↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Current 3D medical image segmentation heavily relies on isotropic 3D convolutions for feature extraction. However, this approach is inefficient due to varying information density across different axes in medical images like CT and MRI. This disparity leads to underutilization of axial-slice plane features compared to time-axial features.
To tackle this issue, the researchers proposed a novel U-shaped Connection (uC) that leverages simplified 2D U-Nets to enhance axial-slice plane feature extraction. They integrated uC into a 3D U-Net backbone to create uC 3DU-Net. Extensive experiments on five public datasets demonstrated that uC 3DU-Net outperforms existing state-of-the-art methods, achieving superior performance with significantly fewer parameters and lower computational cost. The study highlights the potential of combining 2D and 3D convolutions to address limitations of traditional volumetric segmentation methods.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image segmentation as it introduces a novel method (uC 3DU-Net) that significantly improves the accuracy and efficiency of 3D medical image segmentation. Its focus on addressing the inherent limitations of traditional 3D convolution methods, particularly for anisotropic data, opens up new avenues for future research and development in this critical area of medical image analysis. The method’s superior parameter-to-performance ratio also makes it highly practical for real-world applications.
Visual Insights#

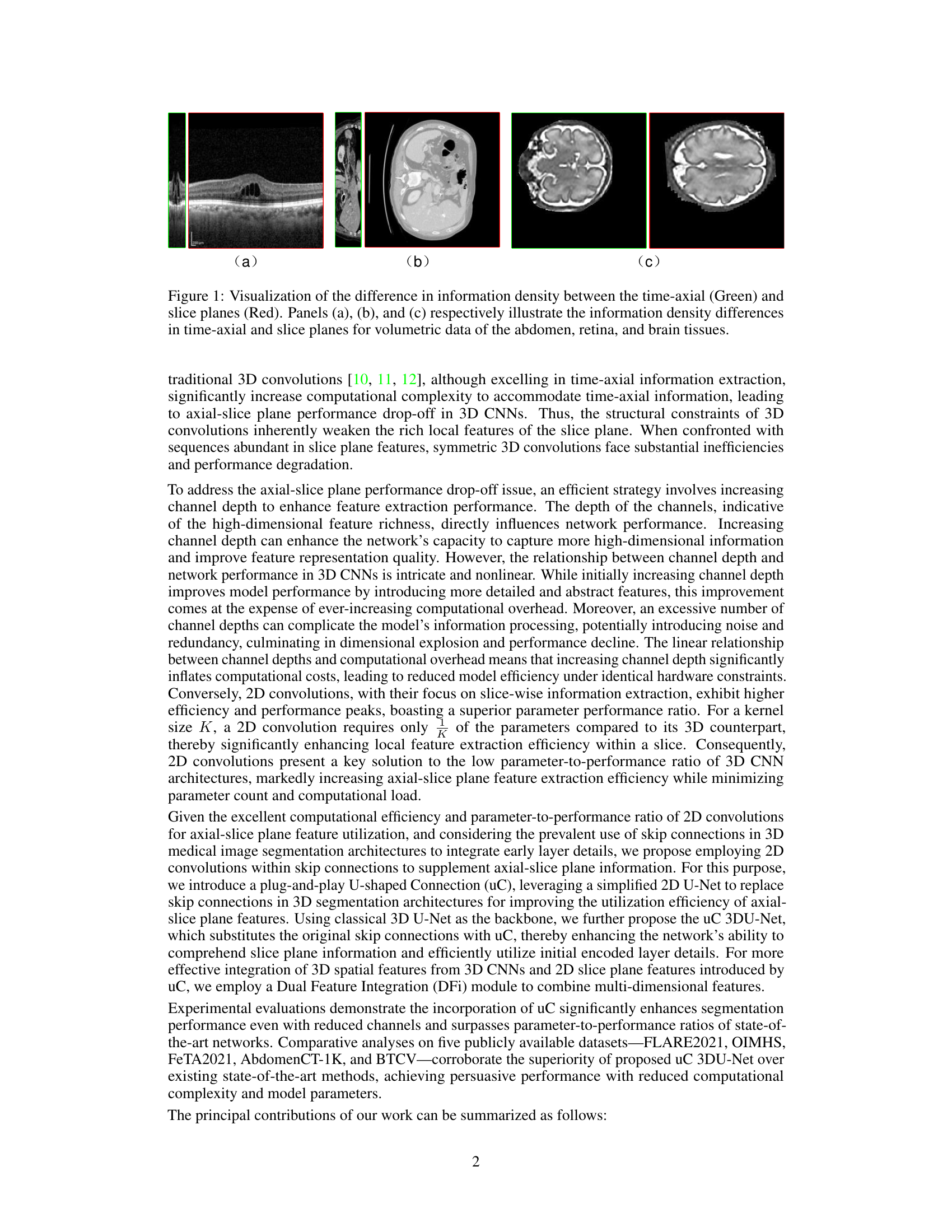
This figure visually demonstrates the varying information density across the three orthogonal axes in 3D medical imaging modalities. It highlights that the information density is significantly higher in the axial slice planes compared to the time-axial direction, a factor often overlooked in traditional 3D CNNs for medical image segmentation. The images showcase examples of abdomen, retina, and brain tissues, each highlighting the disparity in information density.
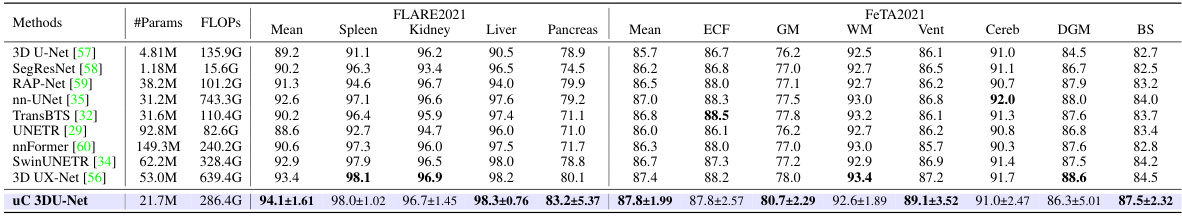
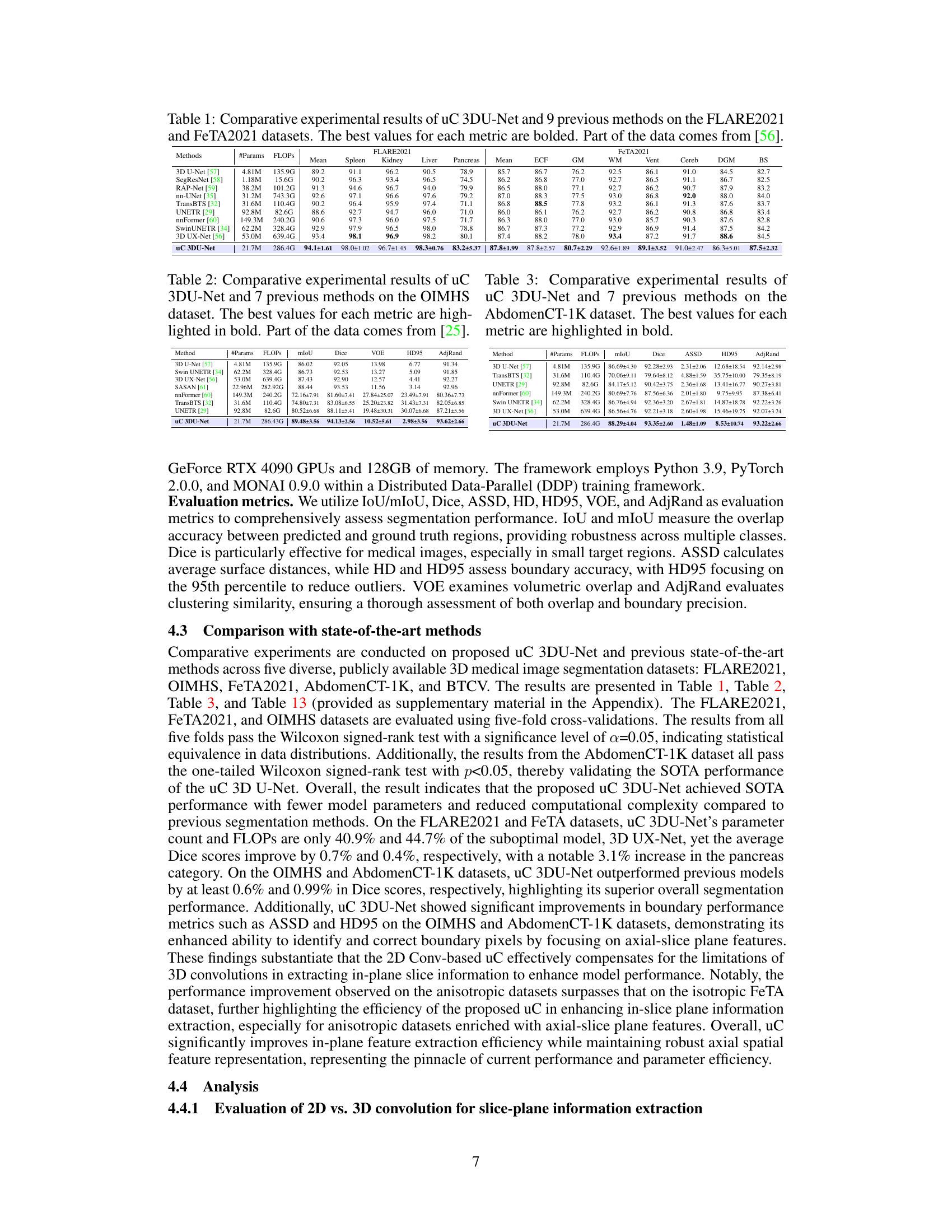
This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two benchmark datasets (FLARE2021 and FeTA2021) for 3D medical image segmentation. The performance is measured using several metrics such as mean Intersection over Union (mIoU), Dice score, and others. The best performance for each metric on each dataset is highlighted in bold. A key aspect to note is the comparison of the number of parameters and FLOPs (floating point operations) required by each model, showing the computational efficiency of the proposed method.
In-depth insights#
2D-Unet in 3D#
The concept of “2D-Unet in 3D” introduces an innovative approach to 3D medical image segmentation by leveraging the strengths of 2D U-Nets within a 3D convolutional neural network (CNN) architecture. This hybrid approach directly addresses the inherent anisotropy of medical images, where information density varies significantly across different spatial axes. Traditional 3D CNNs often struggle with this, leading to inefficiencies and suboptimal performance. By integrating 2D U-Nets, particularly in skip connections, the method aims to enhance the extraction of crucial features from axial slices, where information is usually richer, while simultaneously retaining the contextual awareness provided by 3D convolutions. The proposed method, thus, seeks to achieve a balance between efficiency and accuracy, potentially reducing computational complexity and improving the overall parameter-to-performance ratio. The effectiveness of the approach rests on the complementary nature of 2D and 3D convolutions, capitalizing on the strengths of each to overcome the limitations of relying solely on 3D operations for volumetric segmentation.
Axial-Slice Focus#
The concept of “Axial-Slice Focus” in 3D medical image segmentation highlights the anisotropic nature of medical imaging data, where information density varies significantly across different axes. Traditional 3D convolutional methods struggle to efficiently leverage the rich detail present in axial slices because of their inherent isotropic processing. An axial-slice focus approach emphasizes techniques that prioritize the extraction and utilization of features from axial slices. This may involve using 2D convolutions within a 3D framework, specialized attention mechanisms, or other strategies to enhance the representation of axial-slice information. Effective axial-slice feature extraction is crucial for improving the accuracy and efficiency of 3D segmentation, especially in tasks involving fine-grained anatomical structures, which are often better captured in high-resolution axial views.
uC 3DU-Net#
