TL;DR#
Medical image segmentation struggles with anomalies like tumors due to their variations. Existing methods using either textual or visual prompts alone are insufficient. The long-tail distribution of medical datasets further complicates the task, as rare anomalies lack sufficient training data.
The paper introduces CAT, a dual-prompt model that uses both anatomical prompts (from 3D cropped images) and textual prompts (enriched with medical domain knowledge). CAT employs a query-based design with a novel ShareRefiner to refine prompts and segmentation queries, achieving state-of-the-art performance in various segmentation tasks, particularly for tumors in different cancer stages. This shows that combining different prompt modalities is highly effective.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image segmentation due to its novel approach of combining anatomical and textual prompts. It directly addresses the limitations of existing methods that rely on a single type of prompt, significantly improving accuracy and generalizability, especially for complex scenarios like tumor segmentation. The proposed method, CAT, opens new avenues for multimodal prompt engineering in medical imaging and offers a strong benchmark for future research.
Visual Insights#

🔼 This figure illustrates two key challenges in medical image segmentation. The left panel shows a long-tailed distribution of organ and tumor categories in medical datasets. There are many rare cases, making it difficult to train robust models. The right panel shows colon tumors at different cancer stages (T-Stage III and T-Stage IV). These tumors exhibit significant variation in shape, size, and density, posing a further challenge for accurate segmentation.
read the caption
Figure 1: Left: Long-tailed curve of the category and the number of available cases that can be obtained in the medical field. Right: Tumors in different cancer staging with diverse shapes and sizes.
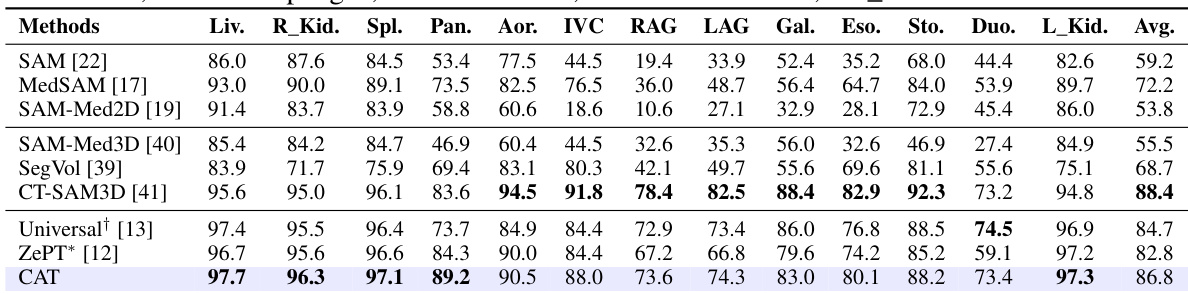
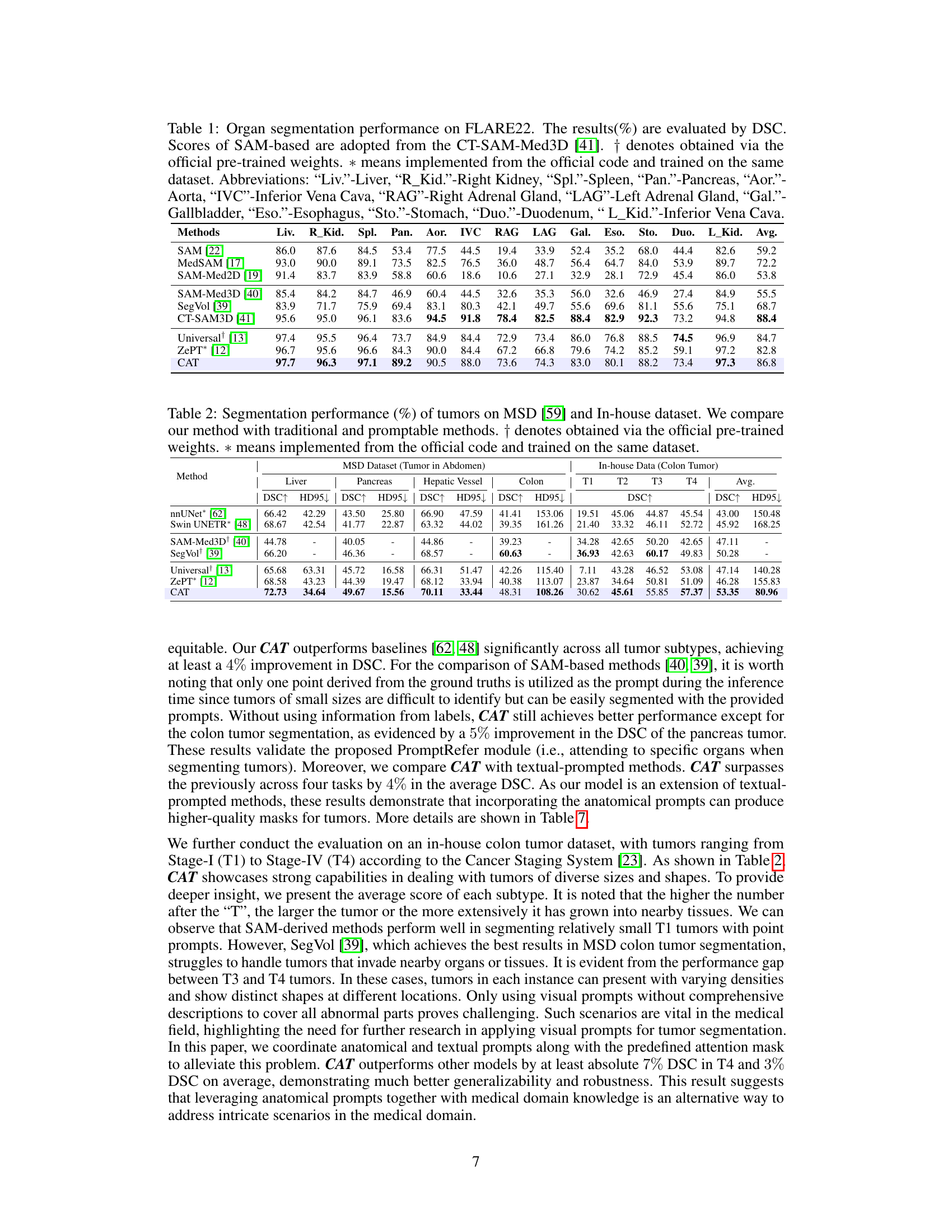
🔼 This table presents a comparison of the organ segmentation performance of the proposed CAT model against several state-of-the-art methods on the FLARE22 dataset. The performance is measured using the Dice Similarity Coefficient (DSC), a common metric for evaluating segmentation accuracy. The table shows DSC scores for 12 different abdominal organs, highlighting CAT’s superior performance compared to other methods, especially those based on adapting the Segment Anything Model (SAM). Abbreviations are provided to clarify the organ names.
read the caption
Table 1: Organ segmentation performance on FLARE22. The results(%) are evaluated by DSC. Scores of SAM-based are adopted from the CT-SAM-Med3D [41]. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset. Abbreviations: “Liv.”-Liver, “R_Kid.”-Right Kidney, “Spl.”-Spleen, “Pan.”-Pancreas, “Aor.”-Aorta, “IVC”-Inferior Vena Cava, “RAG”-Right Adrenal Gland, “LAG”-Left Adrenal Gland, “Gal.”-Gallbladder, “Eso.”-Esophagus, “Sto.”-Stomach, “Duo.”-Duodenum, “L_Kid.”-Inferior Vena Cava.
