↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Geometric Graph Neural Networks (GNNs) are powerful tools for molecular modeling but struggle with long-range interactions in large systems due to their localized nature. This limitation hinders accurate prediction of molecular properties like energy and forces, crucial for various applications. Existing solutions, such as spatial or spectral methods, have limitations in efficiency or flexibility.
To address this, researchers introduce Neural P³M, a novel framework that enhances geometric GNNs by incorporating mesh points alongside atoms. This allows for trainable mathematical operations, effectively modeling long-range interactions. Neural P³M demonstrates superior accuracy in predicting energies and forces compared to existing techniques, showing improvements across diverse molecular systems and architectures. The use of FFT further boosts efficiency. This method offers a significant advancement in molecular modeling, promising more accurate predictions with greater efficiency and flexibility.
Key Takeaways#
Why does it matter?#
This paper is important because it presents Neural P³M, a novel framework that significantly improves the accuracy of geometric GNNs in predicting molecular properties. This is achieved by effectively capturing long-range interactions, a significant challenge in the field. The framework’s versatility and compatibility with various GNN architectures makes it a valuable tool for researchers working on molecular modeling and simulation, paving the way for more accurate predictions and potentially speeding up materials discovery.
Visual Insights#

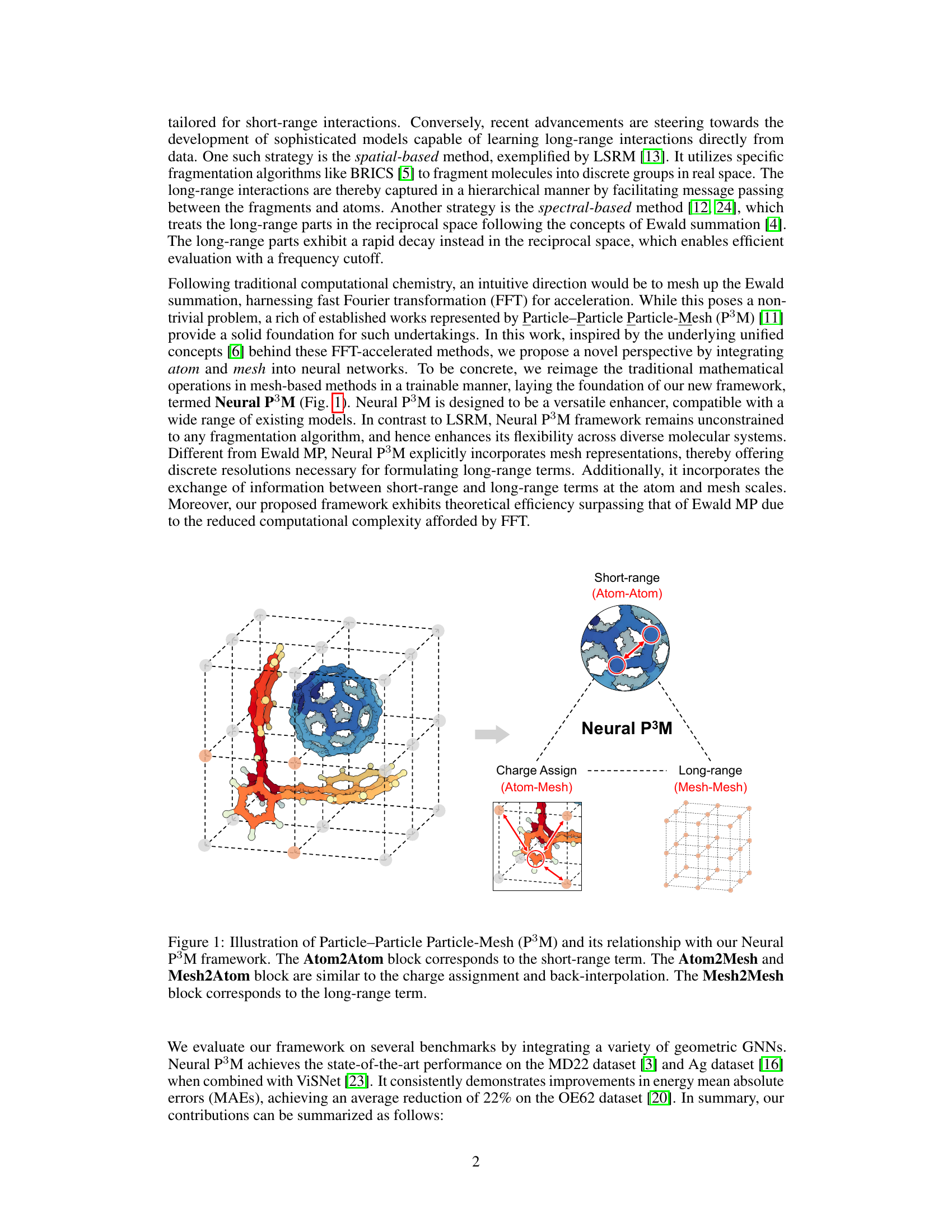
This figure illustrates the Neural P³M framework, which enhances geometric GNNs by incorporating mesh points alongside atoms to model long-range interactions. It shows how the framework integrates short-range (Atom-Atom) and long-range (Mesh-Mesh) interactions through charge assignment (Atom-Mesh) and back-interpolation (Mesh2Atom), improving the accuracy of predicting energies and forces in molecular systems.
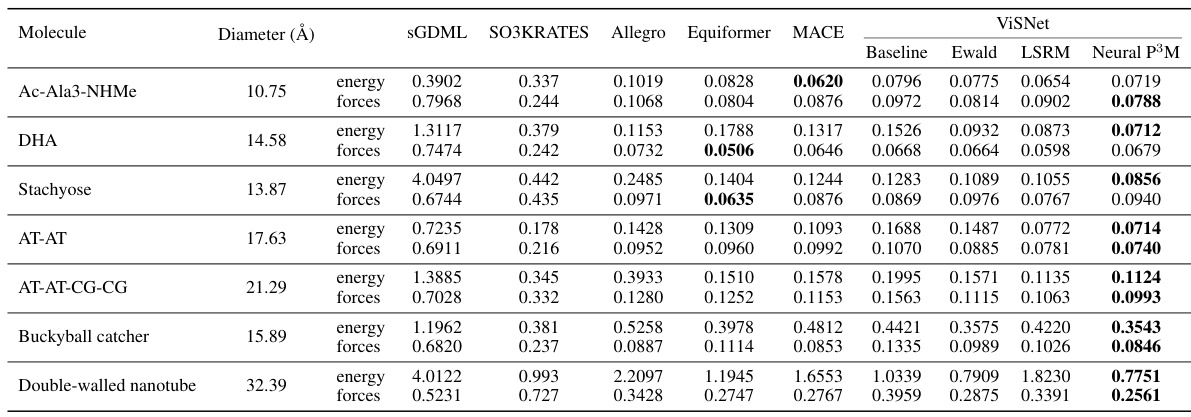
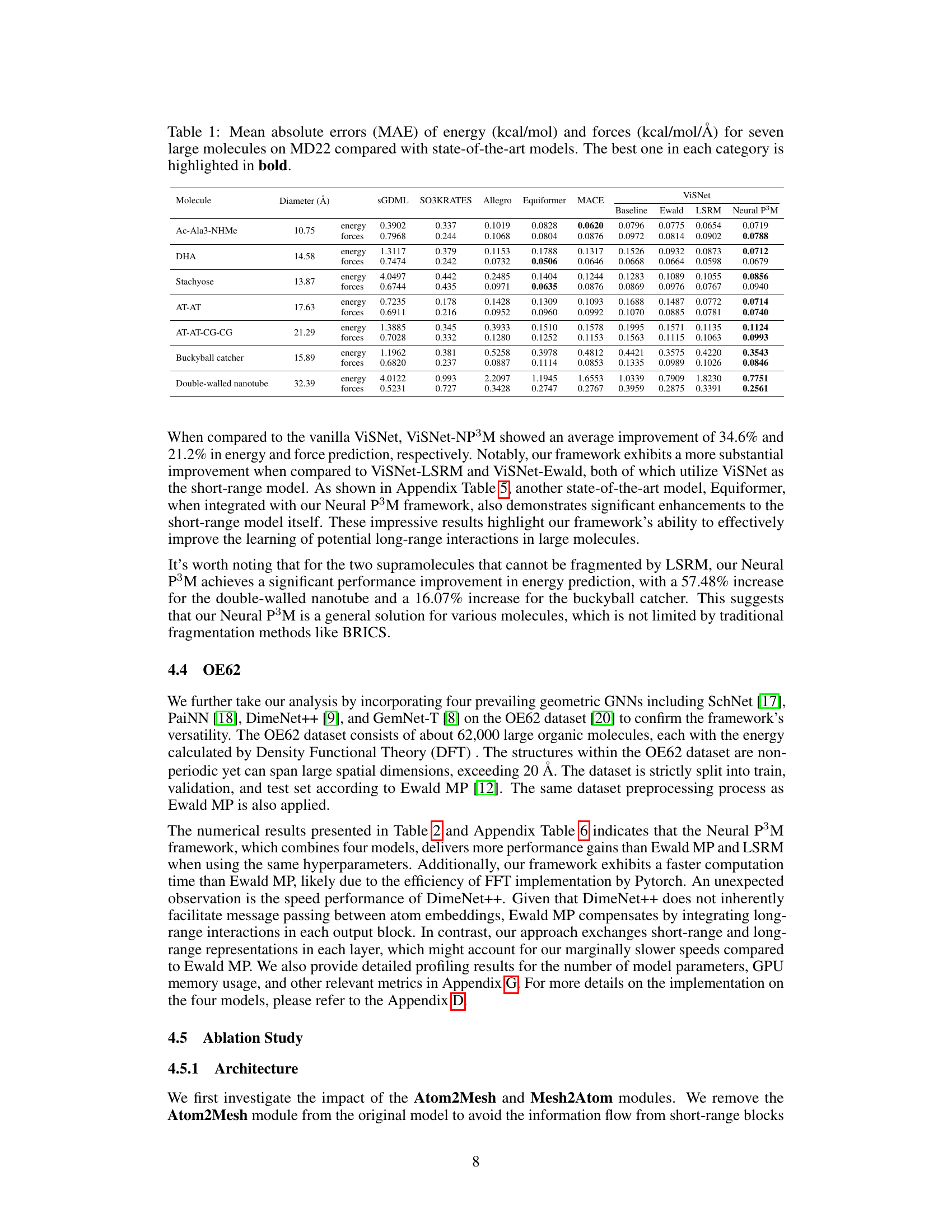
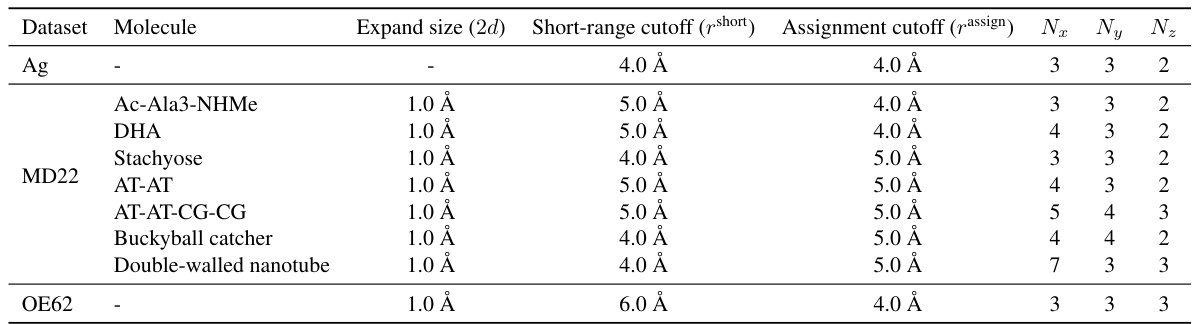
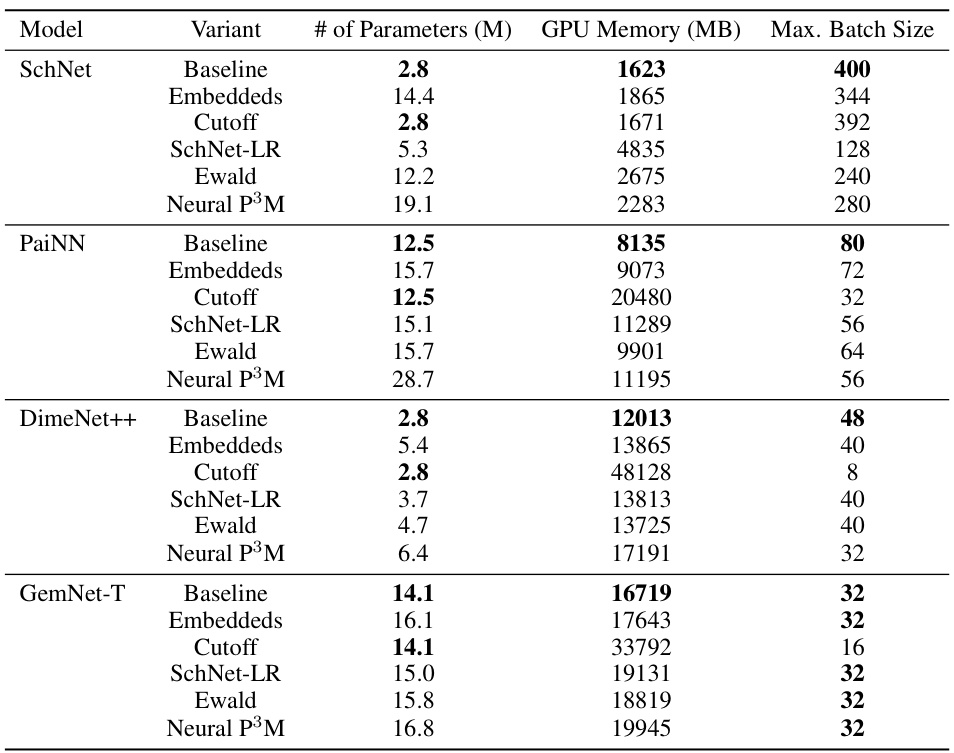
