↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Medical image registration, aligning images from different sources, is crucial in medicine. Two main approaches exist: classical optimization and deep learning. Classical methods are robust and generalizable across different image types, but can be slow. Deep learning methods, while potentially faster and capable of high accuracy, often struggle to generalize and are sensitive to variations in image data. This paper investigates the conditions under which each method performs better.
This study systematically compares classical and deep learning methods’ performance in image registration, focusing on the impact of supervision (label availability). They find that classical methods excel when supervision is limited or when data varies, while deep learning methods perform better with labeled data in consistent data distributions. Their findings help resolve the ongoing debate by highlighting the contexts where each approach is more effective, advancing medical image registration. The research also shows that deep learning methods’ ability to learn image features doesn’t always translate to better generalization across different datasets.
Key Takeaways#
Why does it matter?#
This paper is crucial because it directly addresses the ongoing debate on the superiority of classical vs. deep learning methods in medical image registration. It provides a much-needed empirical comparison, highlighting the strengths and limitations of each approach under various conditions (supervision, training time, domain shift). This clarifies the conditions under which each method excels, impacting the design of future registration systems and research directions.
Visual Insights#
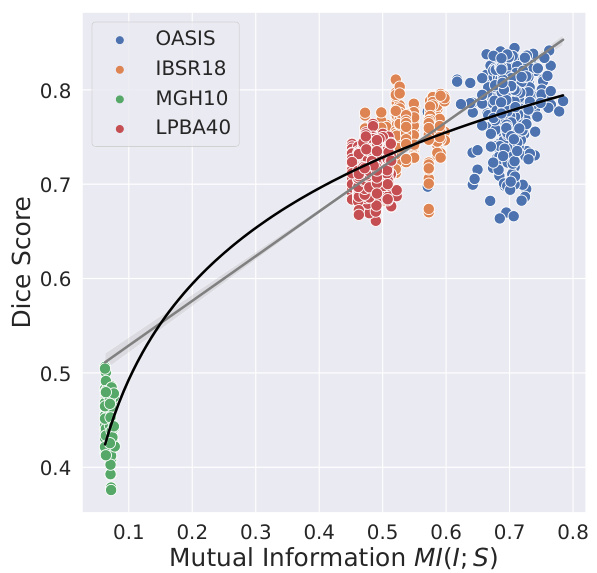
This figure shows the strong correlation between the Dice Score (a measure of registration accuracy) and the mutual information (MI) between the image intensity and label maps for four different brain datasets. The Dice Score measures how well the automatically aligned labels match the ground truth labels, while the mutual information quantifies the amount of shared information between the image intensity and the labels. A higher mutual information indicates that the image intensity provides more information about the labels, leading to better registration accuracy. The strong correlation suggests that the performance of classical registration methods, which primarily rely on image intensity, is strongly linked to the amount of information the image provides about the labels. This suggests that simply improving the architecture of learning-based methods may not significantly improve the performance of such methods, as these architectures are unlikely to impact the underlying relationship between image intensities and labels.