The proposed architecture, “uC 3DU-Net,” innovatively integrates 2D U-Net skip connections into a 3D U-Net backbone. This addresses the inherent limitation of traditional 3D CNNs in medical image segmentation, where the varying information density across three orthogonal axes leads to inefficiency. The 2D U-Net components within uC 3DU-Net enhance axial-slice plane feature extraction, improving the utilization of often underutilized information in this plane. This is done while retaining the volumetric context afforded by the 3D convolutional layers, which are crucial for understanding the full 3D structure. A key innovation is the Dual Feature Integration (DFi) module which effectively merges the 2D and 3D features for optimal segmentation. The result is a model that demonstrates superior performance compared to other state-of-the-art methods across various datasets, with the added benefit of reduced computational complexity and fewer parameters. This highlights the potential of combining 2D and 3D convolutional approaches for more efficient 3D medical image analysis.
DFi Module#
The Dual Feature Integration (DFi) module is a crucial component, designed to effectively fuse features extracted from two distinct pathways: the 3D convolutional backbone and the 2D U-Net based U-shaped connections. Its primary function is to address the inherent anisotropy of 3D medical images, where axial slices often possess richer detail than time-axial planes. The DFi module cleverly combines these features, leveraging a concatenation strategy followed by 1x1 convolutions and attention mechanisms to learn the relative importance of features from each pathway. This approach goes beyond simpler concatenation or addition methods, dynamically weighting the contributions from both 2D and 3D sources to optimize the feature representation for improved segmentation accuracy. By integrating these different levels of information, DFi enhances the model’s ability to capture fine-grained detail while maintaining a broader contextual understanding of the 3D structure. This sophisticated integration likely contributes significantly to the uC 3DU-Net’s superior performance, achieving a balance between preserving high-resolution details and robust spatial context.
Future Work#
The research paper’s ‘Future Work’ section would ideally delve into several promising avenues. Extending the U-shaped Connection (uC) to other 3D medical image segmentation architectures beyond the 3D U-Net backbone is crucial to establish its generalizability and effectiveness. Investigating the optimal configuration of uC, such as the number and placement of 2D U-Nets within the skip connections, could further enhance performance and efficiency. A thorough exploration of the DFi module’s parameter space is needed to refine its ability to seamlessly integrate 2D and 3D features. Furthermore, the study could explore different 2D architectures beyond the simplified 2D U-Net used in uC to discover if other architectures yield better results. Finally, a rigorous analysis of the proposed method’s sensitivity to various noise levels, imaging artifacts, and data variations could help establish its robustness for real-world clinical applications. Exploring the potential of uC in combination with other advanced techniques, such as attention mechanisms and transformers, is another intriguing direction for future research. This will reveal the true potential of the uC for advancing medical image analysis.
More visual insights#
More on figures
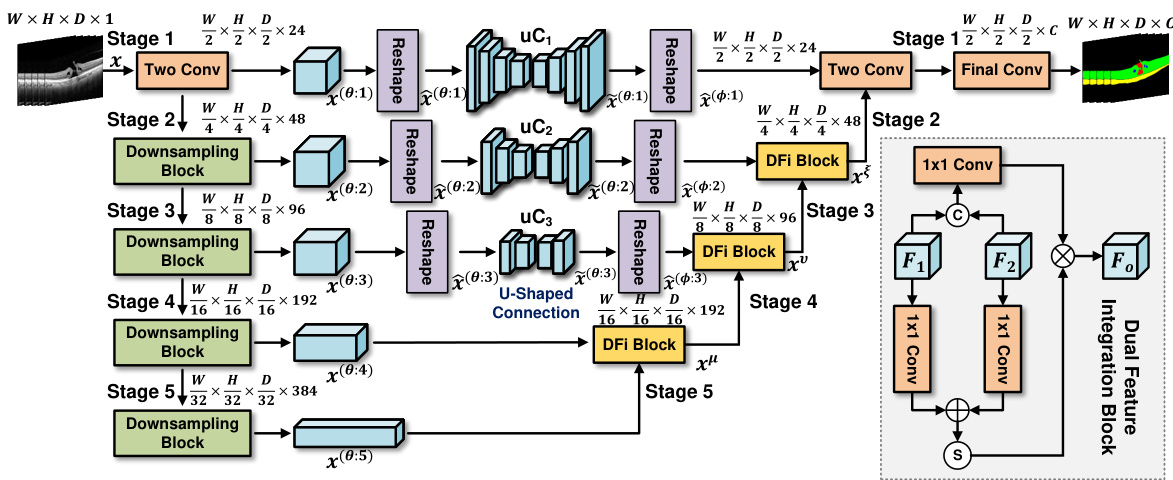
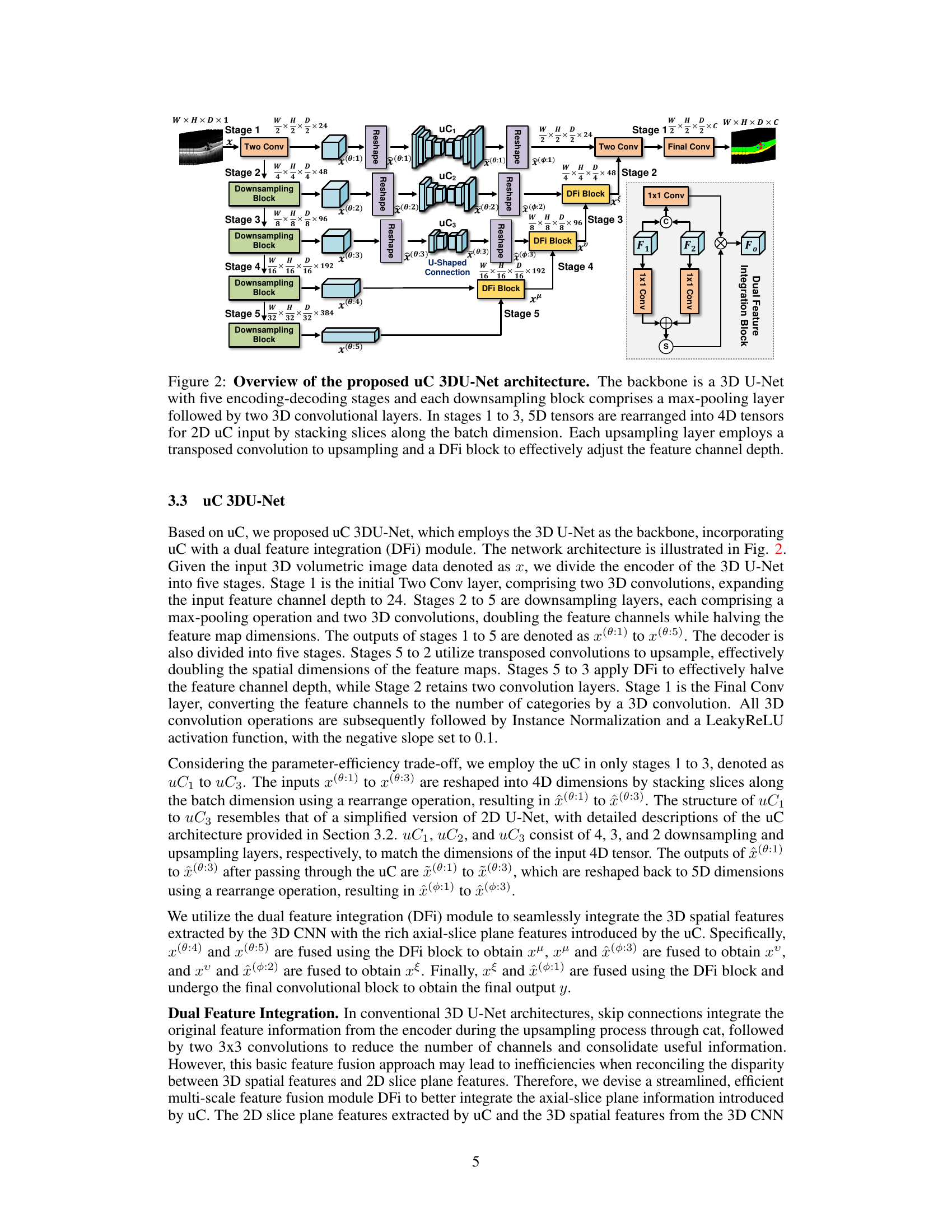
This figure illustrates the architecture of the proposed uC 3DU-Net. It is a 3D U-Net based model. The encoder consists of five stages, each with a downsampling block (max-pooling + two 3D convolutions). Stages 1-3 use a modified skip connection called ‘U-shaped Connection’ (uC) which leverages a simplified 2D U-Net to extract axial-slice plane features. The 5D tensors are reshaped to 4D for processing by uC. The decoder also has five stages with upsampling layers (transposed convolution) and Dual Feature Integration (DFi) blocks to combine 2D and 3D features, effectively adjusting the feature channel depth. The final layer converts to the required number of output classes.
This figure shows the architecture of the proposed uC 3DU-Net. It uses a 3D U-Net as its base, but modifies the skip connections. The skip connections are replaced with the proposed ‘U-shaped Connections’ (uC) which are simplified 2D U-Nets. The figure highlights the five encoding and decoding stages, showing how the 5D tensors are reshaped into 4D tensors for processing by the 2D U-Nets. Finally, a ‘Dual Feature Integration’ (DFi) block is shown which is used to combine the features extracted by the 3D and 2D convolutional layers.