In-depth insights#
Dual Prompt Fusion#
A dual prompt fusion approach in a research paper likely involves intelligently combining two different types of prompts to enhance performance. This might be visual prompts (like images or image patches) combined with textual prompts (keywords or descriptions), or other complementary prompt modalities. The core idea is that each prompt type provides unique information, and their fusion leverages these strengths synergistically. Effective fusion strategies are key; this could involve concatenating embeddings, using attention mechanisms to weigh prompt contributions, or employing more complex fusion architectures like multi-layer perceptrons. Successful dual prompt fusion should lead to improved accuracy, robustness, and generalizability compared to using either prompt type alone, especially in complex scenarios like medical image segmentation where visual details and contextual understanding are crucial. Careful consideration must be given to how the prompts are designed and the weighting of their relative importance in the fusion process, as this can significantly affect model performance.
ShareRefiner Network#
A hypothetical ‘ShareRefiner Network’ in a medical image segmentation context would likely involve a multi-stage process. It would begin by receiving both anatomical (e.g., cropped 3D volumes) and textual (e.g., medical descriptions) prompts. These disparate inputs would be processed through separate encoders, generating distinct feature representations. The core of the network would then involve a shared refinement module, which uses a cross-attention mechanism to enable interaction between the anatomical and textual features. This interaction is crucial for resolving ambiguities. The module might disentangle the prompts, possibly using separate attention heads or pathways for anatomical and textual information, allowing each prompt type to contribute uniquely to the final segmentation mask. Hard and soft assignment strategies could be implemented to assign features selectively to each prompt type for optimal feature refinement and to ensure that anatomical and textual prompt queries are disentangled. Finally, refined prompt features would be combined to generate refined segmentation queries, which would then be used to predict the segmentation mask. The overall design emphasizes synergizing the strengths of both anatomical and textual prompts to achieve highly accurate and robust segmentation, particularly in complex medical scenarios where visual information alone might be insufficient. The success of this network hinges upon the effective integration and disentanglement of multi-modal prompts, as well as the learning of a robust representation that combines the distinct but complementary information sources.
