↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Cryogenic electron microscopy (cryo-EM) image processing faces challenges due to high noise levels and the need for multiple downstream tasks. Existing methods often struggle with noise robustness and generalizability across diverse datasets. The limitations of current self-supervised learning methods, which overlook severe image corruption, are also highlighted.
To overcome these limitations, the researchers developed DRACO, a denoising-reconstruction autoencoder pre-trained on a massive, curated cryo-EM dataset. DRACO’s innovative hybrid training scheme, inspired by Noise2Noise, uses paired odd and even images to learn robust denoising, even without clean reference images. This approach, coupled with the high-quality dataset, allows DRACO to outperform current state-of-the-art methods in denoising, micrograph curation, and particle picking, demonstrating excellent generalizability.
Key Takeaways#
Why does it matter?#
This paper is important because it introduces DRACO, a novel foundation model for cryo-EM, significantly improving performance in image denoising, micrograph curation, and particle picking. Its large-scale curated dataset and hybrid training scheme are valuable contributions to the field, opening new avenues for research and development in cryo-EM image processing.
Visual Insights#
This figure provides a high-level overview of the DRACO architecture and its training process. It shows how a large-scale curated dataset of cryo-EM movies/micrographs is used for pre-training a denoising-reconstruction autoencoder. The pre-trained model (DRACO) is then shown to be adaptable to various downstream cryo-EM tasks, including micrograph curation and particle picking.
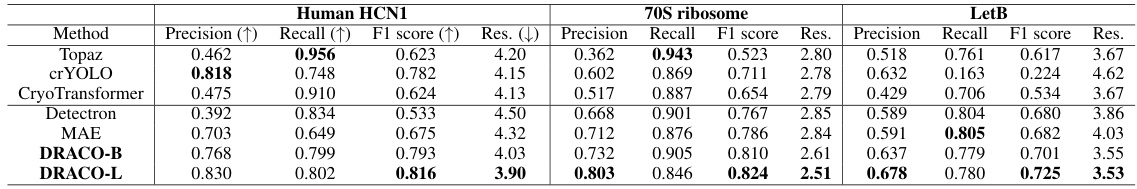
This table presents a comparison of different methods for particle picking on three datasets: Human HCN1, 70S Ribosome, and LetB. For each dataset and method, the precision, recall, F1 score (harmonic mean of precision and recall), and resolution of the 3D reconstruction are shown. Higher precision, recall, and F1 scores are better, indicating more accurate particle picking. Lower resolution is better, reflecting a higher quality 3D reconstruction from accurately picked particles. The resolution is determined using the standard cryoSPARC workflow.
In-depth insights#
Cryo-EM Denoising#
Cryo-EM denoising is a crucial preprocessing step in cryo-electron microscopy (cryo-EM) for obtaining high-resolution 3D structures of macromolecules. Noise significantly impacts image quality, hindering downstream analysis steps such as particle picking and reconstruction. Traditional methods often use simple filtering techniques which may oversimplify noise patterns and lead to information loss. DRACO, a denoising-reconstruction autoencoder, is presented as a novel approach. Inspired by Noise2Noise, DRACO utilizes a hybrid training scheme that leverages both odd and even frames from cryo-EM movies, treating them as noisy observations. This method allows for denoising without requiring clean reference images. The results show that DRACO outperforms existing methods, providing a significant improvement in SNR and detail preservation.
DRACO Autoencoder#
The DRACO Autoencoder, a novel denoising-reconstruction model, addresses the significant challenge of noise in cryo-EM images. Its innovative hybrid training scheme, inspired by Noise2Noise, leverages paired odd and even frames from cryo-EM movies as independent noisy observations. This approach cleverly circumvents the need for clean reference images, a major limitation in cryo-EM. By masking portions of these paired images, DRACO simultaneously tackles denoising and reconstruction tasks. The model’s performance is further enhanced by its pre-training on a massive, high-quality curated dataset exceeding 270,000 micrographs, thereby ensuring robustness and generalizability across various cryo-EM downstream applications, such as micrograph curation and particle picking. DRACO’s exceptional results, outperforming existing state-of-the-art methods in these tasks, highlight the effectiveness of its design and data-driven approach. The architecture showcases the benefits of combining self-supervised learning with tailored loss functions designed to excel in the specific challenges of cryo-EM data. This work represents a significant advancement in developing foundation models specifically tailored for biological imaging.