This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by different machine learning models on seven large molecules from the MD22 dataset. The models compared include various geometric graph neural networks (GNNs) with and without the proposed Neural P³M enhancement. The table highlights the best performing model for each molecule and metric (energy and force). The diameter of each molecule is also provided as a contextual factor.
In-depth insights#
Neural P³M Framework#
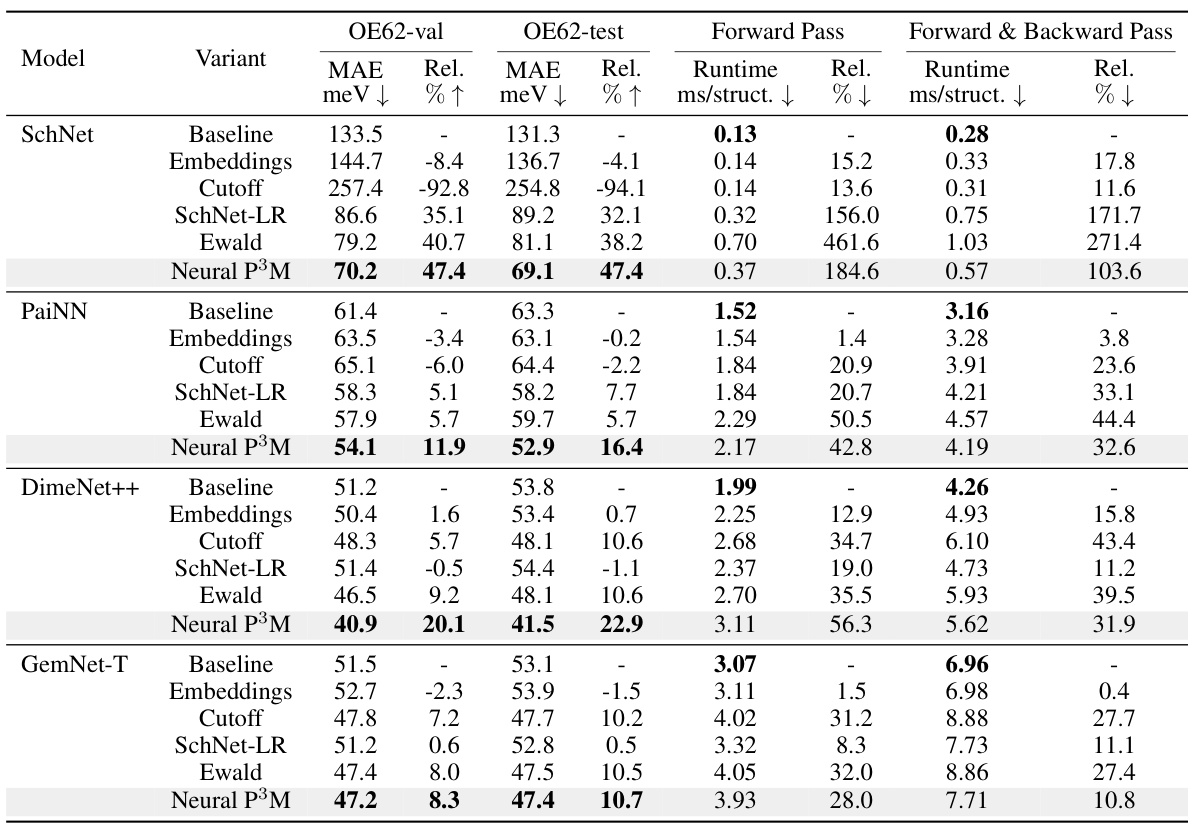
The core of this research is the novel Neural P³M framework, designed to significantly enhance Geometric Graph Neural Networks (GNNs) by effectively modeling long-range interactions within molecular systems. Neural P³M cleverly integrates mesh points alongside atoms, moving beyond the traditional atom-only approach of GNNs. This integration allows for a more nuanced representation of molecular geometry, especially crucial in large systems where long-range forces play a significant role. The framework’s innovation lies not only in the mesh integration but also in its trainable reimagining