Ablation Study#
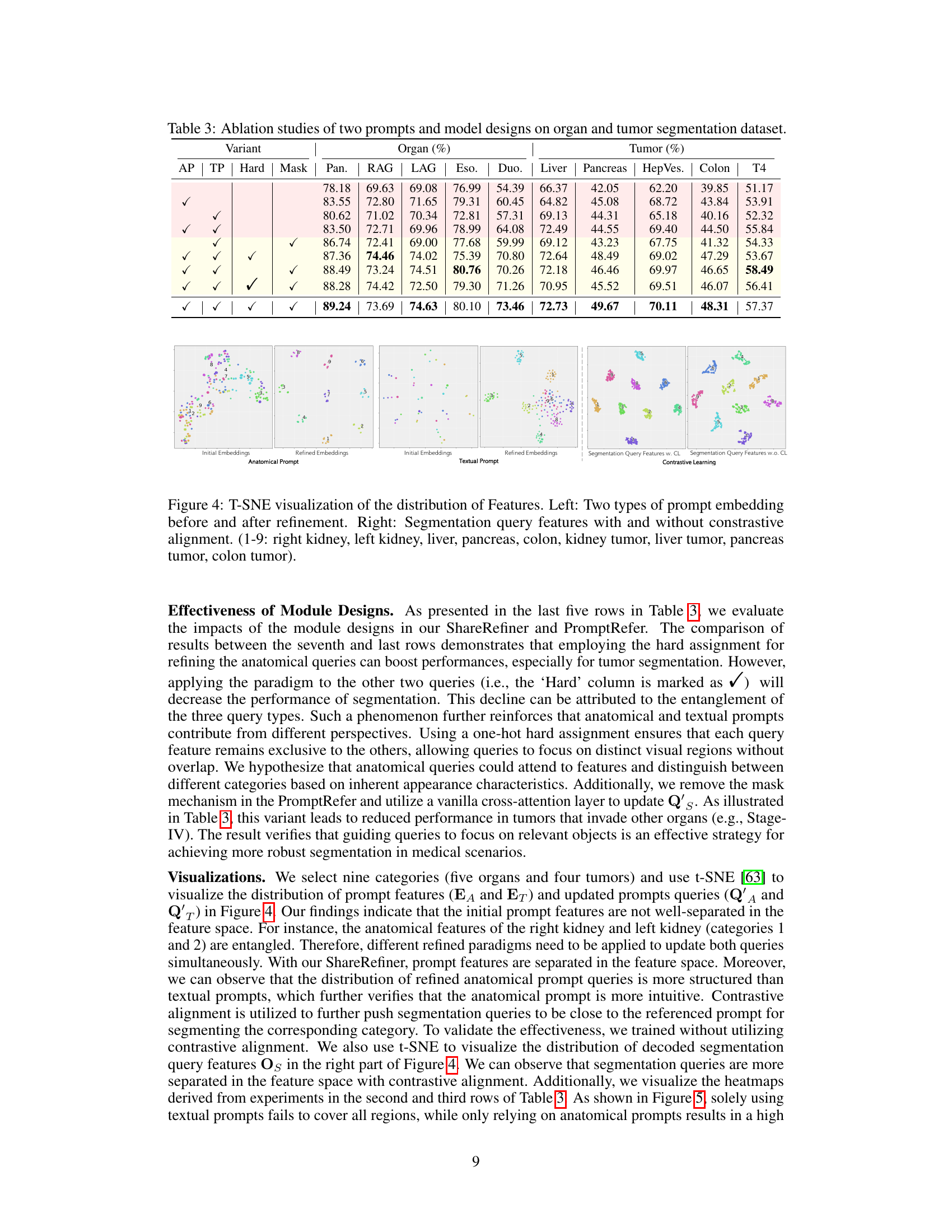
Ablation studies systematically remove components of a model to assess their individual contributions. In this context, it would involve selectively disabling parts of the proposed dual-prompt framework (anatomical prompts, textual prompts, the ShareRefiner module, or the PromptRefer module) to determine how each element affects the overall segmentation performance. By comparing the results of the complete model against the results of models missing each component, researchers gain insights into the importance of every individual module. This method helps isolate the impact of each component, offering valuable evidence to support the claims about the framework’s design choices. For example, if removing the textual prompt significantly reduces performance, it highlights the importance of textual information for accurate tumor segmentation. Conversely, if removing the anatomical prompt does not lead to a substantial drop, it suggests that the model’s primary strength might not stem from visual inputs. Therefore, conducting thorough ablation studies is crucial to validate the model’s design and showcase the contribution of each carefully designed component. The results from this ablation study would help establish the effectiveness and necessity of the proposed framework.
Long-Tail Problem#
The long-tail problem in medical image segmentation presents a significant challenge due to the imbalanced distribution of medical data. Rare diseases or anomalies, such as specific tumor types or stages, are underrepresented, hindering the training of robust and generalizable models. Data augmentation techniques can help mitigate this issue by artificially increasing the number of samples in the underrepresented categories. However, this must be done carefully to avoid introducing artifacts or biases that negatively affect model performance. Transfer learning from large, general datasets, or the use of semi-supervised or self-supervised learning methods, could allow models to learn from a limited number of rare cases while leveraging information from more common ones. Multimodal learning, integrating visual and textual data, offers a promising avenue to address the long-tail issue by incorporating detailed descriptions and clinical knowledge that can compensate for limited visual examples. Furthermore, prompt engineering methods, particularly the coordination of anatomical and textual prompts, may assist in improving accuracy in the rare categories.