Downstream Tasks#
The research paper explores various downstream tasks enabled by the developed model, demonstrating its versatility beyond the core denoising functionality. Particle picking, a crucial step in cryo-EM analysis, is significantly improved by the model’s ability to accurately identify particles within noisy micrographs. The model achieves this by leveraging its pre-trained denoising capabilities, directly addressing the challenge of low signal-to-noise ratios in cryo-EM data. Micrograph curation is another key area where the model shows promise. Its capacity to classify micrographs as high- or low-quality based on pre-trained features highlights its utility in streamlining the cryo-EM workflow. By efficiently filtering out low-quality images, the model reduces the computational burden and improves the overall quality of subsequent analysis steps. The results from these downstream tasks showcase the model’s generalizability and effectiveness across multiple stages of cryo-EM processing. Micrograph denoising, the foundation upon which other downstream applications are built, also demonstrates strong performance. The model outperforms existing methods in terms of signal-to-noise ratio improvements and particle structure preservation, ultimately leading to higher-resolution reconstructions. Overall, these downstream applications validate the model’s potential as a robust and versatile tool for advancing cryo-electron microscopy research.
Dataset Creation#
The creation of a high-quality dataset is crucial for training robust and effective deep learning models. This paper emphasizes the importance of dataset curation, acknowledging that publicly available datasets often suffer from inconsistencies in quality, format, and annotation. A key contribution is the creation of a large-scale, curated dataset consisting of over 270,000 cryo-EM movies or micrographs from 529 protein datasets. This meticulous process involved filtering and verifying data, ensuring high resolution and completeness. This carefully curated dataset addresses limitations of existing methods that often rely on smaller, less diverse datasets, leading to improved model performance and generalizability. The detailed description of the dataset creation workflow highlights the significant effort invested in data cleaning, annotation, and quality control, which is often overlooked but critical to the success of subsequent research. The availability of this new dataset will enable future advances in cryo-EM image analysis. The rigorous approach to dataset development showcases a commitment to transparency and reproducibility, promoting greater trust in the findings.
Future Directions#
Future research could explore improving DRACO’s robustness to various types of noise and artifacts present in cryo-EM images, potentially incorporating more sophisticated noise models or incorporating data augmentation strategies that specifically target these issues. Another avenue for exploration is enhancing DRACO’s generalizability across different cryo-EM instruments and experimental conditions, possibly through transfer learning techniques or by training on a more diverse and larger dataset. The effectiveness of the denoising-reconstruction hybrid training scheme should be investigated further, and the optimal balance between these two tasks needs to be explored for various downstream applications. Finally, extending DRACO to handle other cryo-EM modalities, such as cryo-electron tomography, and exploring its capabilities in other single-particle analysis tasks beyond micrograph curation, denoising, and particle picking, would broaden its utility and impact within the field.