of traditional mathematical operations, such as those found in Particle-Particle Particle-Mesh (P3M) methods. This trainable aspect allows Neural P³M to adapt and learn the optimal way to handle long-range interactions, offering flexibility and adaptability across various molecular systems and GNN architectures. The integration of short and long-range interaction terms is achieved through a sophisticated system of information exchange between atom and mesh-based representations. The framework’s efficiency comes from the application of Fast Fourier Transformations (FFTs), offering a theoretical advantage in computational speed over traditional Ewald methods. Neural P³M’s versatility is highlighted by its seamless integration with several existing GNN models, showing consistent improvement in energy and force prediction across diverse benchmark datasets. This adaptability underscores its potential as a widely applicable tool for advanced molecular simulations.
Long-Range Modeling#
Long-range interaction modeling in molecular systems presents a significant challenge for geometric graph neural networks (GNNs). Traditional GNNs excel at capturing local interactions but struggle with long-range effects crucial for accurate prediction of molecular properties. Approaches to address this limitation often involve either explicitly incorporating long-range forces (such as Coulomb’s law) or employing techniques to enhance the receptive field of GNNs. Spatial-based methods, like LSRM, divide molecules into fragments to propagate messages across longer distances. Conversely, spectral-based methods utilize Fourier transforms to accelerate the handling of interactions in reciprocal space. The novelty of Neural P³M lies in its integration of Particle-Particle Particle-Mesh (P3M) methods, offering a trainable framework that effectively blends short-range and long-range information. By combining traditional P3M techniques with trainable neural network components, Neural P³M achieves significant improvements in prediction accuracy, surpassing existing state-of-the-art methods. The key is its ability to incorporate mesh points alongside atoms, allowing for a more efficient representation and computation of long-range interactions.