This figure illustrates the architecture of the proposed uC 3DU-Net, which uses a 3D U-Net as its backbone. The encoder consists of five downsampling stages, each with a max-pooling layer and two 3D convolutional layers. The decoder uses five upsampling stages; in stages 1-3, 2D U-Nets (uC) are used to process the axial slice information; a Dual Feature Integration (DFi) module merges the 2D and 3D features to enhance feature extraction; finally, a 1x1 convolutional layer outputs the segmentation results.
This figure shows the architecture of the proposed uC 3DU-Net. It uses a 3D U-Net as the backbone and integrates the U-shaped Connection (uC) module in stages 1 to 3. The 5D input tensors are reshaped to 4D before being fed into the 2D uC modules, and a Dual Feature Integration (DFi) module is used in the upsampling layers to combine 2D and 3D features.
This figure illustrates the architecture of the proposed uC 3DU-Net model. It uses a 3D U-Net as its base, but modifies the skip connections with a simplified 2D U-Net (called uC). The figure highlights the five encoding and decoding stages. In the encoding stages, max-pooling and 3D convolutions reduce spatial dimensions and increase channel depth. In stages 1-3, the input 5D tensors are converted to 4D tensors to allow for processing by the 2D uC. The decoding stages use transposed convolutions to upsample, and DFi blocks to effectively adjust the channel depth. The model combines 2D and 3D features to improve segmentation performance.

This figure provides a detailed illustration of the uC 3DU-Net architecture, which is a modified 3D U-Net that integrates a simplified 2D U-Net (the U-shaped connection, or uC) into its skip connections. The figure shows the five encoding and decoding stages. The key innovation is the use of the uC module in stages 1-3, where the input 5D tensors are reshaped to 4D tensors for processing by the 2D U-Net. The DFi block is also highlighted, showing how features from the 2D and 3D parts of the network are combined.
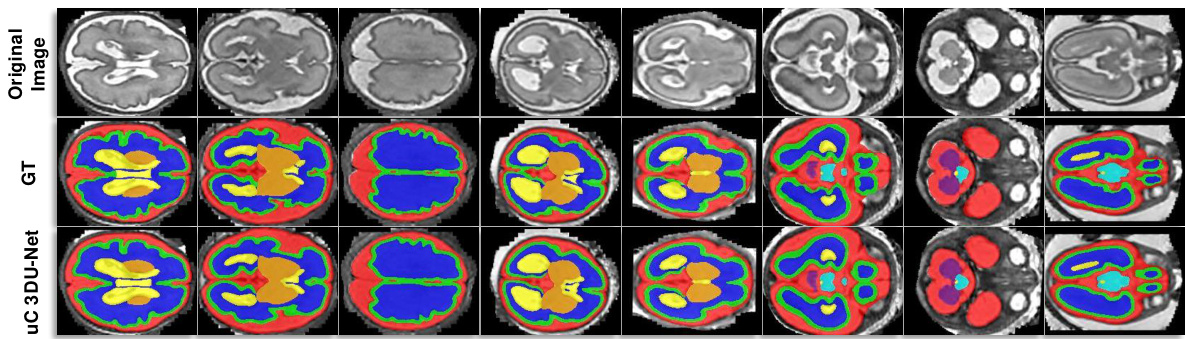
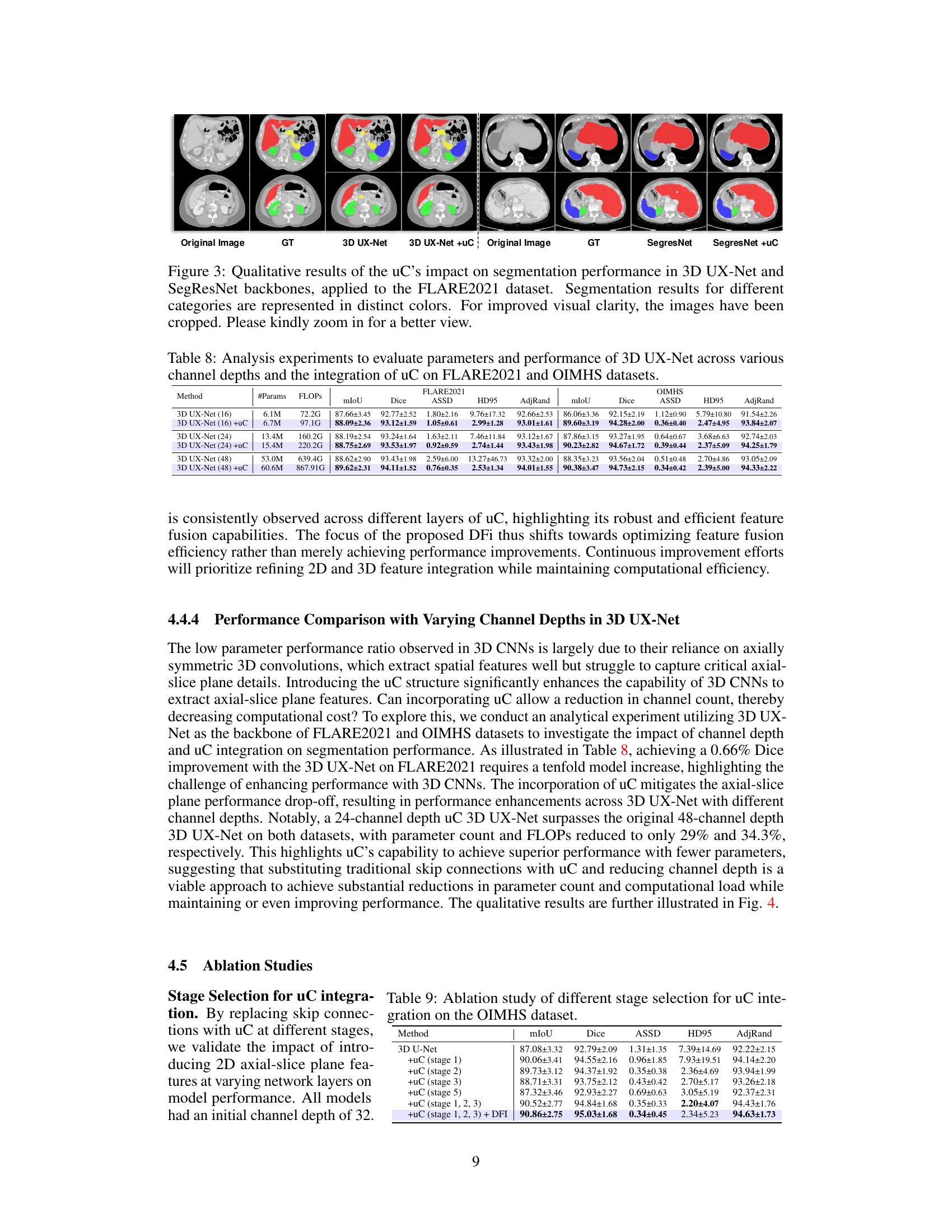
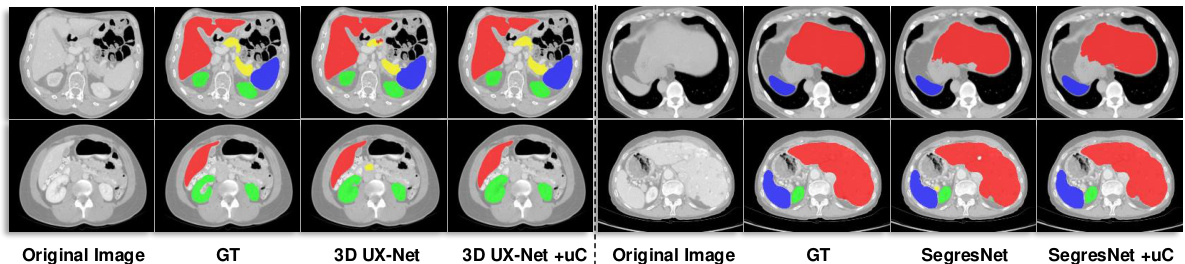
This figure shows the qualitative comparison of segmentation results for 3D UX-Net and SegResNet with and without the proposed U-shaped Connection (uC) method. The results are visually presented for different organ classes on the FLARE2021 dataset. The uC method improves the segmentation results by enhancing the extraction of axial-slice plane features. The images have been cropped for improved visual clarity.