More visual insights#
More on figures
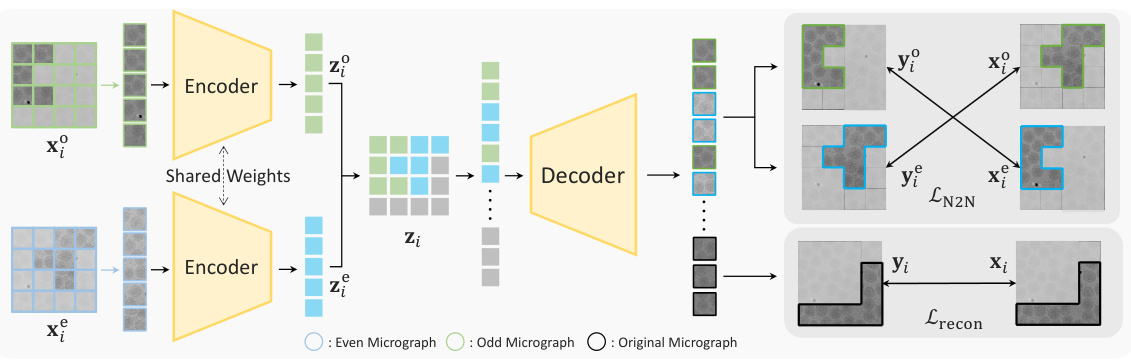
This figure illustrates the architecture of DRACO, a denoising-reconstruction autoencoder for cryo-EM. It shows how the model processes odd and even micrographs, uses masking to create denoising and reconstruction tasks, and applies both Noise2Noise (N2N) and reconstruction losses for training. The encoder extracts features from visible patches, and the decoder reconstructs the entire micrograph using both visible and masked patches, with additional supervision from the original micrograph for masked patches. The diagram also clearly shows the flow of data and the application of different loss functions.

This figure visualizes the performance of DRACO and several baseline methods on particle picking for three different protein datasets: LetB transport protein, 70S ribosome, and Human HCN1 channel protein. Each image shows a micrograph with particles identified by different colored circles: blue circles indicate correctly identified particles (true positives), red circles show incorrectly identified particles (false positives), and yellow circles represent missed particles (false negatives). The figure highlights the superior performance of DRACO in accurately identifying particles compared to the baselines, especially when dealing with challenging datasets containing diverse particle sizes and shapes.
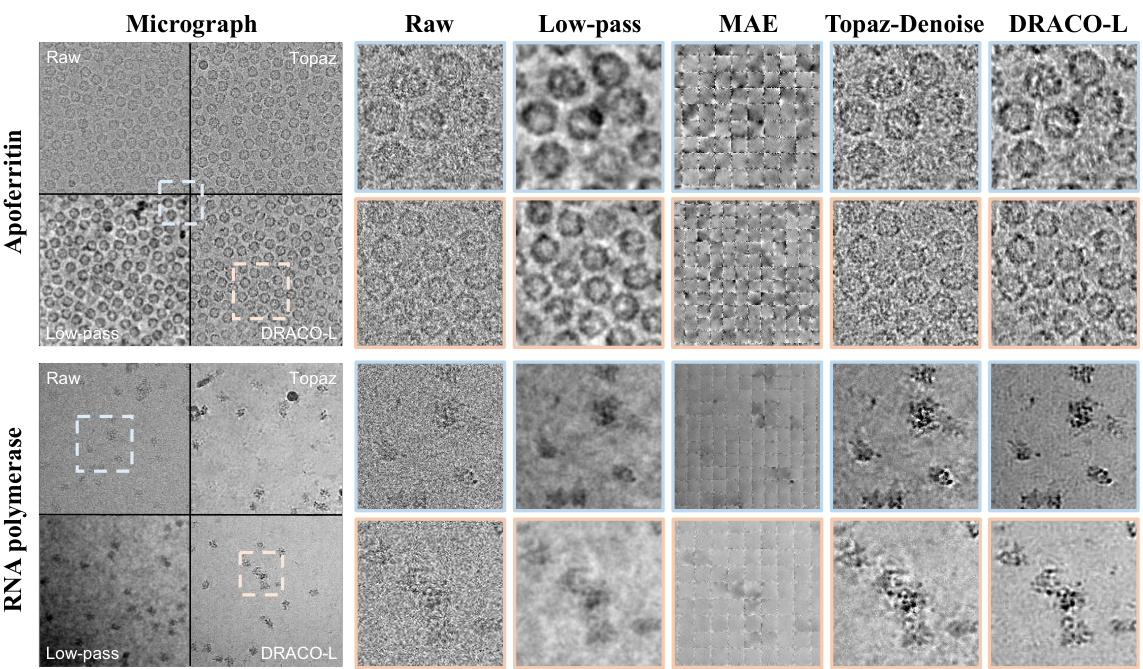
This figure shows a qualitative comparison of micrograph denoising results using DRACO and several state-of-the-art methods. It highlights DRACO’s ability to improve the signal-to-noise ratio (SNR) while preserving fine details of the particle structures, unlike other methods which either cause significant blurring or introduce unwanted artifacts.