Future Directions#
Future research could explore several promising avenues. Improving the robustness and generalizability of the model across diverse datasets and imaging modalities is crucial. This includes addressing the challenges posed by variations in image quality, artifacts, and patient-specific pathologies. Developing more sophisticated prompt engineering techniques that can effectively integrate different types of prompts and guide the model towards accurate segmentation is vital. Furthermore, investigating the potential of incorporating additional data sources, such as patient clinical information or longitudinal imaging, to enhance segmentation accuracy warrants further investigation. Exploring different model architectures, beyond the query-based design, may reveal superior performance. Finally, rigorous validation on large, diverse, and clinically relevant datasets is needed to demonstrate the clinical utility and trustworthiness of the proposed method.
More visual insights#
More on figures
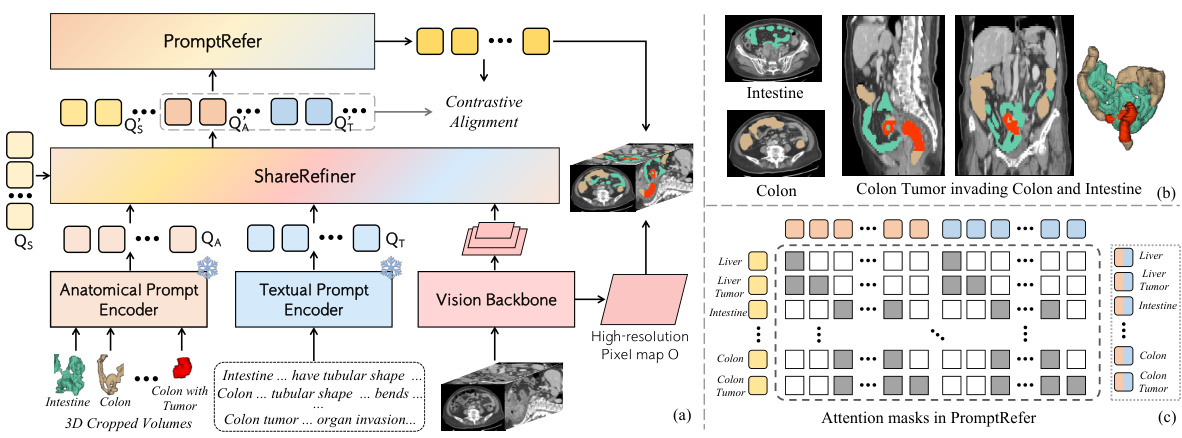
🔼 This figure illustrates the CAT model’s architecture and workflow. Panel (a) shows the overall architecture, highlighting the use of both anatomical (3D cropped volumes) and textual prompts (enhanced with medical knowledge) which are processed by separate encoders to generate prompt queries. These queries, along with segmentation queries, are refined by the Share Refiner module before being integrated by the PromptRefer module for final mask prediction. Panel (b) shows an example case of a Stage-IV colon tumor invading the intestine, illustrating a complex scenario the model handles. Panel (c) visualizes the attention masks within PromptRefer, demonstrating how it selectively assigns prompt queries to influence specific segmentation queries.
read the caption
Figure 2: (a) CAT follows the query-based segmentation architecture. 3D cropped volumes according to the anatomical structure are utilized as anatomical prompts. Texts enhanced by professional knowledge are adopted as textual prompts. Learnable queries and both prompts are utilized for the final prediction via ShareRefiner and PromptRefer. (b) The case of colon tumor in Stage-IV invading the intestine. (c) Attention masks in PromptRefer for assigning specific prompts to queries.