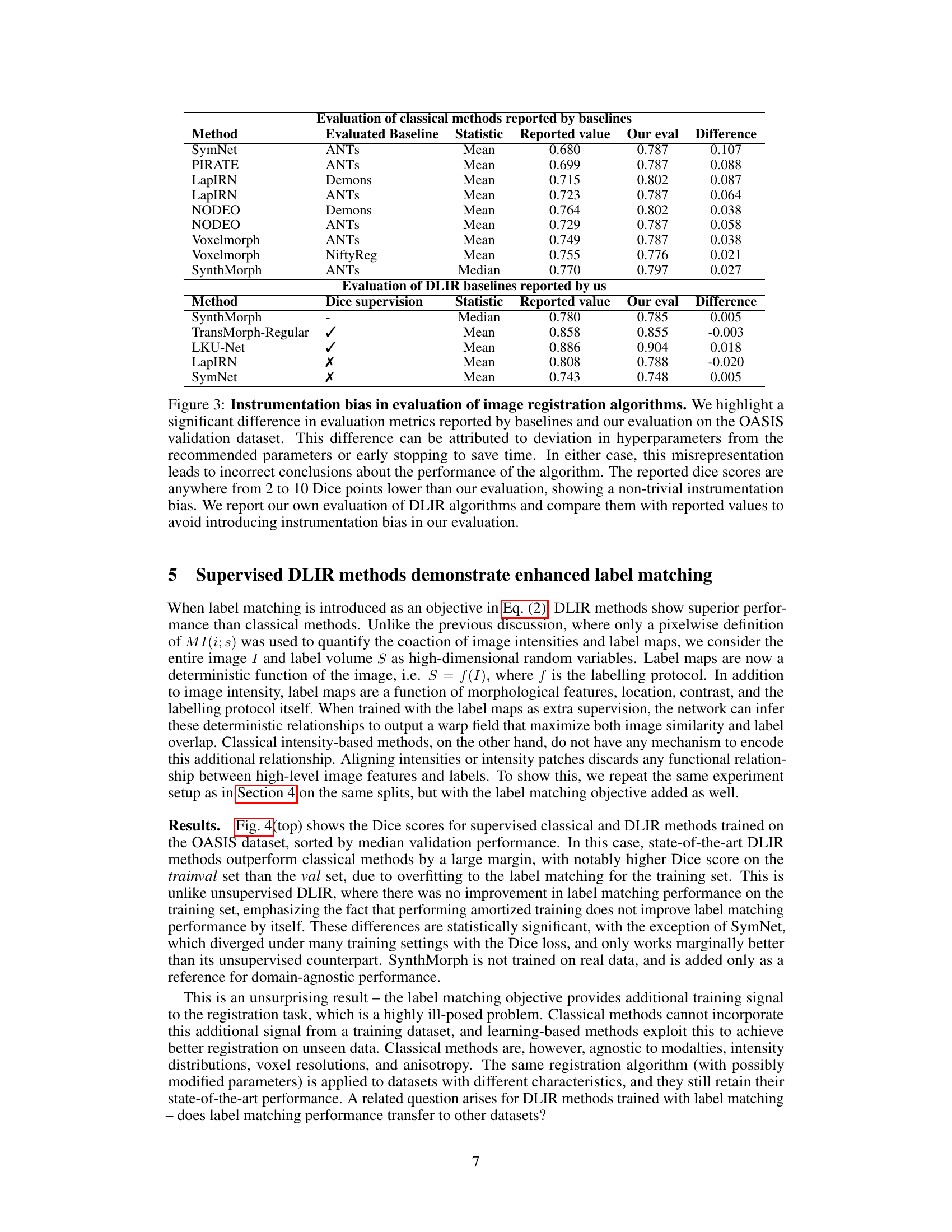
This table compares the performance of classical and deep learning image registration methods reported in existing literature against the author’s own evaluation. It highlights a significant discrepancy, termed as ‘instrumentation bias’, attributed to differences in hyperparameter choices and early stopping criteria used in prior studies. The authors’ re-evaluation shows that classical methods perform significantly better than previously reported and often surpasses Deep Learning based methods.
In-depth insights#
DLIR vs. Classical DIR#
The comparative analysis of DLIR and classical DIR methods reveals complementary strengths and weaknesses. Classical methods, while slower, offer robustness and generalizability across diverse data, leveraging well-established optimization techniques and inherent insensitivity to domain shifts. DLIR methods, conversely, demonstrate superior performance when sufficient labeled data are available, allowing for efficient learning of complex relationships between image intensity and anatomical labels. This advantage manifests as significantly improved label matching accuracy. However, DLIR’s performance is sensitive to domain shift, exhibiting decreased generalization to unseen datasets. The study underscores that the ideal choice hinges on the availability of labeled data and the importance of generalizability versus peak performance in specific applications. Neither method universally outperforms the other; rather, their effectiveness is context-dependent.