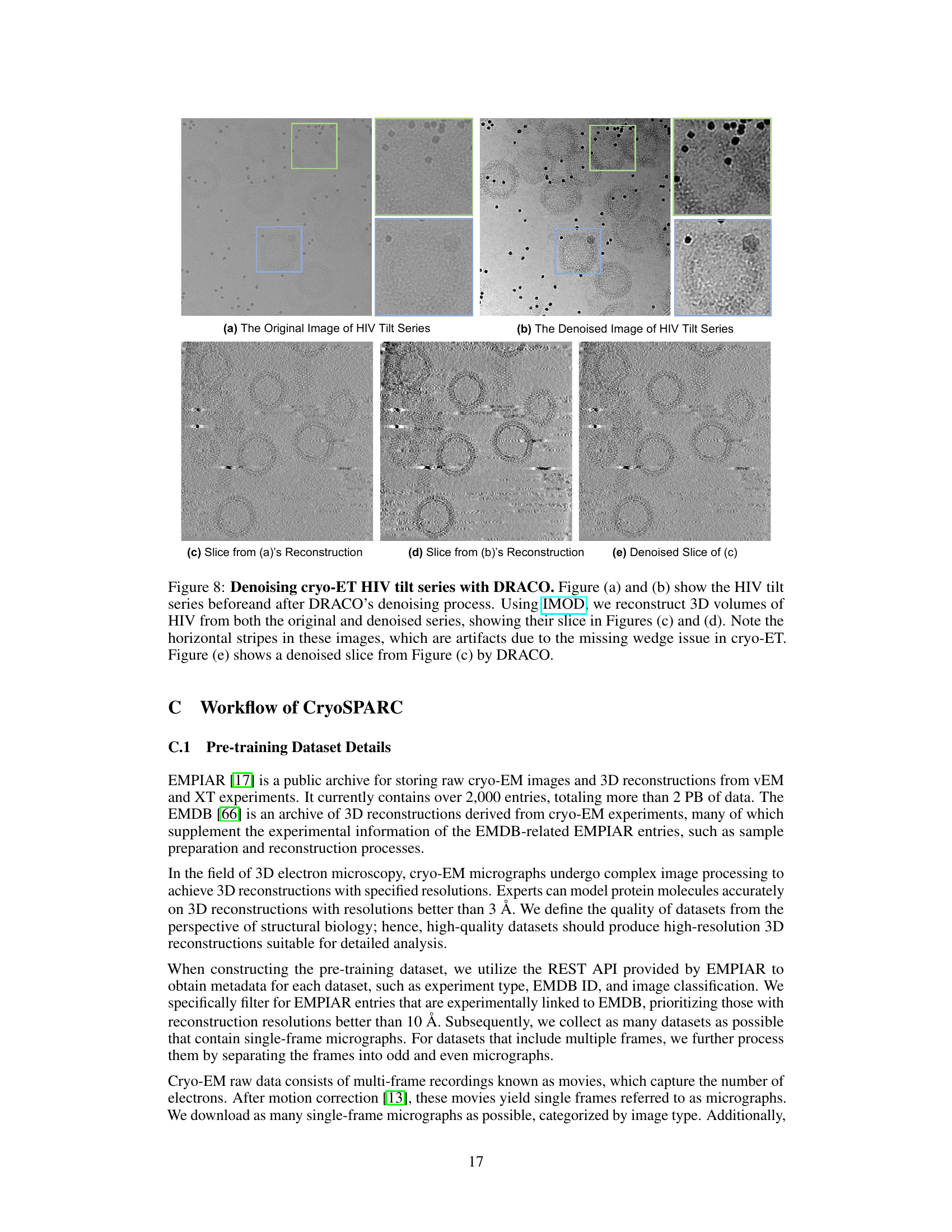
This figure shows the qualitative results of DRACO and other denoising methods on two different types of cryo-EM micrographs: HA Trimer and Phage MS2. The results demonstrate that DRACO achieves the highest visual denoising quality in terms of both signal preservation and noise reduction, better than Low-pass filtering, MAE, and Topaz-Denoise. The image showcases the raw micrographs, their low-pass filtered versions, the results using MAE, Topaz-Denoise, and DRACO-L respectively. The results show that DRACO effectively reduces noise while preserving fine details of the particles. In contrast, Low-pass filtering significantly blurs the images; MAE introduces some patch-wise artifacts; and Topaz-Denoise shows some minor improvements or blurred results.