GNN Integration#
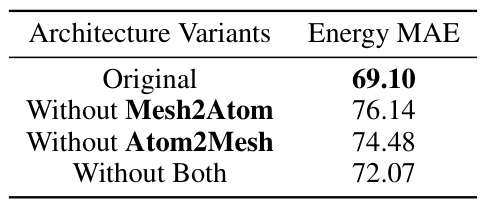
The integration of geometric graph neural networks (GNNs) is a critical aspect of the proposed Neural P³M framework. The paper doesn’t explicitly use ‘GNN Integration’ as a heading, but the methodology extensively discusses the seamless merging of GNNs with the novel mesh-based long-range interaction model. Different GNN architectures are explored, highlighting the versatility of Neural P³M in enhancing various existing GNNs. The integration strategy appears sophisticated, leveraging short-range GNNs to capture local atomic interactions, and long-range components (FNO) to model global effects. A key strength lies in the flexible architecture, allowing Neural P³M to enhance the performance of various GNNs without significant modification. The representation assignment block plays a vital role, efficiently exchanging information between short-range atomic and long-range mesh representations. Overall, the GNN integration approach in Neural P³M showcases a clear understanding of the strengths and limitations of different GNN architectures, resulting in a powerful and adaptable framework for molecular modeling.
Benchmark Results#
A thorough analysis of benchmark results is crucial for evaluating the effectiveness of a new model. This section should meticulously detail the datasets used, metrics employed (e.g., MAE, RMSE), and a comparison against relevant state-of-the-art baselines. Quantitative results, presented in tables and figures, are essential, clearly indicating the model’s performance across different datasets and metrics. It’s important to highlight not only overall performance gains but also any strengths or weaknesses that the new model exhibits on specific datasets. For instance, does it excel on certain molecular system types but falter on others? A discussion of these nuances, along with potential explanations, deepens the analysis. Furthermore, the choice of baselines significantly impacts the assessment. The baselines must be appropriately selected and their performance accurately reported, enabling a fair comparison. Finally, statistical significance should be assessed, and the robustness of the results should be evaluated by exploring variations in hyperparameters and experimental settings.