This figure illustrates the architecture of the proposed uC 3DU-Net, a modified 3D U-Net that incorporates 2D U-Nets within its skip connections. The figure highlights the five encoding and decoding stages, showing how the 3D convolutional layers are combined with 2D U-Net layers to improve the extraction of features from axial slices. The use of a Dual Feature Integration (DFi) module is also highlighted to effectively merge the 2D and 3D features.

This figure provides a detailed illustration of the uC 3DU-Net architecture. It shows the 3D U-Net backbone with its five encoding-decoding stages. Each downsampling stage uses max-pooling followed by two 3D convolutional layers. Stages 1-3 have a unique modification where 5D tensors (W, H, D, C) are reshaped to 4D tensors to be processed by the 2D U-net skip connection (uC). Each upsampling stage employs a transposed convolution and a dual feature integration (DFi) block to integrate features from the 2D and 3D pathways and adjust the feature channel depth. The figure highlights the key components of the architecture and how they interact to achieve efficient axial-slice plane feature utilization.

This figure illustrates the architecture of the proposed uC 3DU-Net, a 3D U-Net based model that integrates 2D U-Net skip connections (uC) to improve axial-slice plane feature utilization. The figure details the encoder and decoder stages, highlighting the use of 3D convolutions, max-pooling, transposed convolutions, and the dual feature integration (DFi) module. The schematic shows how the 5D input tensors are transformed into 4D tensors for processing by the 2D U-Nets within the skip connections. It also shows how the DFi module integrates 2D and 3D features at each upsampling layer.
More on tables
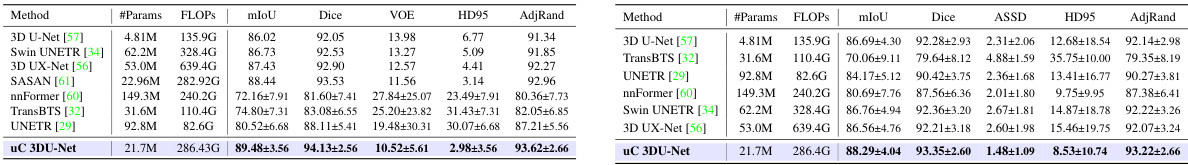
This table presents a comparison of the proposed uC 3DU-Net against nine other methods for medical image segmentation on two datasets (FLARE2021 and FeTA2021). It shows the number of parameters and FLOPs (floating point operations) for each method, along with several key performance metrics (Mean, Spleen, Kidney, Liver, Pancreas, ECF, GM, WM, Vent, Cereb, DGM, BS) to evaluate segmentation accuracy. The best performance for each metric is highlighted in bold. Part of the data used in this comparison is sourced from reference [56].