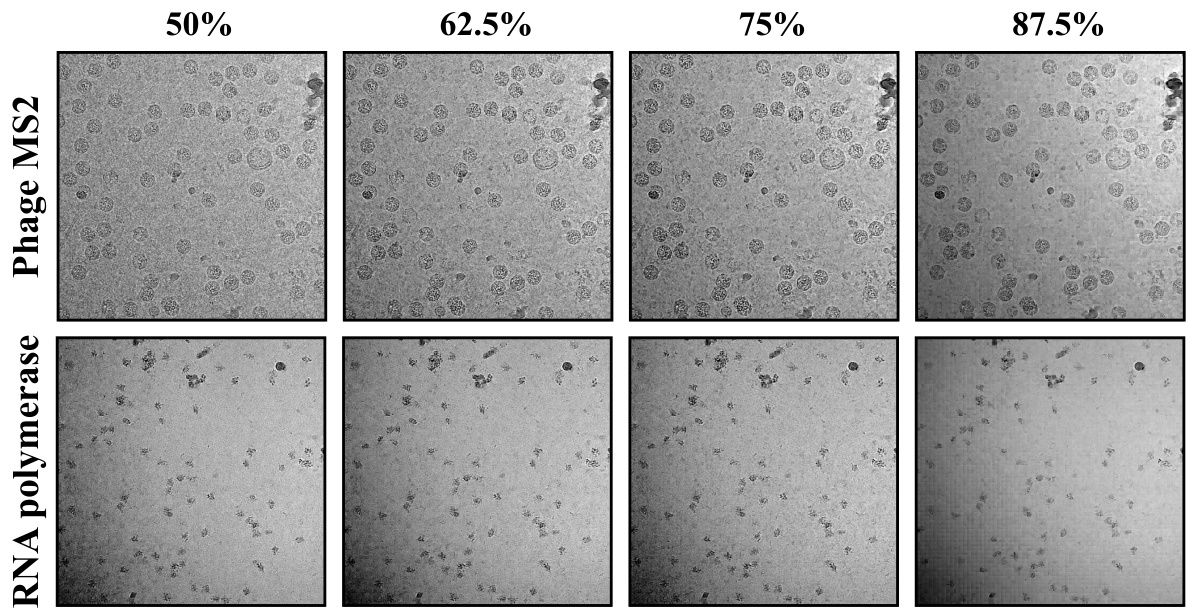
This figure shows the results of DRACO’s denoising performance with different mask ratios (50%, 62.5%, 75%, 87.5%). The results indicate that a mask ratio of 0.75 provides the optimal balance between preserving important signal details and removing background noise. The top row displays the results for Phage MS2, while the bottom row shows the results for RNA polymerase. The images are cryo-EM micrographs, and the improvement in clarity and detail with the 0.75 mask ratio is evident.