Future Enhancements#
Future enhancements to Neural P³M could explore several promising avenues. Improving the efficiency of the long-range interaction modeling is crucial, potentially through optimized FFT algorithms or alternative approaches that reduce computational complexity. Expanding the framework to handle larger and more complex systems would involve investigating more advanced mesh generation techniques and efficient data structures for handling the increased number of atoms and meshes. Addressing the challenges posed by diverse molecular systems requires further investigation into the selection and parameterization of the short-range GNNs, allowing the framework to adapt flexibly to different chemical environments. Finally, integrating advanced features such as incorporating explicit solvent effects, exploring more sophisticated charge assignment methods, and developing efficient training strategies for larger datasets represent important avenues for future development. These enhancements would further broaden the applicability and accuracy of Neural P³M.
More visual insights#
More on figures

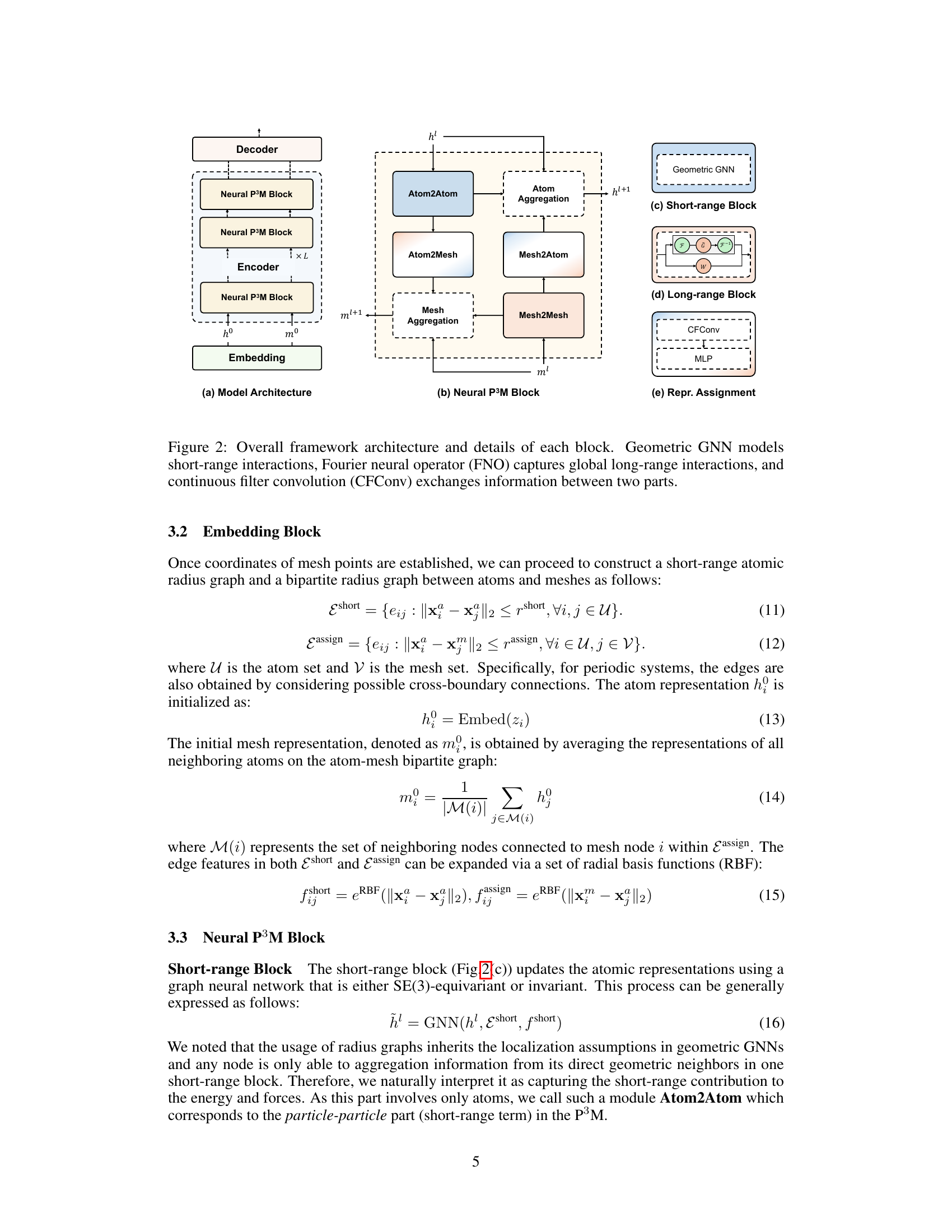
This figure shows the overall architecture of the Neural P³M framework, detailing its different components. Panel (a) provides a high-level view of the model’s structure, illustrating the flow of information from the input embeddings through multiple Neural P³M blocks to the final decoder that outputs energy and forces. Panel (b) zooms in on a single Neural P³M block, illustrating its internal workings which involve distinct modules for handling short-range (Atom2Atom) and long-range interactions (Mesh2Mesh) using techniques such as Atom2Mesh and Mesh2Atom to exchange information between atom and mesh representations, and an aggregation step to combine information from different parts of the Neural P³M block. Panels (c), (d), and (e) illustrate the specific details of the short-range block (using geometric GNNs), the long-range block (employing a Fourier Neural Operator), and the representation assignment, respectively. The figure clearly demonstrates the interplay between short-range and long-range interactions in the proposed framework.