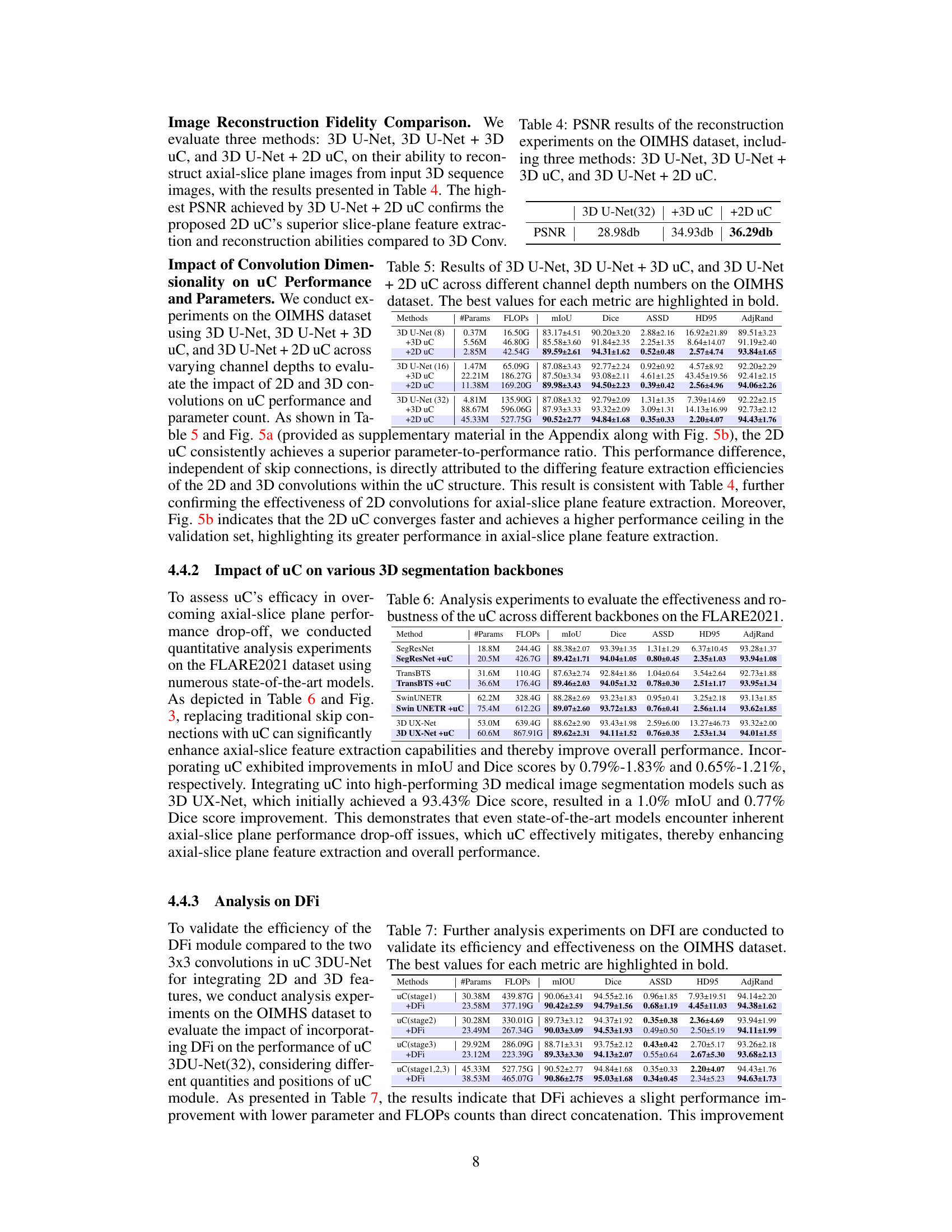
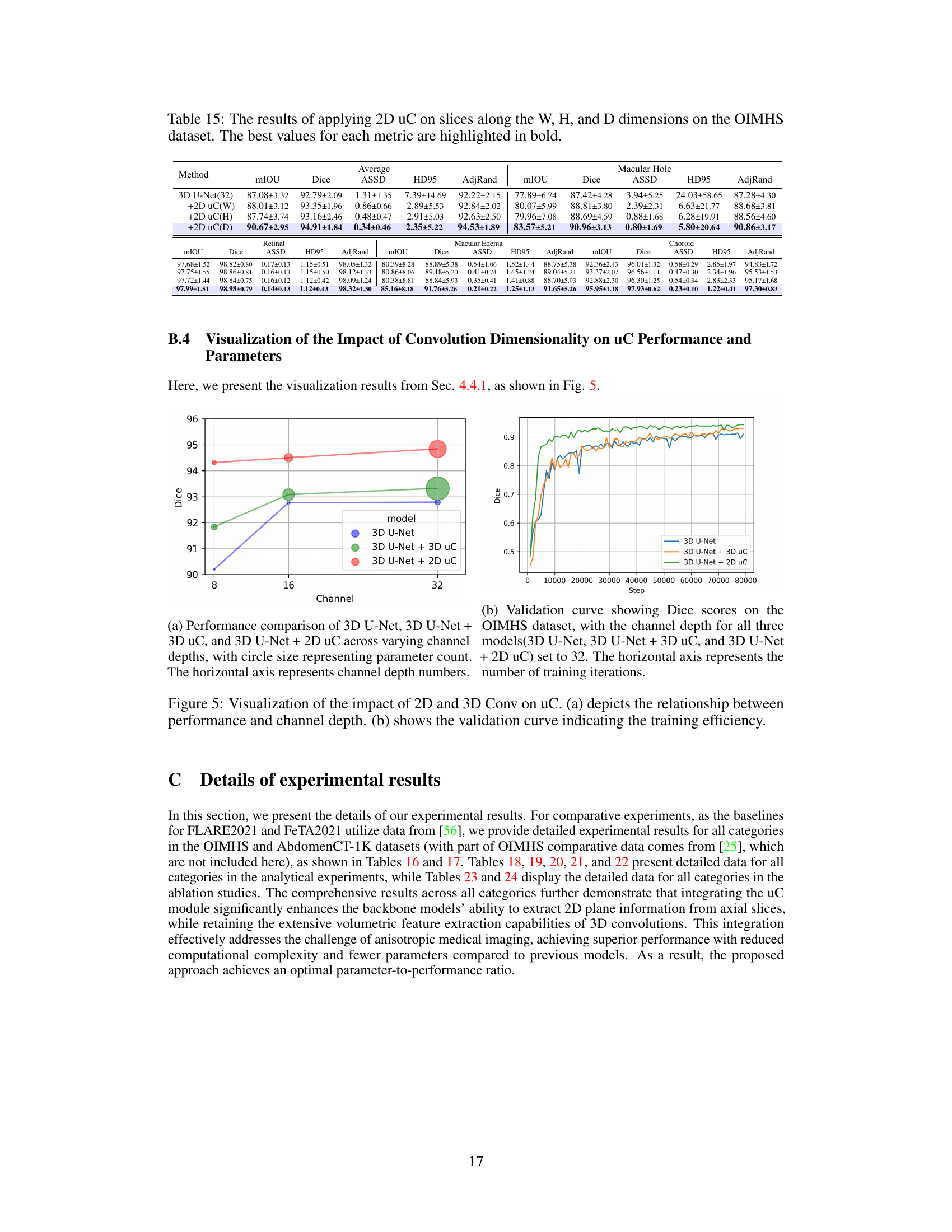
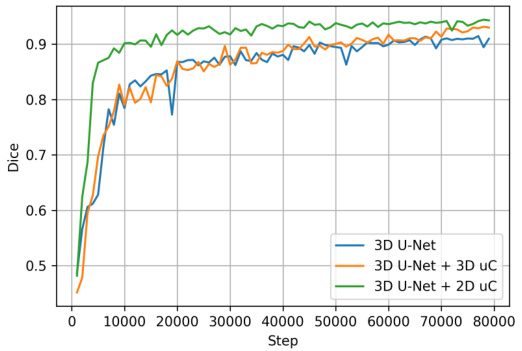
This table presents the Peak Signal-to-Noise Ratio (PSNR) results for image reconstruction experiments conducted on the OIMHS dataset. Three different methods were compared: the baseline 3D U-Net, 3D U-Net with 3D U-shaped connections (+3D uC), and 3D U-Net with 2D U-shaped connections (+2D uC). The PSNR values reflect the quality of the reconstructed axial-slice plane images, with higher values indicating better reconstruction quality. The table demonstrates the improvement in reconstruction fidelity achieved by incorporating the U-shaped connections, particularly the 2D variant.
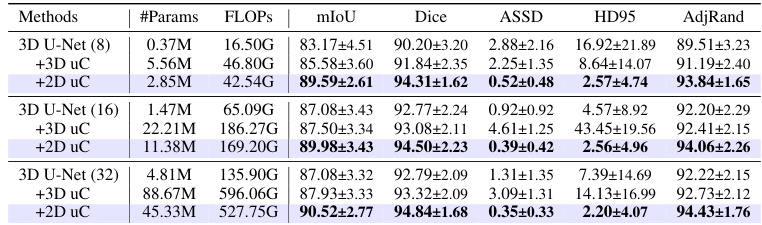
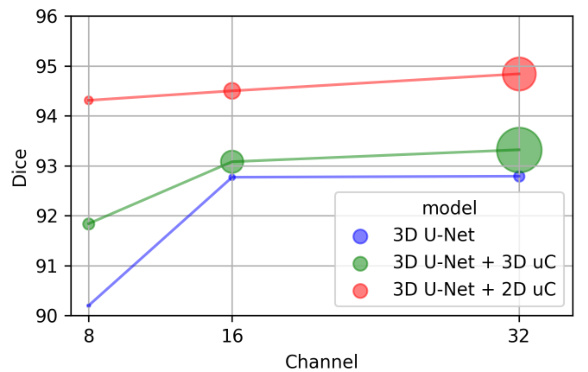
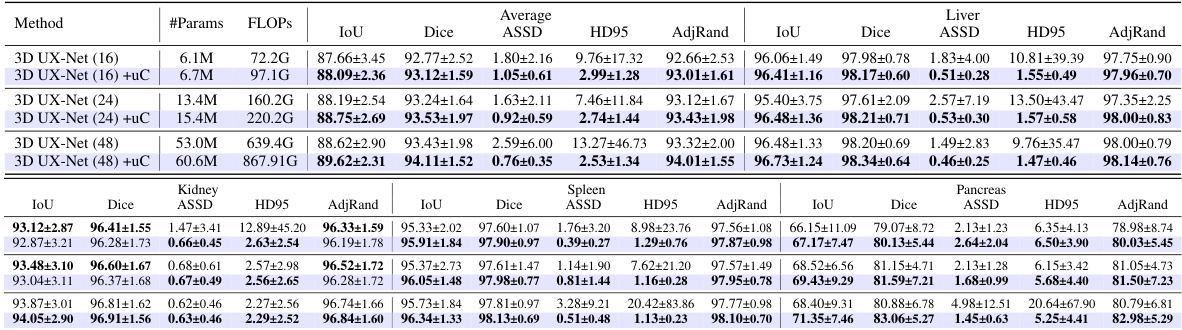
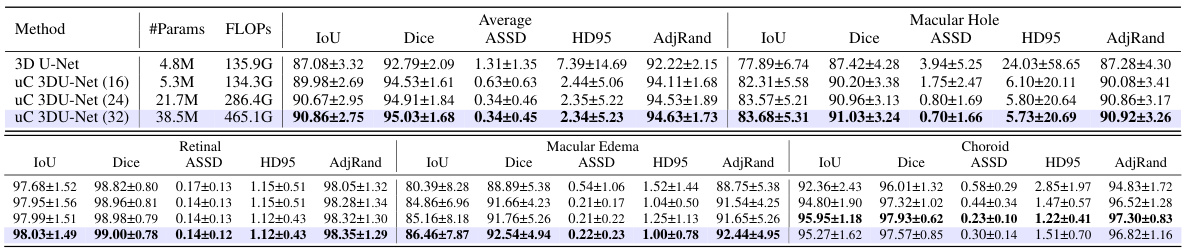
This table presents a comparison of the performance of three different models (3D U-Net, 3D U-Net + 3D uC, and 3D U-Net + 2D uC) on the OIMHS dataset across varying channel depths. The metrics used for comparison include mIoU (mean Intersection over Union), Dice, ASSD (Average Symmetric Surface Distance), HD95 (95th percentile Hausdorff Distance), and AdjRand (Adjusted Rand Index). The table shows how the performance and parameter counts of each model change as the channel depth increases. This allows for an analysis of how the dimensionality of the convolution operation (2D vs. 3D) impacts efficiency and performance when used in conjunction with the uC.
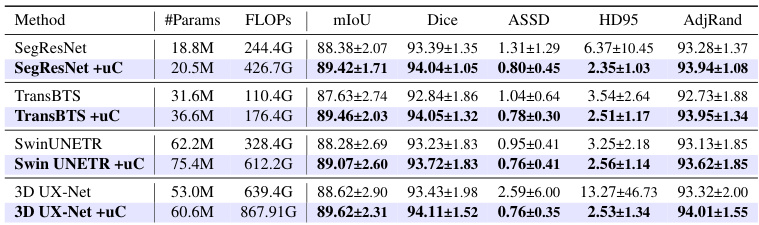
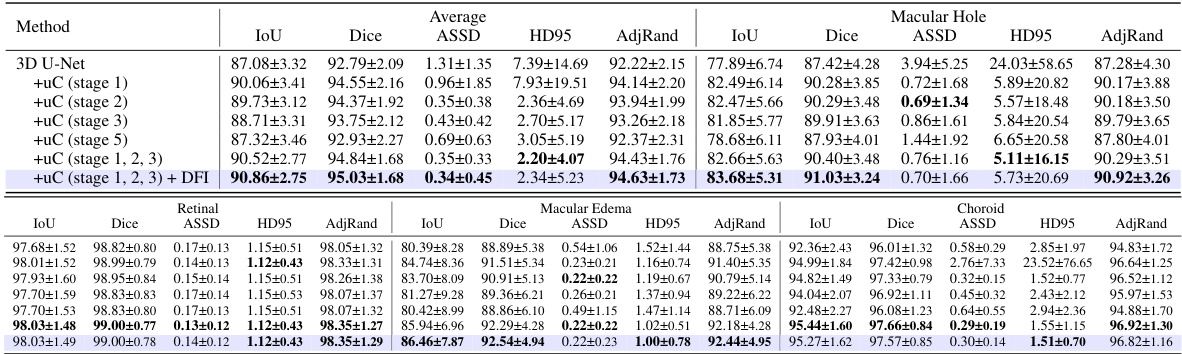
This table presents a comparison of various state-of-the-art 3D medical image segmentation models’ performance on the FLARE2021 dataset, both with and without the integration of the proposed U-shaped Connection (uC) module. It demonstrates the improvement in performance metrics (mIoU, Dice, ASSD, HD95, AdjRand) achieved by incorporating the uC into different backbones. The table highlights that uC consistently improves performance across various architectures, indicating its robustness and generalizability.
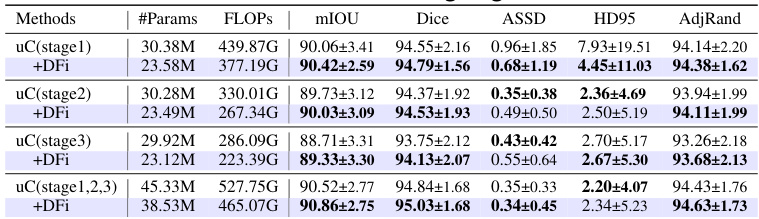
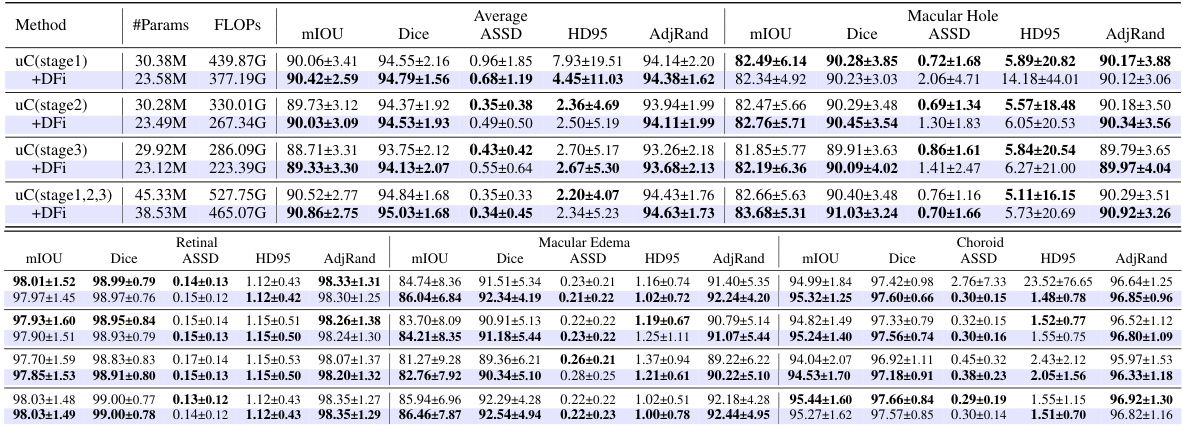
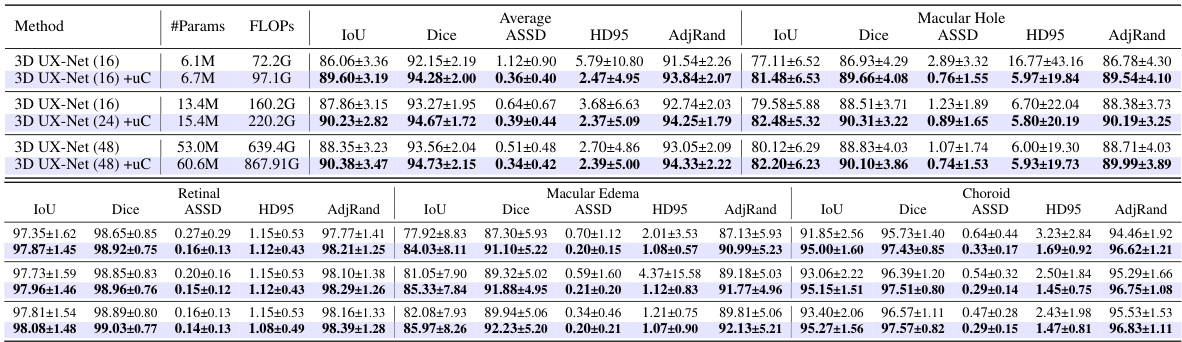
This table presents a detailed breakdown of the Dual Feature Integration (DFi) module’s performance on the OIMHS dataset. It compares different configurations of DFI, including applying it to different stages of the U-Net architecture, against a baseline (no DFI). The results are presented for several evaluation metrics such as mIoU, Dice, ASSD, HD95, and AdjRand, for each of the four classes of the OIMHS dataset (Average, Macular Hole, Macular Edema, Choroid, Retinal). The best values for each metric are highlighted to show the optimal DFI configuration.