This figure shows the reconstruction results of DRACO and MAE at different resolutions, using a constant mask ratio of 0.75. It highlights DRACO’s superior ability to preserve fine details in the visible (unmasked) regions of the image compared to MAE.

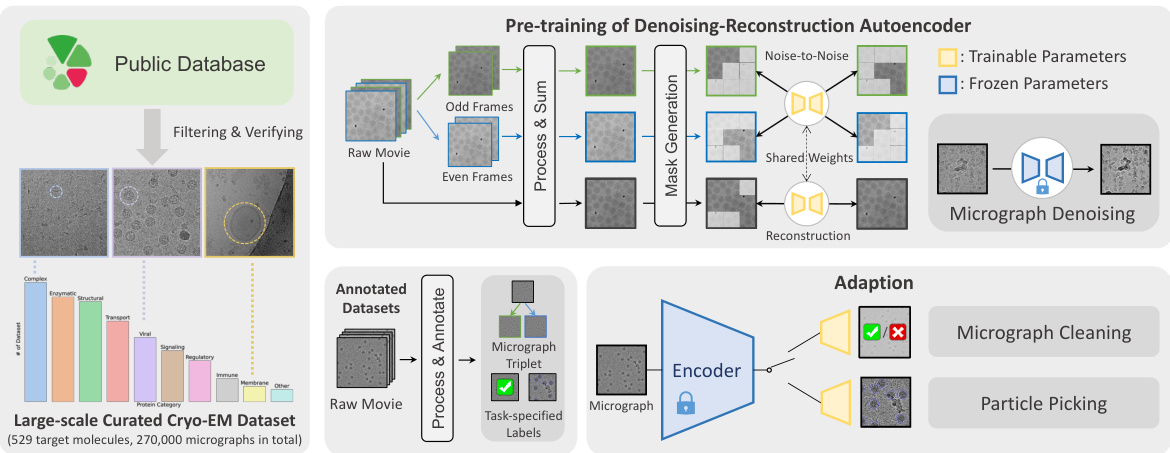
This figure demonstrates DRACO’s ability to denoise cryo-ET data. It shows a comparison of original and denoised HIV tilt series images (a, b). The denoising improves the quality of the 3D reconstruction (c, d), with the denoised reconstruction showing a clearer image than the reconstruction done with the original image (e).
More on tables

This table presents a quantitative comparison of the signal-to-noise ratio (SNR) achieved by different denoising methods on a test dataset. The SNR is calculated using Equation 11 described in the paper. The table compares the SNR of the raw micrographs, micrographs denoised using low-pass filtering, Topaz-Denoise, MAE, DRACO-B, and DRACO-L. The results show that DRACO-L achieves the highest SNR, demonstrating its superior performance in denoising cryo-EM images.