Label Map Learning#
Label map learning in medical image registration explores how deep learning models can leverage label information to improve registration accuracy. A key aspect is the implicit learning of relationships between image intensities and labels, enabling the network to predict transformations that align not only image intensities but also anatomical structures represented by labels. This contrasts with classical methods that primarily rely on intensity-based similarity metrics. The effectiveness of label map learning is highly dependent on the quality and availability of labeled data, with larger, well-annotated datasets generally resulting in better performance. However, a significant challenge lies in generalization to unseen data, as models trained on one dataset may not perform well on another due to differences in imaging protocols, anatomical variations, or labeling conventions. Weakly supervised methods aim to mitigate the need for large labeled datasets, but achieving robust performance remains an area of active research. Addressing the domain shift problem and exploring strategies to improve generalization are critical for advancing the practical application of label map learning in medical image registration.
Domain Shift Effects#
The concept of “Domain Shift Effects” in the context of medical image registration using deep learning methods is crucial. Deep learning models are highly susceptible to variations in data distribution, exhibiting reduced performance when faced with unseen data that deviates from the training set. This vulnerability stems from the inherent nature of deep learning models to learn specific data characteristics. If the test data’s imaging modality, acquisition parameters, or anatomical variations differ, the learned model may fail to generalize effectively, leading to significant performance degradation. Addressing domain shift necessitates strategies such as data augmentation, domain adaptation techniques, or the development of more robust and invariant models. Investigating this issue requires careful examination of how different data characteristics influence model accuracy. Moreover, identifying the factors that cause performance drops will help to develop more generalizable models, improving reliability and clinical applicability of deep learning techniques in medical image registration.
MI(i;s) Correlation#
The mutual information between image intensity (i) and label maps (s), denoted as MI(i;s), is crucially important in determining the performance of classical image registration methods. A strong correlation exists between MI(i;s) and the Dice score, a common metric for evaluating registration accuracy. This suggests that classical methods’ success hinges on the level of information about labels contained within the image intensity. The intensity’s fidelity, directly impacting MI(i;s), becomes a limiting factor. Deep learning methods, although employing more sophisticated architectures, do not inherently improve this fundamental relationship. This implies that improvements in DLIR methods’ performance will be constrained by the information contained in the images themselves, rather than solely improvements in the architecture or learning strategy. Ultimately, this analysis highlights the significance of image quality and its direct impact on both classical and deep learning registration approaches.
Generalization Limits#
The concept of “Generalization Limits” in the context of deep learning for medical image registration (DLIR) is crucial. DLIR models, while showing promise in achieving high accuracy on specific datasets, often struggle to generalize to unseen data with even minor domain shifts. This limitation stems from the fact that these models learn highly specific features tailored to the training data’s characteristics (intensity distributions, image resolutions, labeling protocols etc.), hindering their adaptability to new datasets with varying properties. The strong correlation observed between the mutual information of intensity and label distributions and the performance of classical registration methods highlights the inherent challenge. Classical methods, being primarily intensity-based, are less susceptible to domain shifts than DLIR models that implicitly or explicitly utilize label information during training, making them less reliant on specific training data characteristics. While supervised DLIR methods show superior performance in label-matching within the same dataset as the training data, this advantage does not translate to improved performance across different datasets. This underscores the critical need to evaluate the generalization capabilities of DLIR rigorously, considering different data acquisition modalities and processing parameters. This exploration of generalization limits emphasizes the importance of developing more robust, domain-invariant methods for medical image registration.
