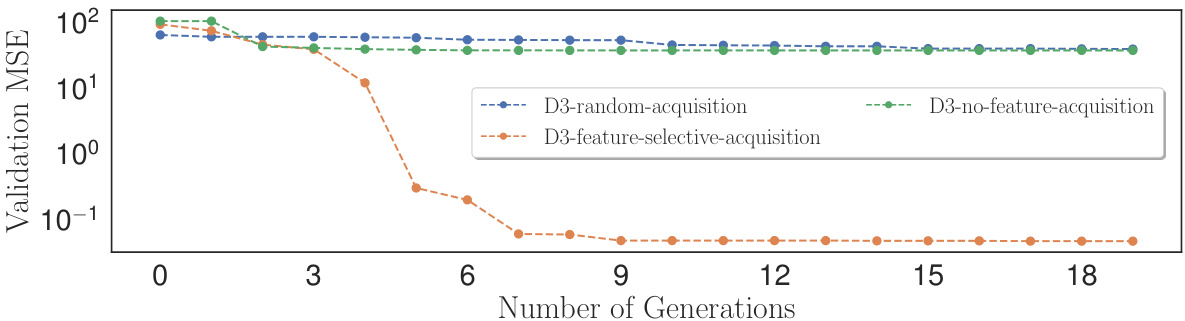
This figure shows the results of an experiment comparing three different feature acquisition methods for the Lung Cancer (with Chemo. & Radio.) dataset. The three methods are: D3-feature-selective-acquisition (D3’s proposed method), D3-random-acquisition (randomly selecting features), and D3-no-feature-acquisition (using only the initial features). The y-axis represents the validation MSE, and the x-axis represents the number of generations (iterations of the model improvement loop). The graph demonstrates that D3’s feature-selective acquisition consistently achieves the lowest validation MSE, indicating that its approach to intelligently selecting new features leads to better model performance than random selection or using only the initial features.
This table presents the mean squared error (MSE) achieved by different methods (DyNODE, SINDY, ZeroShot, ZeroOptim, RNN, Transformer, D3-white-box, D3-hybrid) on six different datasets (Lung Cancer, Lung Cancer (with Chemo.), Lung Cancer (with Chemo. & Radio.), Plankton Microcosm, COVID-19, Warfarin PK). The results are averaged over ten runs with different random seeds, and 95% confidence intervals are included to show the variability in performance. The table highlights that D3 consistently achieves the lowest MSE across all datasets.
In-depth insights#
LLM-driven PK Modeling#
LLM-driven PK modeling represents a paradigm shift in pharmacokinetic analysis. By leveraging the capabilities of large language models, this approach automates the traditionally manual and expert-driven process of PK model discovery and refinement. LLMs can generate and evaluate numerous model candidates, accelerating the identification of optimal models. This automation addresses the limitations of human expertise, reducing the time and cost associated with PK model development. Furthermore, the ability of LLMs to process and integrate diverse data types, including unstructured information from literature, promises to enhance the accuracy and interpretability of PK models. However, challenges remain in ensuring the reliability and trustworthiness of LLM-generated models, necessitating rigorous validation and expert review. The ethical implications of relying on AI for critical clinical decisions must also be carefully considered. Future research should focus on enhancing the transparency and explainability of LLM-driven PK modeling and establishing robust validation protocols. This will be pivotal for establishing confidence in using this technology for personalized medicine.