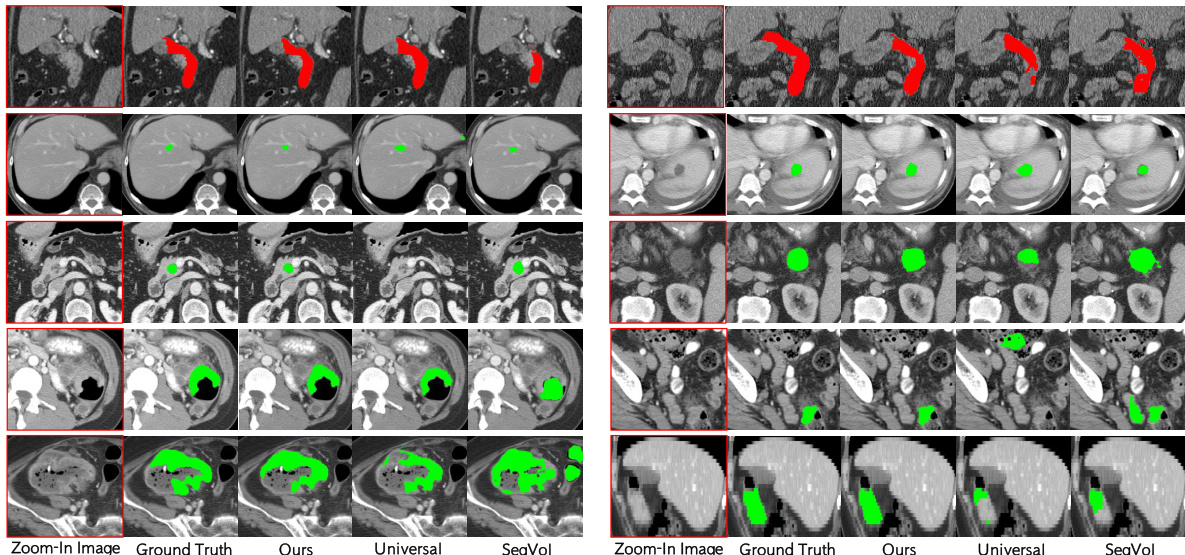
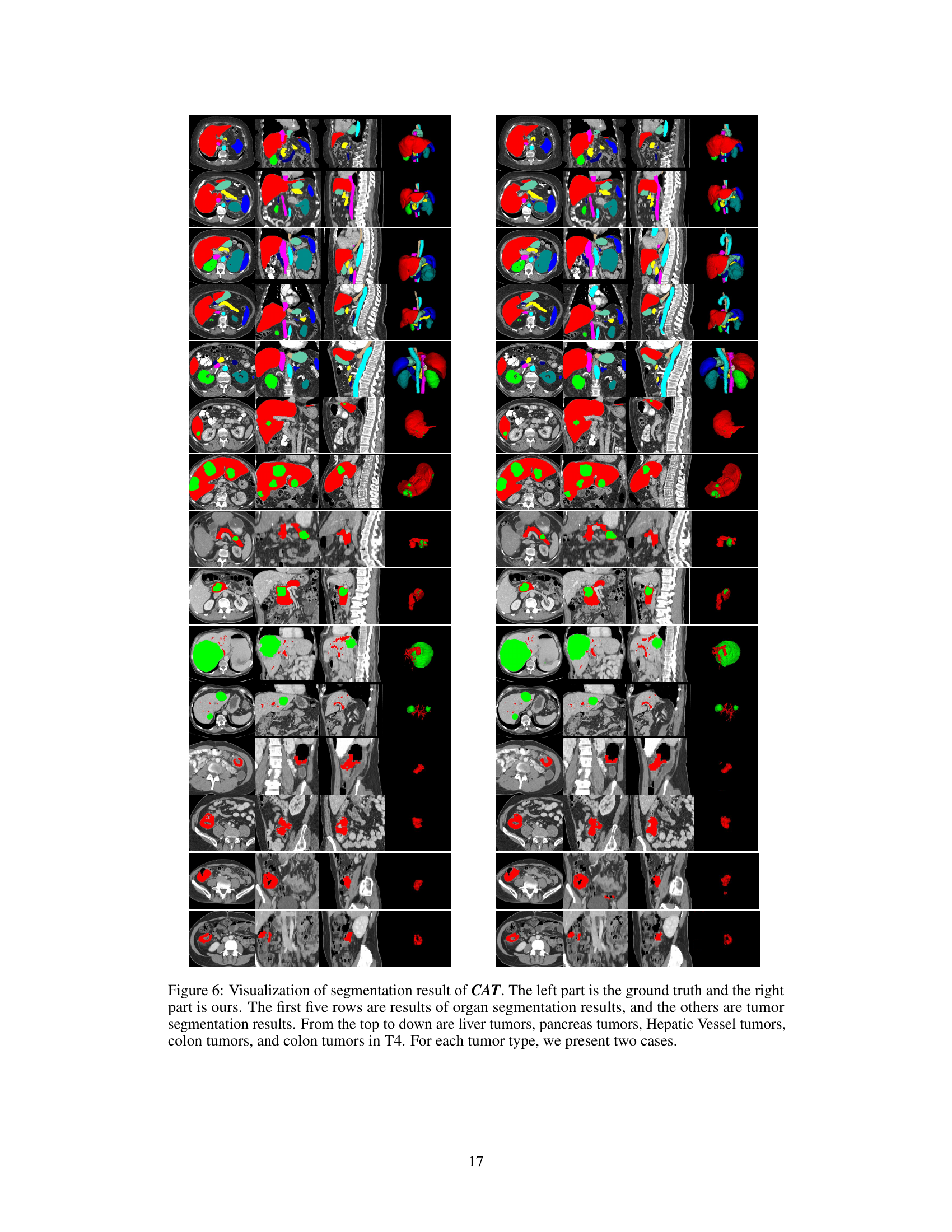
🔼 This figure shows a qualitative comparison of the proposed CAT model’s segmentation performance against other state-of-the-art methods. Each row displays a different organ or tumor type (duodenum, liver tumor, pancreas tumor, colon tumor, Stage IV colon tumor) and shows ground truth segmentations alongside results from CAT, Universal, and SegVol. The visual comparison highlights CAT’s superior ability to accurately segment the target structures, particularly in complex scenarios like Stage IV colon tumors where the tumor invades nearby tissue.
read the caption
Figure 3: Qualitative visualizations of the proposed model and other prompting methods on organ/tumor segmentation. The segmentation results presented from rows one to five correspond, in order, to the duodenum, liver tumors, pancreas tumors, colon tumors, and colon tumors in Stage-IV.