More visual insights#
More on figures
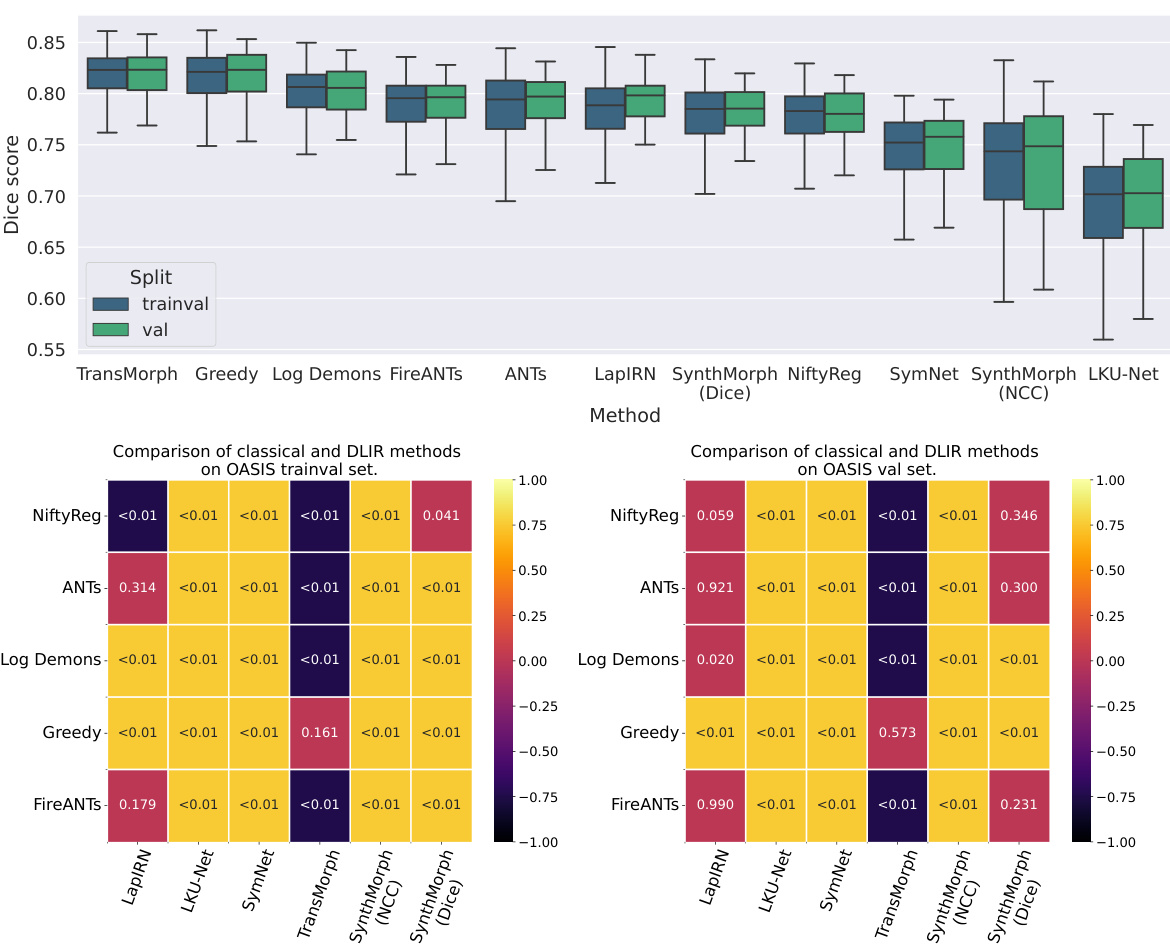
This figure compares the performance of classical and unsupervised deep learning-based image registration (DLIR) methods on the OASIS dataset. The top part shows boxplots illustrating the Dice scores achieved by each method on both the train/validation split and the validation split. The bottom part presents a table of p-values obtained from pairwise two-sided t-tests comparing the performance of classical and DLIR methods. The results indicate that classical methods generally outperform unsupervised DLIR methods, and the performance of unsupervised DLIR methods does not noticeably improve with more training data.