D3 Framework#
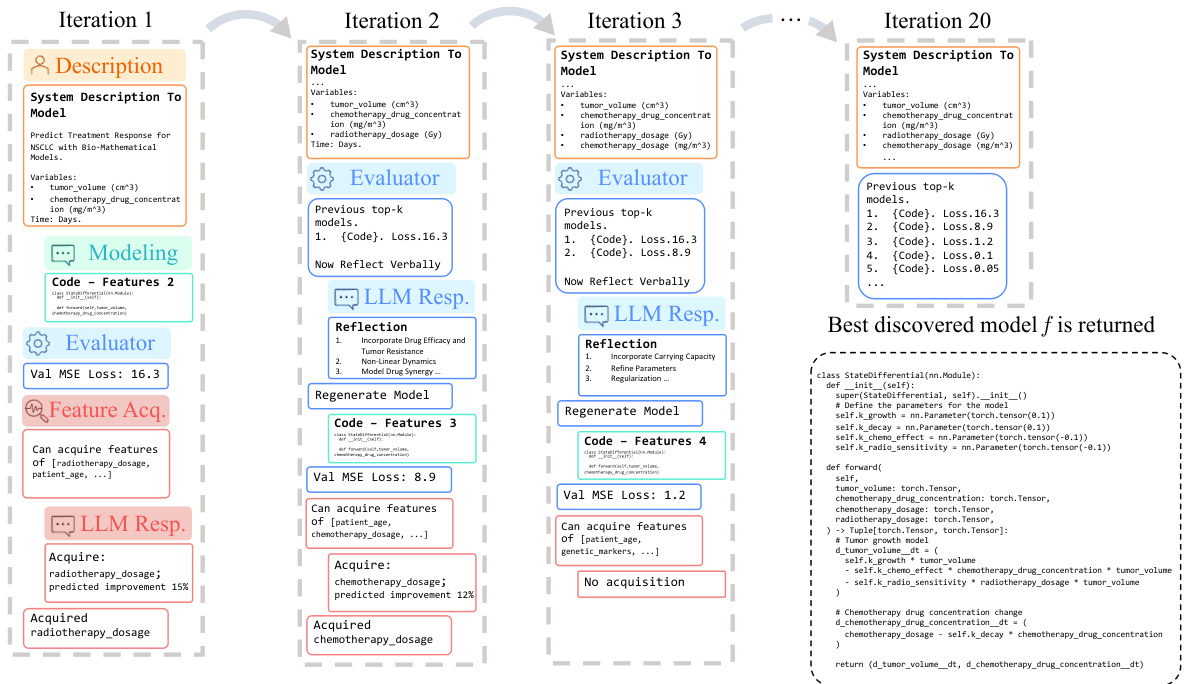
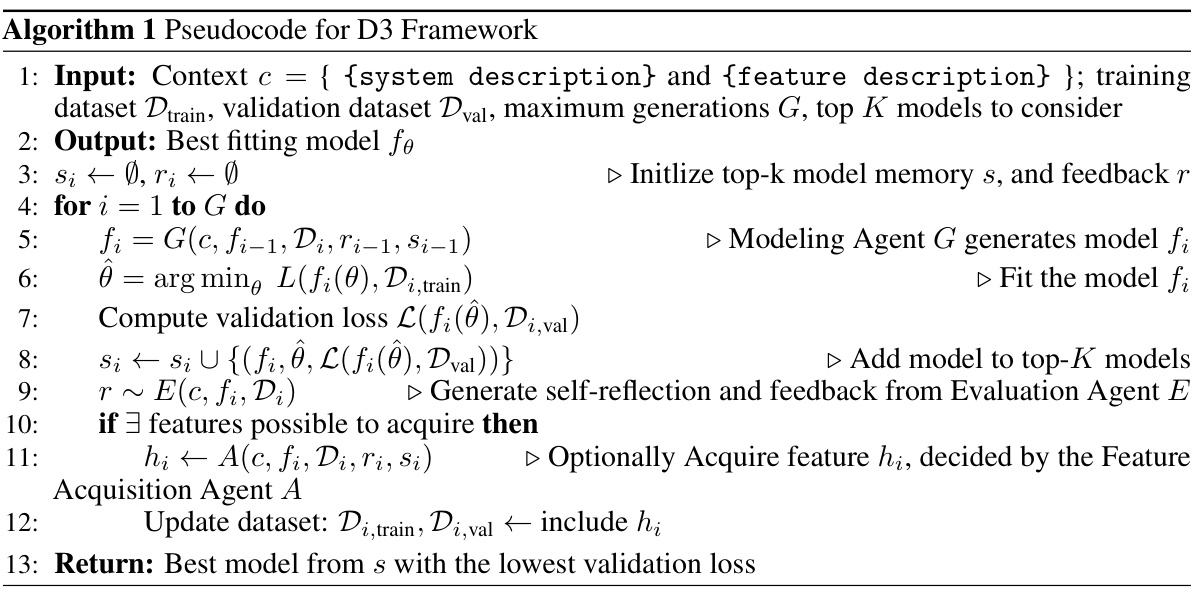
The D3 framework, a data-driven discovery system, leverages large language models (LLMs) to iteratively refine dynamical system models. Its modular design incorporates three key agents: a Modeling Agent generating hypothetical models as code, a Feature Acquisition Agent strategically selecting additional features using value of information calculations, and an Evaluation Agent providing feedback via refined metrics and verbal insights. This iterative process allows D3 to explore a vast model space, uncovering interpretable models that outperform traditional methods and black-box alternatives, as demonstrated in the Warfarin case study. LLM capabilities in code generation, natural language understanding, and few-shot learning are crucial to D3’s success, enabling efficient model refinement and feature acquisition. The system’s ability to integrate unstructured data and prior knowledge further enhances its ability to uncover accurate and clinically relevant models in various domains.