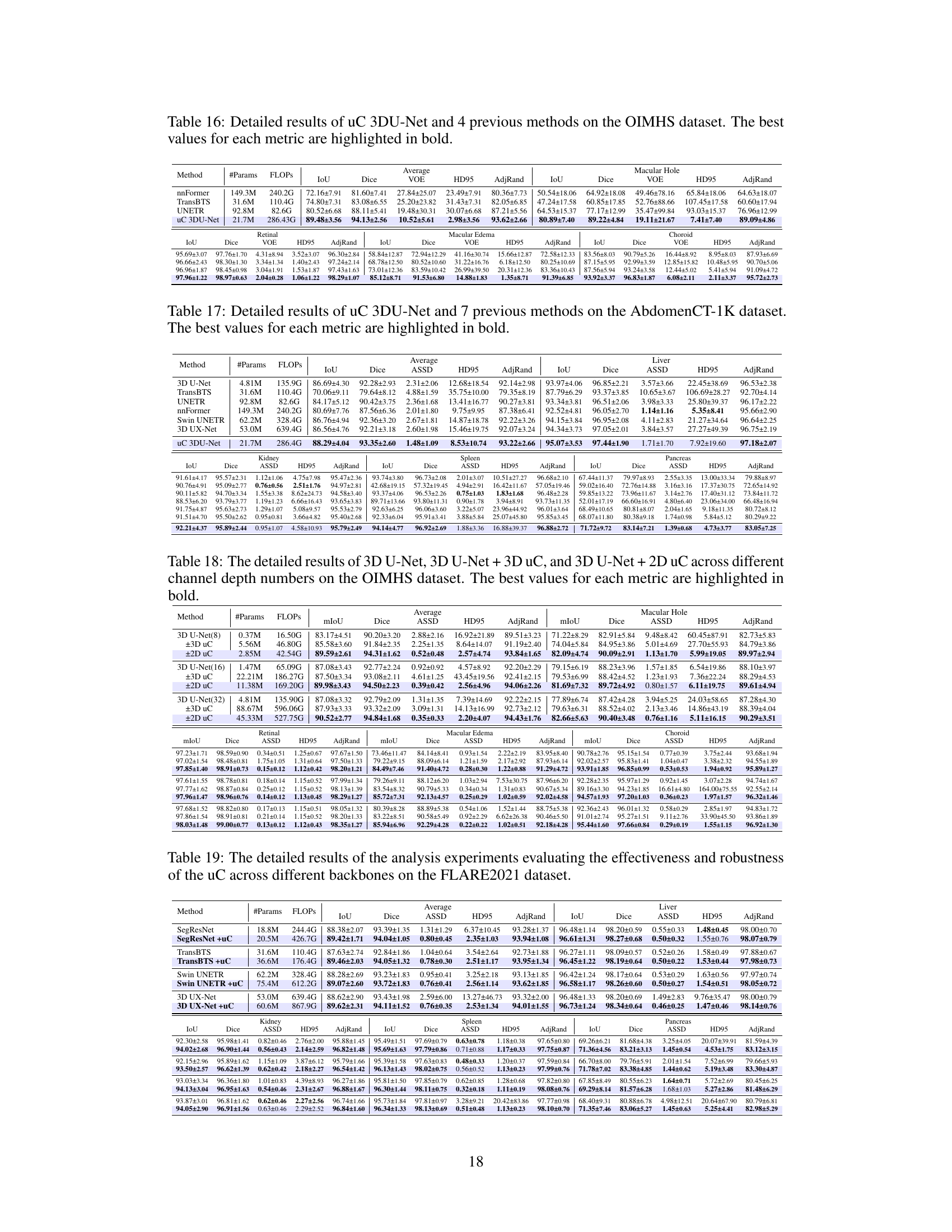
This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other state-of-the-art methods on two benchmark datasets for medical image segmentation: FLARE2021 and FeTA2021. The metrics used to evaluate performance include multiple measures relevant to the accuracy and efficiency of segmentation. The best performance is highlighted in bold for each metric, allowing for easy identification of the top performing models and comparison across different evaluation metrics. Part of the data is taken from reference [56].
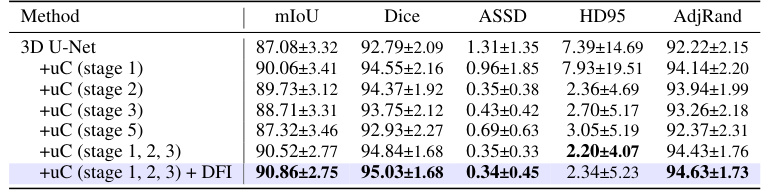
This table presents the ablation study results on the OIMHS dataset, showing the impact of integrating the U-shaped Connection (uC) at different stages of the 3D U-Net architecture. It compares the performance metrics (mIoU, Dice, ASSD, HD95, AdjRand) across various configurations, demonstrating the effectiveness of the uC module in enhancing the model’s ability to capture axial-slice plane features. The results show how the choice of uC integration stage significantly influences the overall performance. The row with uC integration in stages 1, 2, and 3, combined with the Dual Feature Integration (DFi) module, achieves the best results.
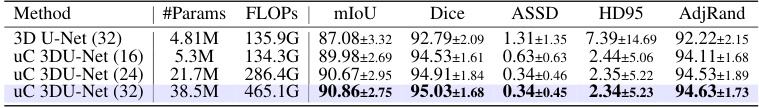
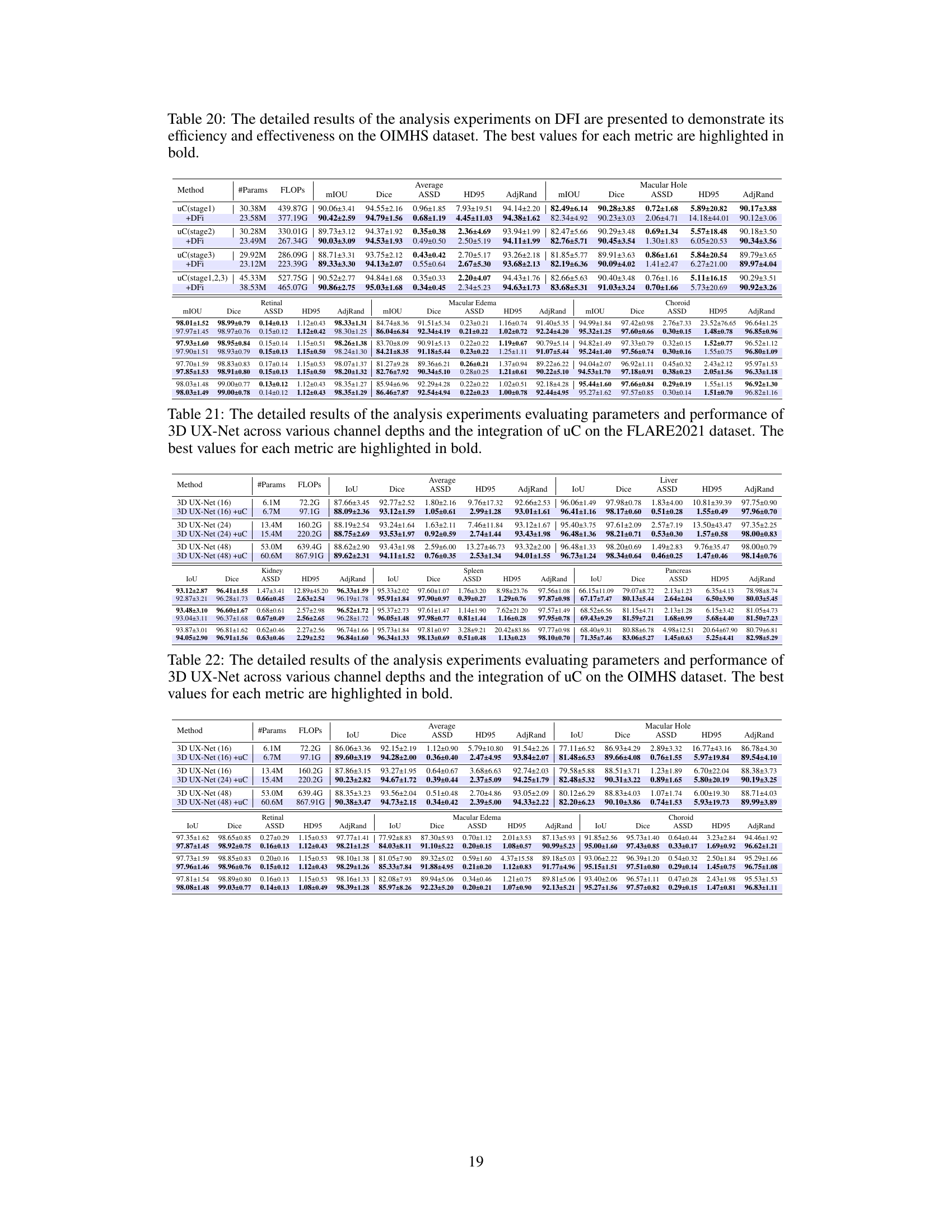
This table presents a comparison of the performance of three different models (3D U-Net, 3D U-Net + 3D uC, and 3D U-Net + 2D uC) across various channel depths on the OIMHS dataset. The table shows the number of parameters, FLOPs, mIoU, Dice, ASSD, HD95, and AdjRand for each model and channel depth. The best results for each metric are highlighted in bold, allowing for easy comparison of model performance and efficiency.
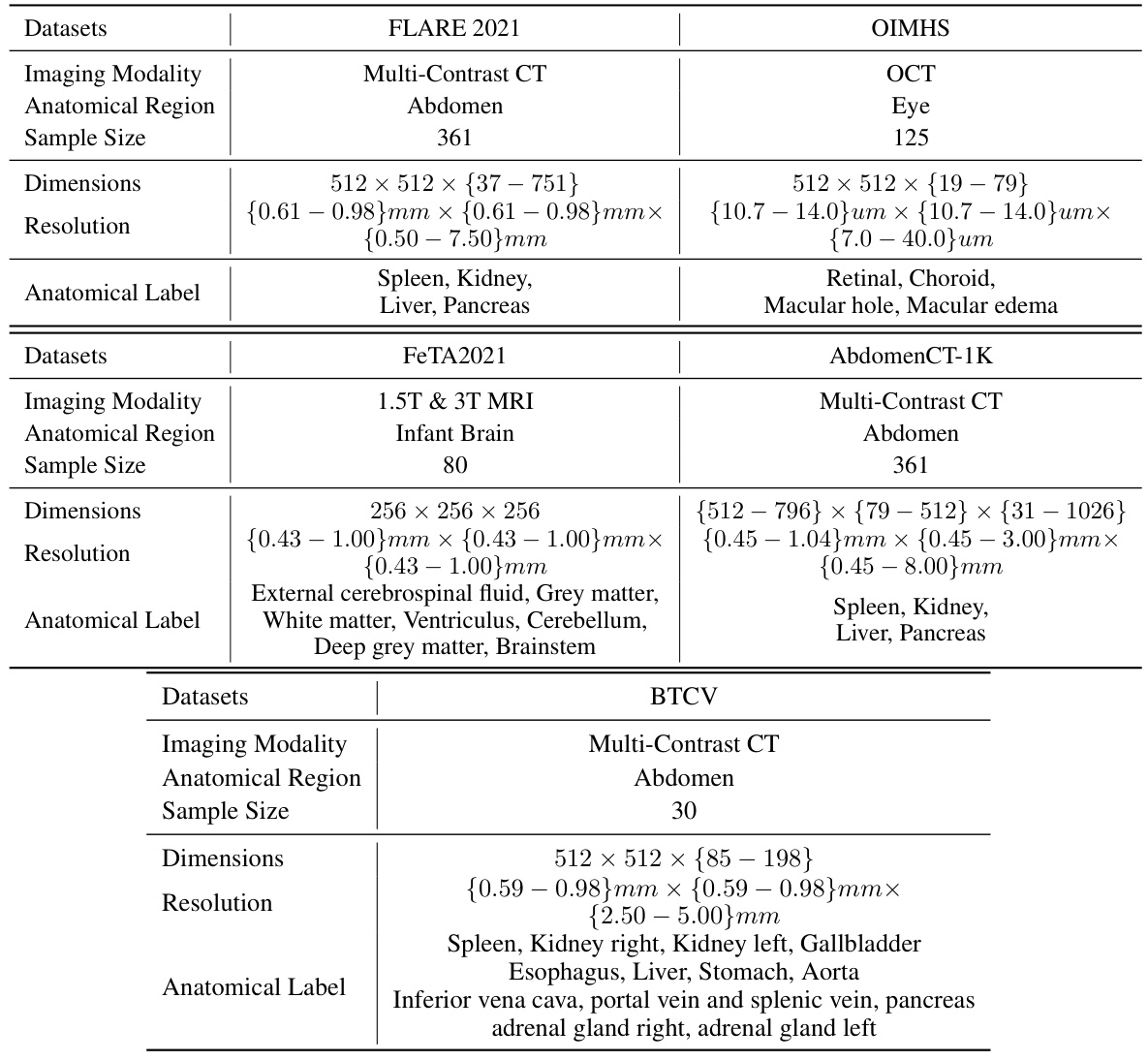
This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two datasets: FLARE2021 and FeTA2021. The comparison includes the number of parameters and FLOPs (floating point operations), as well as several evaluation metrics for each organ or tissue in each dataset. The best-performing method for each metric is highlighted in bold.