🔼 This figure uses t-SNE to visualize the distribution of features before and after applying different refinement techniques to the prompt embeddings. The left panels show the initial and refined embeddings for anatomical and textual prompts separately, illustrating how the refinement process improves the separation of features in the embedding space. The right panels compare the segmentation query features with and without contrastive learning. The visualization helps to demonstrate that the refinement and contrastive learning steps effectively disentangle the different feature representations, which enhances the model’s ability to accurately segment organs and tumors.
read the caption
Figure 4: T-SNE visualization of the distribution of Features. Left: Two types of prompt embedding before and after refinement. Right: Segmentation query features with and without contrastive alignment. (1-9: right kidney, left kidney, liver, pancreas, colon, kidney tumor, liver tumor, pancreas tumor, colon tumor).

🔼 This figure shows a qualitative comparison of the proposed CAT model’s segmentation performance against other prompting methods (Universal, SegVol) on various organ and tumor types. Each row displays a different organ or tumor, illustrating the model’s ability to segment challenging cases such as tumors invading adjacent organs. The results highlight CAT’s superior performance in accurately segmenting complex and diverse anatomical structures compared to alternative approaches.
read the caption
Figure 3: Qualitative visualizations of the proposed model and other prompting methods on organ/tumor segmentation. The segmentation results presented from rows one to five correspond, in order, to the duodenum, liver tumors, pancreas tumors, colon tumors, and colon tumors in Stage-IV.
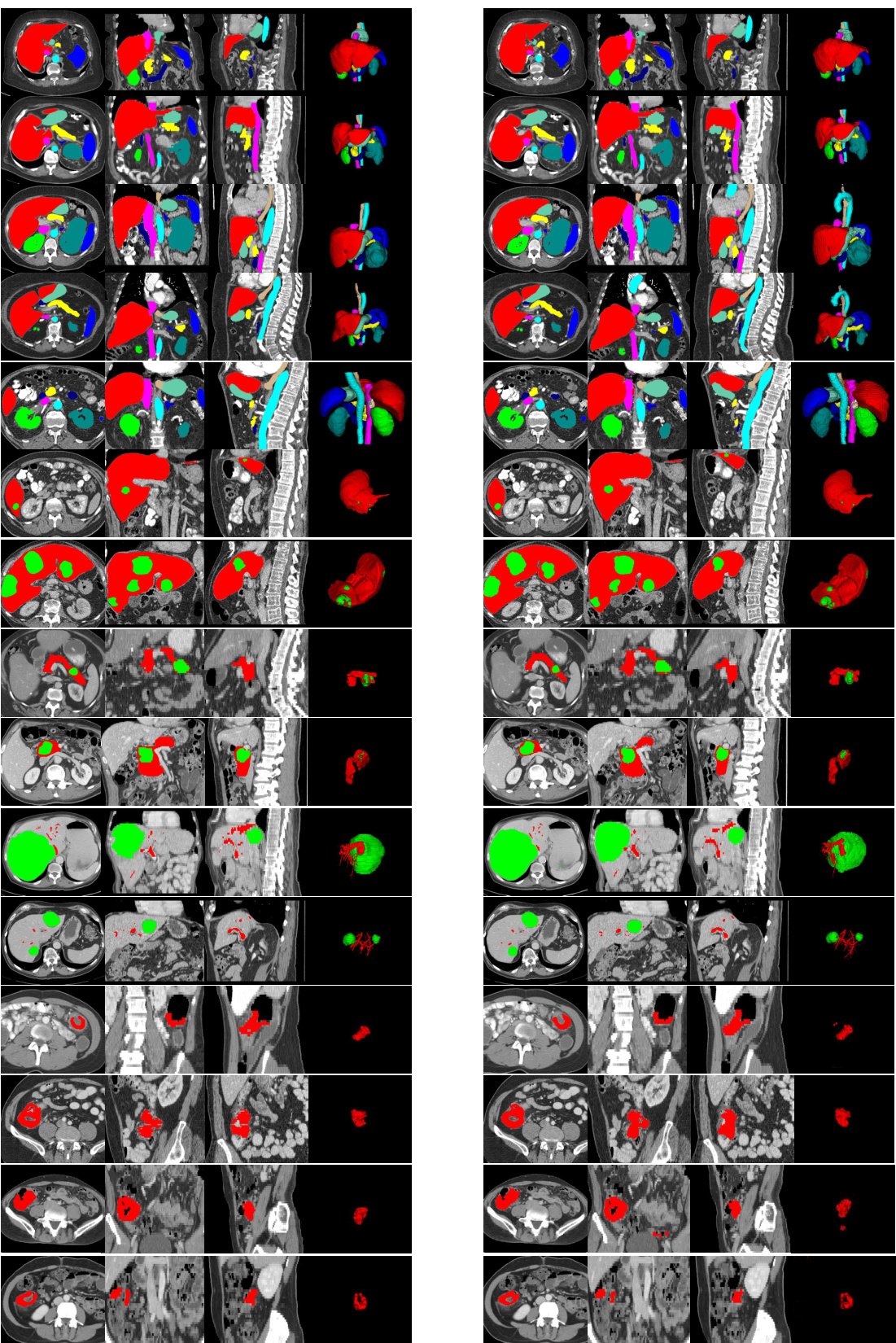
🔼 This figure presents a qualitative comparison of the proposed CAT model’s performance against other state-of-the-art prompting methods for organ and tumor segmentation. It showcases the results for five different cases: duodenum, liver tumors, pancreas tumors, colon tumors, and stage IV colon tumors. Each row shows the ground truth segmentation alongside the results from CAT and other methods (Universal and SegVol). The comparison highlights CAT’s superior ability to accurately segment various organs and tumors with complex shapes and varying appearances, particularly in challenging scenarios involving tumor invasion into nearby tissues.