Warfarin Case Study#
The Warfarin Case Study section presents a compelling example of the Data-Driven Discovery (D3) framework’s capabilities. D3 successfully identifies a novel, more accurate pharmacokinetic (PK) model for Warfarin, a drug crucial for treating various cardiovascular conditions. This new model improves upon existing models by incorporating additional parameters that capture complex interactions between patient characteristics (age, sex) and drug dosage, leading to more precise dosing predictions. The model’s improved accuracy, as demonstrated by a lower test MSE, highlights D3’s capacity to not only discover but also refine interpretable dynamics models in pharmacological applications. The involvement of expert pharmacologists in validating the new model underscores the clinical relevance and practicality of the D3 framework. This validates the significance of the D3’s ability to uncover clinically plausible models. The case study also showcases the iterative refinement process of D3, where the model was iteratively improved based on LLM-generated feedback, leading to a superior model. This iterative approach is a key strength of the D3 framework, enabling it to efficiently explore the vast space of possible models and ultimately discover more accurate and informative models. The successful application to a real-world clinical dataset further solidifies the potential of D3 for advancing PK modeling and personalized medicine.
Feature Acquisition#
The paper introduces a novel Feature Acquisition agent, a crucial component of its Data-Driven Discovery (D3) framework. This agent tackles the challenge of strategically selecting new features for improving model accuracy. Unlike traditional methods, D3 uses LLMs to estimate the value of acquiring a new feature, leveraging the LLM’s capabilities in zero-shot and few-shot learning. This approach cleverly bypasses the need for pre-existing features, a major limitation in existing methods. The agent predicts the value of a feature based on available information (e.g., summary statistics, textual descriptions), using the LLM’s understanding of unstructured information to propose informed model improvements. This iterative process, combined with feedback from the Evaluation Agent, guides D3’s model refinement, allowing it to uncover more accurate dynamical system models with minimal parameter tuning. The value of information framework provides a principled way to assess the potential benefit of feature acquisition, balancing the improvement in model performance against the cost of data acquisition. The LLM’s ability to handle unstructured data and optimize data acquisition based on summary statistics is a significant advancement over traditional manual methods, enabling efficient and scalable exploration of the model space.
Future Directions#
Future research could explore several avenues. Improving the efficiency and scalability of the D3 framework is crucial. This might involve optimizing the LLM interactions or employing more efficient model search strategies to reduce computational costs and increase speed. Investigating the robustness of D3 to different data types and noise levels is another key area. Applying D3 to a wider range of pharmacological contexts (beyond warfarin) and other domains like epidemiology and ecology, where dynamical systems are prevalent, would further demonstrate the versatility of the approach. Expanding the model space considered by D3, including more complex ODE models or incorporating mechanistic insights from human experts, could improve the accuracy and interpretability of the models generated. Development of robust methods for handling missing data and outliers in the dataset is also necessary for real-world applications. Finally, incorporating explainability techniques into the model output would add valuable transparency, fostering trust and facilitating broader adoption.