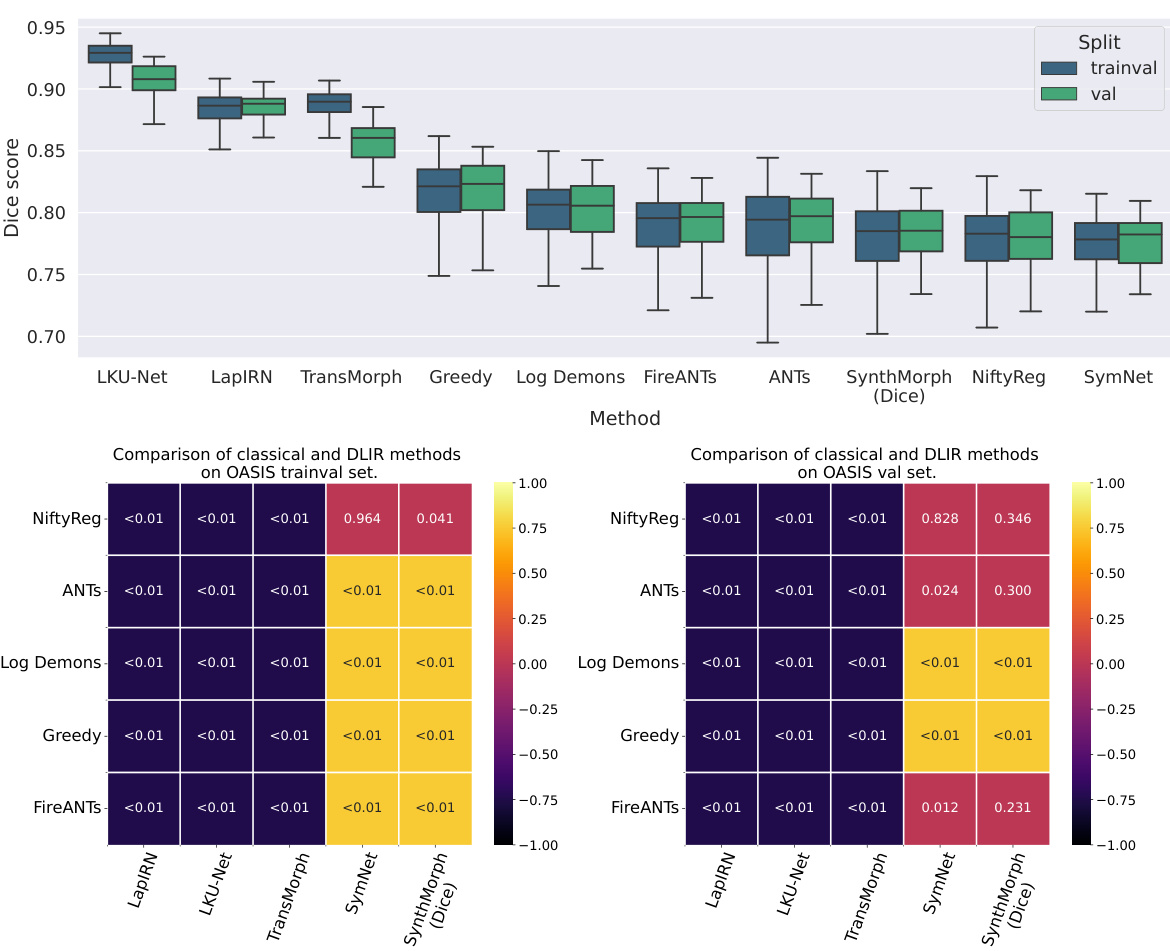
This figure compares the performance of classical and unsupervised deep learning-based image registration (DLIR) methods on the OASIS dataset. The top part shows box plots illustrating the Dice scores for different methods on both trainval (training and validation) and validation sets. It highlights that classical methods generally outperform unsupervised DLIR methods. The lack of improvement in unsupervised DLIR methods on the trainval set compared to the validation set suggests that deep learning doesn’t inherently offer an advantage in label alignment in this unsupervised setting. The bottom part presents tables of p-values from pairwise two-sided t-tests comparing the performance of each classical method against each DLIR method on both datasets. The color-coding emphasizes which method performed significantly better in each comparison (classical or DLIR). Most comparisons show classical methods significantly outperform DLIR methods.