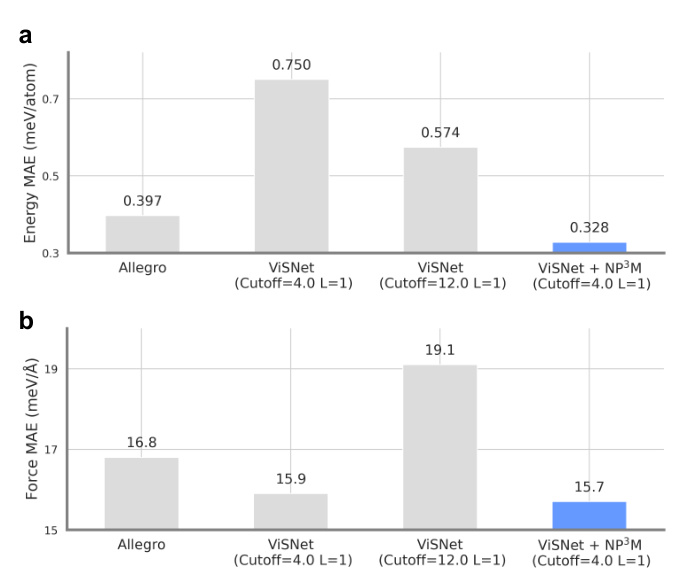
This figure compares the performance of three different models on the Ag dataset in terms of mean absolute error (MAE) for energy and force prediction. The three models are Allegro, ViSNet (with a cutoff of 4.0 Å and 1 layer), ViSNet (with a cutoff of 12.0 Å and 1 layer), and ViSNet integrated with the Neural P³M framework (with a cutoff of 4.0 Å and 1 layer). The results show that ViSNet with Neural P³M significantly outperforms the other models, demonstrating the effectiveness of the proposed method in capturing long-range interactions.

This figure shows the architecture of the Neural P³M framework. It consists of three main blocks: an embedding block, a Neural P³M block, and a decoder block. The embedding block creates representations of atoms and meshes. The Neural P³M block updates these representations by incorporating short-range and long-range interactions, and information exchange between atoms and meshes using GNNs, FNOs, and CFConvs. The decoder block predicts energies and forces based on the updated representations.
More on tables
This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by different models on seven large molecules from the MD22 dataset. The models compared include ViSNet, SGDML, SO3KRATES, Allegro, Equiformer, MACE, baseline Ewald summation, LSRM, and the proposed Neural P³M. The table highlights the best-performing model for each molecule and metric (energy and force). The diameters of the molecules are also provided for context.
This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by several state-of-the-art models on seven large molecules from the MD22 dataset. The models include ViSNet, SGDML, SO3KRATES, Allegro, Equiformer, MACE, Ewald, LSRM, and Neural P³M. The table highlights the best-performing model for each molecule and metric (energy and force). The diameter of each molecule is also included, indicating that larger molecules pose a more significant challenge to these models.

This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by different models on seven large molecules from the MD22 dataset. The models compared include ViSNet, SGDML, SO3KRATES, Allegro, Equiformer, MACE, Baseline Ewald, LSRM, and Neural P³M. The table highlights the best-performing model for each molecule and metric (energy and force). The diameter of each molecule is also provided to give context to the results.

This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by different models on seven large molecules from the MD22 dataset. The models compared include ViSNet, SGDML, SO3KRATES, Allegro, Equiformer, and MACE, along with baselines using Ewald summation and LSRM. The table highlights the best-performing model for each molecule and metric (energy and forces), showing the effectiveness of the Neural P³M framework in improving accuracy.

This table presents a comparison of the mean absolute errors (MAE) for energy and forces predicted by various state-of-the-art models on seven large molecules from the MD22 dataset. The models compared include ViSNet, SGDML, SO3KRATES, Allegro, Equiformer, MACE, and ViSNet integrated with Neural P³M (baseline). The table highlights the best performing model for energy and force prediction for each molecule, demonstrating the superior performance of Neural P³M in most cases.
This table presents the mean absolute errors (MAE) for energy and forces predicted by several state-of-the-art models and the proposed Neural P³M model on seven large molecules from the MD22 dataset. The results are compared to show that Neural P³M achieves the best performance on most of the molecules, particularly on the larger ones, highlighting its ability to accurately model long-range interactions.
This table presents a comparison of the mean absolute errors (MAE) in energy and force predictions for seven large molecules from the MD22 dataset. It compares the performance of the ViSNet model enhanced with Neural P³M against several state-of-the-art methods (SGDML, SO3KRATES, Allegro, Equiformer, and MACE). The best performing method for each molecule (energy and force) is highlighted in bold, showcasing the effectiveness of the Neural P³M enhancement.
Full paper#