More visual insights#
More on tables
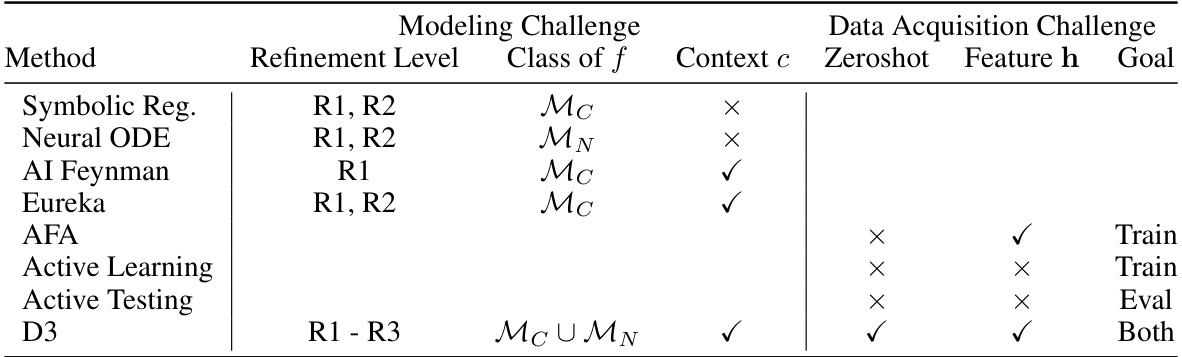
This table compares D3 with other related works in terms of their abilities to address the modeling and data acquisition challenges in discovering and refining dynamical systems models. It assesses each method based on several factors, including the level of refinement it can achieve, the type of ODE model it uses (closed-form or neural network), whether it leverages contextual information, whether it can acquire new samples or features, and the goal of its data acquisition (training or evaluation).
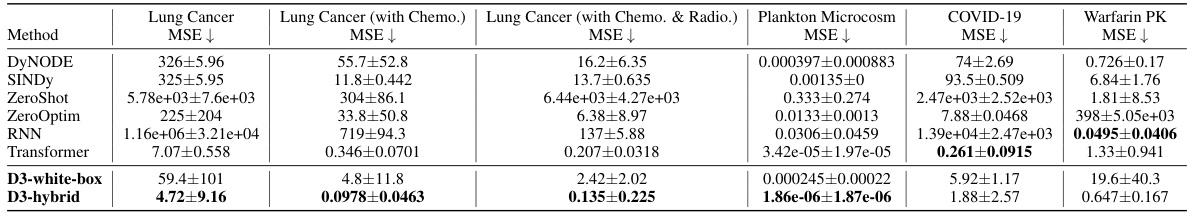
This table presents the mean squared error (MSE) achieved by different methods on six real-world datasets. The methods include various ODE solving techniques and neural network models. The results show the average MSE and 95% confidence interval across ten runs for each method and dataset. The table highlights that the D3 framework consistently outperforms other methods in achieving the lowest MSE, showcasing its effectiveness in discovering accurate models for diverse dynamical systems.
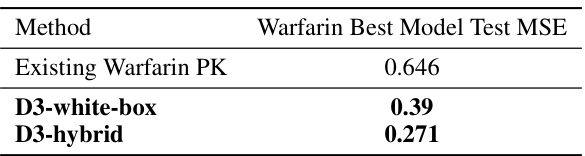
The table compares the test mean squared error (MSE) of three different Warfarin PK models: the existing Warfarin PK model, the D3-white-box model (a white-box model discovered by the D3 framework), and the D3-hybrid model (a hybrid model discovered by the D3 framework). The results show that both D3 models significantly outperform the existing model, indicating the effectiveness of the D3 framework in discovering more accurate and interpretable PK models for Warfarin.
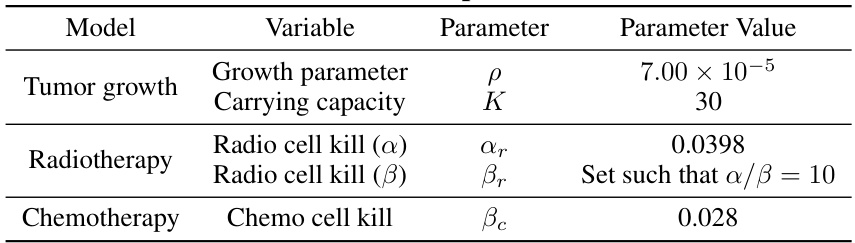
This table lists the parameter values used in the Cancer PKPD model for simulating lung cancer tumor growth. The model incorporates parameters for tumor growth (growth parameter, carrying capacity), radiotherapy (radio cell kill α and β), and chemotherapy (chemo cell kill). The α/β ratio for radiotherapy is set to 10.
This table compares the paper’s proposed method (D3) with other related works in the field of discovering and refining interpretable dynamical systems models. It highlights the differences in terms of the refinement level achieved (R1-R3), the type of ODE model used (closed-form or neural network), the use of context information, the ability to acquire new samples or features, and the ultimate goal of data acquisition (training, evaluation, or both). This comparison helps to showcase the novelty and advantages of the D3 framework.
Full paper#