This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two datasets (FLARE2021 and FeTA2021). The metrics used to evaluate the models include the number of parameters (#Params), floating point operations (FLOPs), mean Intersection over Union (mIoU), Dice coefficient, and the Hausdorff distance at 95th percentile (HD95). The best performing model for each metric on each dataset is highlighted in bold. Note that some of the data included in the table is from a previous publication.
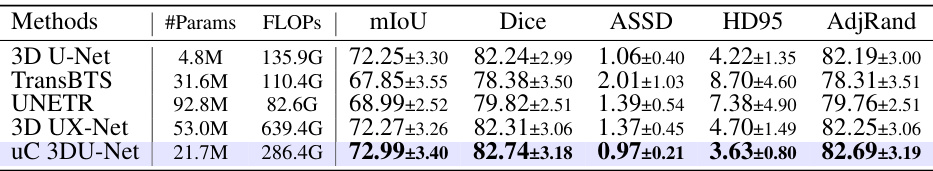
This table presents a comparison of the uC 3DU-Net’s performance against four other methods on the BTCV dataset for medical image segmentation. The metrics used for comparison include mIoU (mean Intersection over Union), Dice coefficient, Average Surface Distance (ASSD), Hausdorff Distance (HD95), and Adjusted Rand Index (AdjRand). The table shows the performance of each method in terms of these metrics, with the best performance for each metric highlighted in bold. This allows for a direct comparison of the proposed method’s effectiveness against established techniques on a challenging dataset with 13 classes.