read the caption
Figure 3: Qualitative visualizations of the proposed model and other prompting methods on organ/tumor segmentation. The segmentation results presented from rows one to five correspond, in order, to the duodenum, liver tumors, pancreas tumors, colon tumors, and colon tumors in Stage-IV.
More on tables

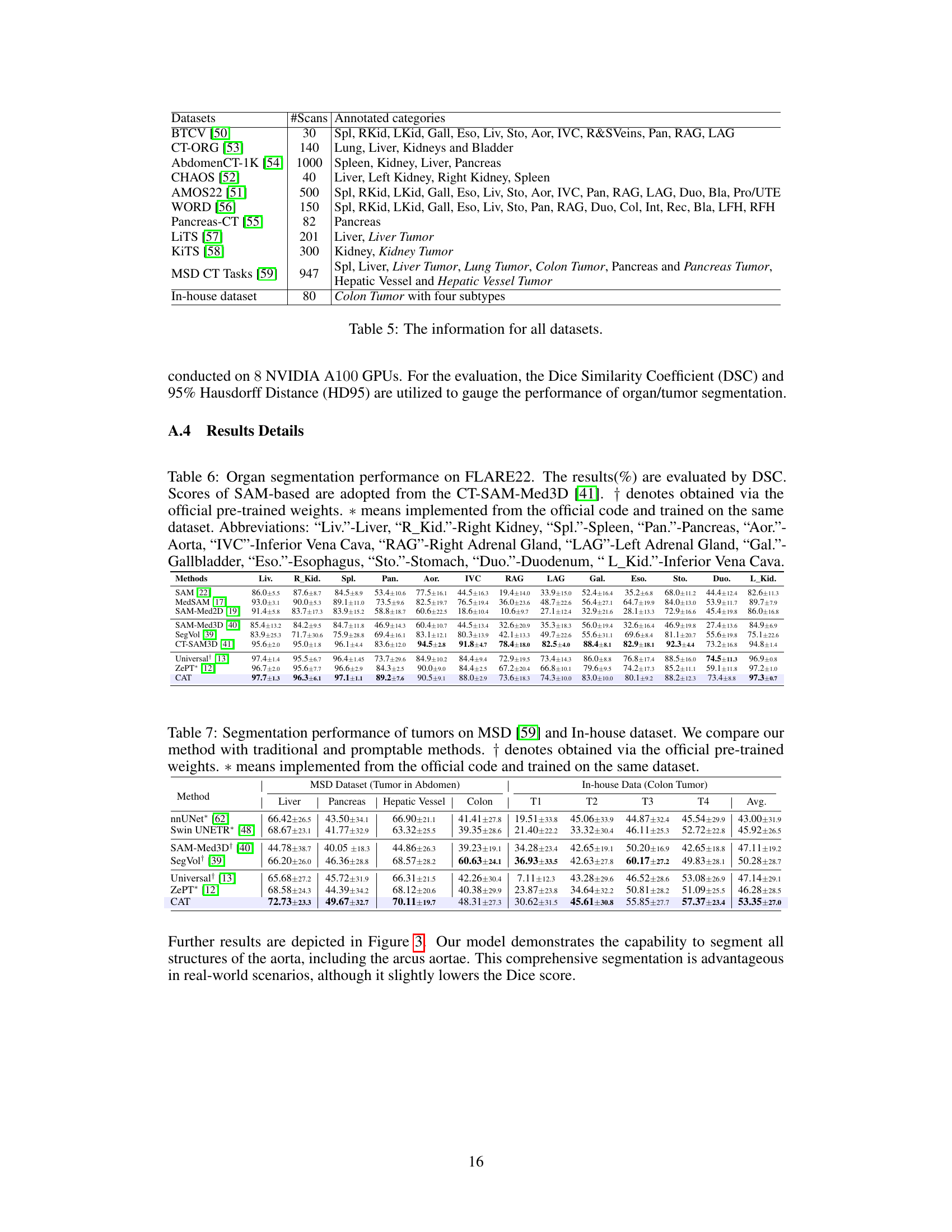
🔼 This table presents a comparison of the segmentation performance of different methods on two datasets: the MSD dataset (tumors in the abdomen) and an in-house dataset (colon tumors). The metrics used are Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD95). The table compares the performance of the proposed CAT model against several baselines, including nnUNet, Swin UNETR, SAM-Med3D+, SegVol, Universal, and ZePT. The results are broken down by tumor type and dataset, allowing for a detailed analysis of the model’s performance across various scenarios.
read the caption
Table 2: Segmentation performance (%) of tumors on MSD [59] and In-house dataset. We compare our method with traditional and promptable methods. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset.