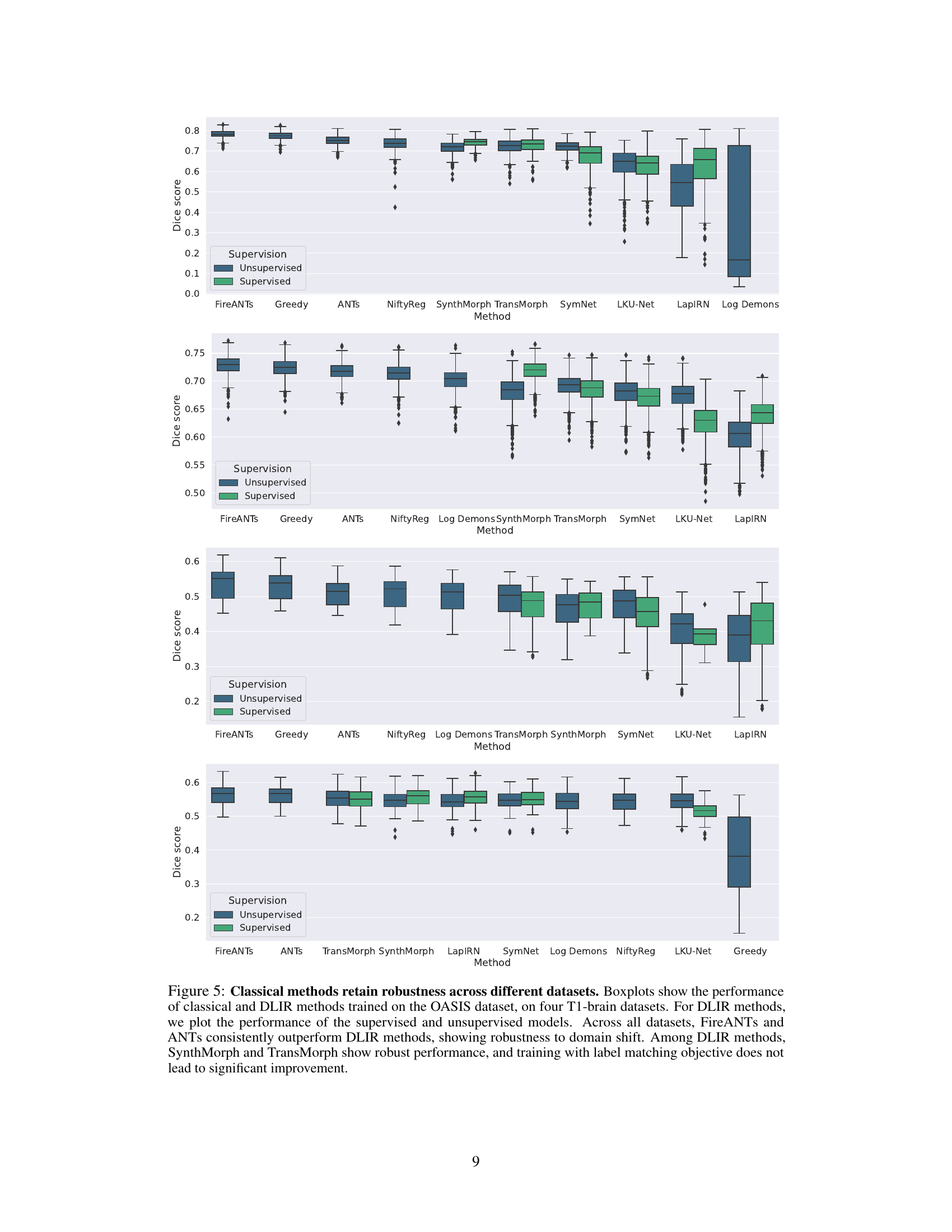
This figure compares the performance of classical and deep learning-based image registration methods across four different brain datasets. The methods were initially trained on the OASIS dataset. The boxplots show the distribution of Dice scores, a measure of registration accuracy, for each method across datasets. The results highlight the robustness of classical methods (FireANTS and ANTs) to domain shift, consistently outperforming deep learning methods even when the latter are trained with label supervision.
Full paper#