This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two datasets: FLARE2021 and FeTA2021. The comparison includes the number of parameters, FLOPS (floating point operations per second), and several evaluation metrics (mean IoU, Dice score, and others) for each organ or tissue type in the datasets. The best performance for each metric is highlighted in bold, indicating the superiority of the proposed method.

This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other methods on two datasets, FLARE2021 and FeTA2021. The metrics used for comparison include various measures of segmentation accuracy (e.g., mean Intersection over Union (mIoU), Dice score). The table also shows the number of parameters and FLOPS (floating point operations per second) for each model, providing insights into computational efficiency. The best performance for each metric on each dataset is highlighted in bold.

This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other methods on two datasets, FLARE2021 and FeTA2021. The comparison includes several metrics (e.g., mean IoU, Dice score) across different organ classes within each dataset. The best performance for each metric is highlighted in bold.

This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other state-of-the-art methods on two publicly available datasets: FLARE2021 and FeTA2021. The results are shown for several key metrics, including the number of parameters and FLOPs (floating-point operations), as well as organ-specific metrics for each dataset (e.g., Dice, mIoU, and HD95 for FLARE2021; similar metrics for FeTA2021). The best performance for each metric across all methods is highlighted in bold, allowing for easy identification of the superior-performing model. The inclusion of data from reference [56] indicates a comparative analysis incorporating results from a previous publication.

This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two datasets, FLARE2021 and FeTA2021, for abdominal and fetal brain segmentation. The table shows the number of parameters and FLOPs (floating point operations) for each method, along with key evaluation metrics such as mean Intersection over Union (mIoU) and Dice similarity coefficient for each organ or tissue class. The best performing method for each metric is highlighted in bold. The results demonstrate that uC 3DU-Net achieves superior performance while using fewer parameters.

This table presents a comparison of the proposed uC 3DU-Net against nine other state-of-the-art methods for medical image segmentation on two datasets: FLARE2021 and FeTA2021. The results are reported for various metrics such as mean Intersection over Union (mIoU), Dice coefficient, and others. The table also includes the number of parameters (#Params) and floating-point operations (FLOPs) for each method, indicating computational complexity.
This table presents a comparison of the proposed uC 3DU-Net against nine other methods on two datasets, FLARE2021 and FeTA2021. It shows the number of parameters (#Params), floating point operations (FLOPS), mean Intersection over Union (mIoU), Dice score, and other relevant metrics (e.g., volume overlap error (VOE), Hausdorff distance (HD95), Adjusted Rand Index (AdjRand)) for each method on the datasets. The best performance for each metric is highlighted in bold, indicating the superiority of the proposed method.
This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other state-of-the-art methods on two datasets: FLARE2021 and FeTA2021. The metrics used for comparison include various performance indicators such as mean Intersection over Union (mIoU), Dice coefficient, Volume Overlap Error (VOE), Hausdorff Distance (HD), and Adjusted Rand Index. The table highlights the best performance achieved by each method for each metric across both datasets. Part of the data in this table is sourced from another publication (reference [56]).
This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other state-of-the-art methods on two datasets: FLARE2021 and FeTA2021. The metrics used for comparison include various segmentation performance measures such as Mean IoU, Dice score, and others. The table highlights the superior performance of uC 3DU-Net, particularly its ability to achieve better performance while using significantly fewer parameters. A portion of the data used for comparison was obtained from a previous study referenced as [56].
This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other methods on two datasets: FLARE2021 and FeTA2021. The table shows key metrics like the number of parameters, FLOPs (floating point operations), mean Intersection over Union (mIoU), Dice similarity coefficient, and other evaluation metrics for each organ or brain region in the datasets. The best performing model for each metric is highlighted in bold.
This table presents a comparison of the proposed uC 3DU-Net model’s performance against nine other state-of-the-art methods on two benchmark datasets for medical image segmentation (FLARE2021 and FeTA2021). The metrics used to evaluate performance include various measures such as Mean IoU, Dice scores, and others for different organ classes. The table highlights the superior performance of the proposed method, which achieves the best results across multiple metrics while also having a reduced number of parameters compared to other methods.
Full paper#