🔼 This table presents the results of ablation studies conducted to evaluate the contribution of different components of the proposed model. It shows the performance of organ and tumor segmentation tasks under various configurations, including the presence or absence of anatomical and textual prompts, and the usage of hard or soft attention mechanisms. The goal is to demonstrate the individual and combined impact of these components on the overall performance of the model.
read the caption
Table 3: Ablation studies of two prompts and model designs on organ and tumor segmentation dataset.

🔼 This table presents a comparison of the organ segmentation performance of the proposed CAT model against several existing methods on the FLARE22 dataset. The performance is measured using the Dice Similarity Coefficient (DSC), a common metric for evaluating segmentation accuracy. The table includes results for 12 different abdominal organs and shows that CAT significantly outperforms existing methods in most cases.
read the caption
Table 1: Organ segmentation performance on FLARE22. The results (%) are evaluated by DSC. Scores of SAM-based are adopted from the CT-SAM-Med3D [41]. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset. Abbreviations: “Liv.”-Liver, “R_Kid.”-Right Kidney, “Spl.”-Spleen, “Pan.”-Pancreas, “Aor.”-Aorta, “IVC”-Inferior Vena Cava, “RAG”-Right Adrenal Gland, “LAG”-Left Adrenal Gland, “Gal.”-Gallbladder, “Eso.”-Esophagus, “Sto.”-Stomach, “Duo.”-Duodenum, “L_Kid.”-Inferior Vena Cava.
🔼 This table presents a comparison of the organ segmentation performance of the proposed CAT model against several other state-of-the-art methods on the FLARE22 dataset. The evaluation metric used is the Dice Similarity Coefficient (DSC), which measures the overlap between the predicted segmentation mask and the ground truth. The table includes results for 12 different abdominal organs, and the abbreviations used for each organ are explicitly defined in the caption.
read the caption
Table 1: Organ segmentation performance on FLARE22. The results (%) are evaluated by DSC. Scores of SAM-based are adopted from the CT-SAM-Med3D [41]. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset. Abbreviations: “Liv.”-Liver, “R_Kid.”-Right Kidney, “Spl.”-Spleen, “Pan.”-Pancreas, “Aor.”-Aorta, “IVC”-Inferior Vena Cava, “RAG”-Right Adrenal Gland, “LAG”-Left Adrenal Gland, “Gal.”-Gallbladder, “Eso.”-Esophagus, “Sto.”-Stomach, “Duo.”-Duodenum, “L_Kid.”-Inferior Vena Cava.

🔼 This table presents the performance comparison of different models on the FLARE22 dataset for organ segmentation. The performance metric used is Dice Similarity Coefficient (DSC). The results show CAT’s superior performance compared to other state-of-the-art methods in most organs.
read the caption
Table 1: Organ segmentation performance on FLARE22. The results (%) are evaluated by DSC. Scores of SAM-based are adopted from the CT-SAM-Med3D [41]. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset. Abbreviations: “Liv.”-Liver, “R_Kid.”-Right Kidney, “Spl.”-Spleen, “Pan.”-Pancreas, “Aor.”-Aorta, “IVC”-Inferior Vena Cava, “RAG”-Right Adrenal Gland, “LAG”-Left Adrenal Gland, “Gal.”-Gallbladder, “Eso.”-Esophagus, “Sto.”-Stomach, “Duo.”-Duodenum, “L_Kid.”-Inferior Vena Cava.

🔼 This table presents the segmentation performance results for various tumor types using different methods. It compares the proposed CAT method with several baselines, including traditional methods (nnUNet, Swin UNETR) and other prompt-based methods (SAM-Med3D, SegVol, Universal, ZePT). The performance is evaluated using the Dice Similarity Coefficient (DSC) metric on two datasets: the MSD dataset (tumors in the abdomen) and an in-house dataset (colon tumors in different stages). The table shows the DSC scores for each method on various tumor types (liver, pancreas, hepatic vessel, colon) for the MSD dataset, as well as for different colon tumor stages (T1-T4) in the in-house dataset. The average DSC across all tumors is also presented for each method.
read the caption
Table 2: Segmentation performance (%) of tumors on MSD [59] and In-house dataset. We compare our method with traditional and promptable methods. † denotes obtained via the official pre-trained weights. * means implemented from the official code and trained on the same dataset.
Full paper#