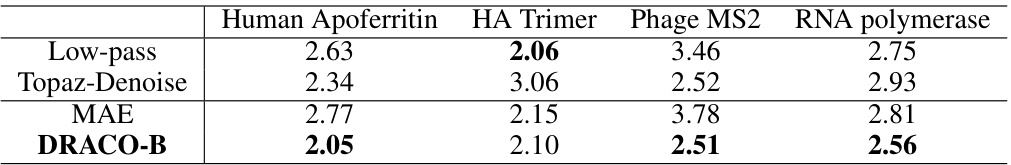
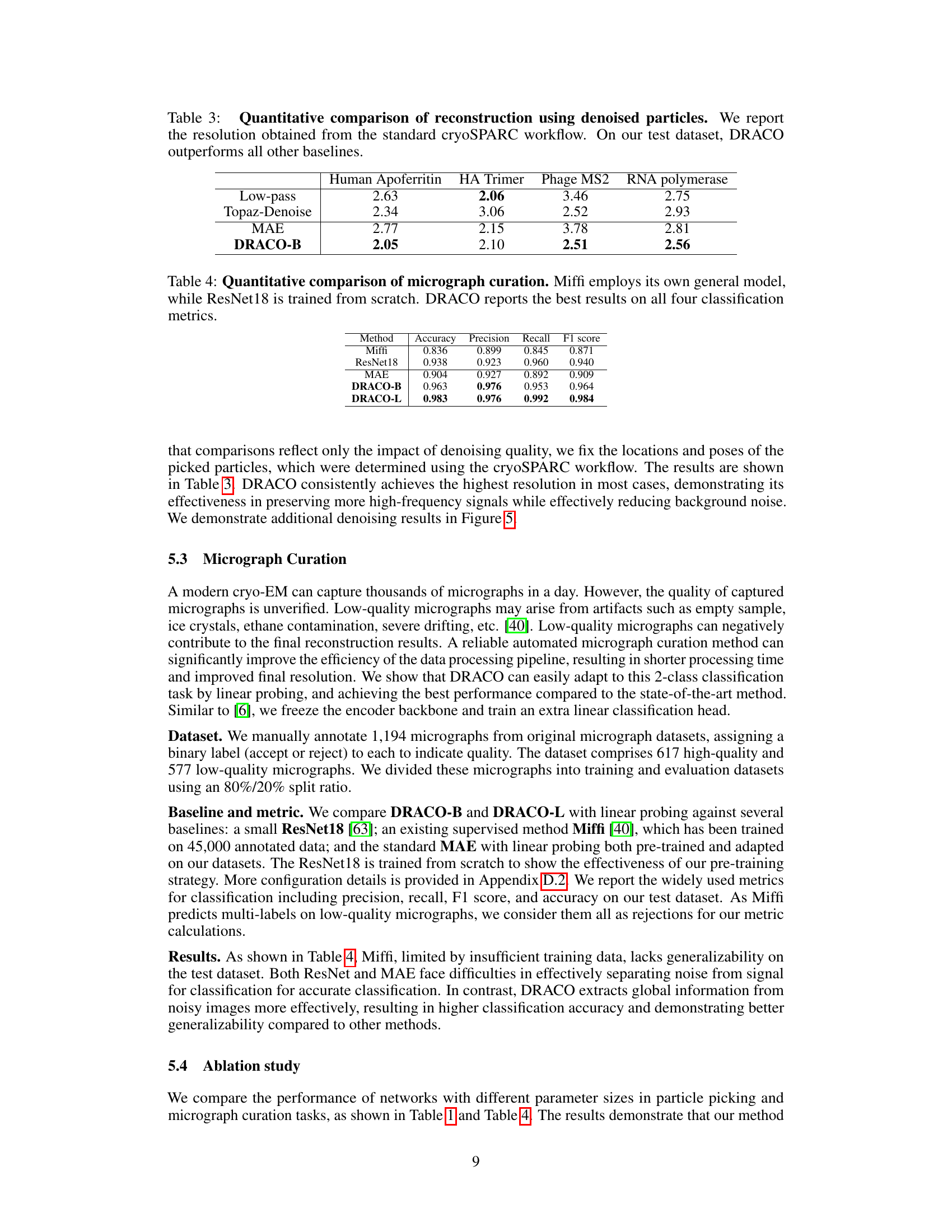
This table presents a quantitative comparison of 3D reconstruction resolution using particles denoised by different methods: Low-pass filtering, Topaz-Denoise, MAE, and DRACO-B. The resolution is determined using the standard cryoSPARC workflow. The results show that DRACO-B achieves the best resolution across all four test datasets (Human Apoferritin, HA Trimer, Phage MS2, and RNA polymerase).
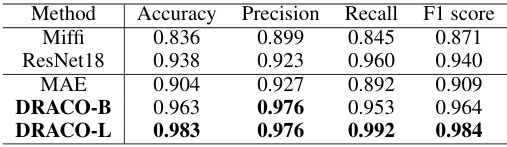
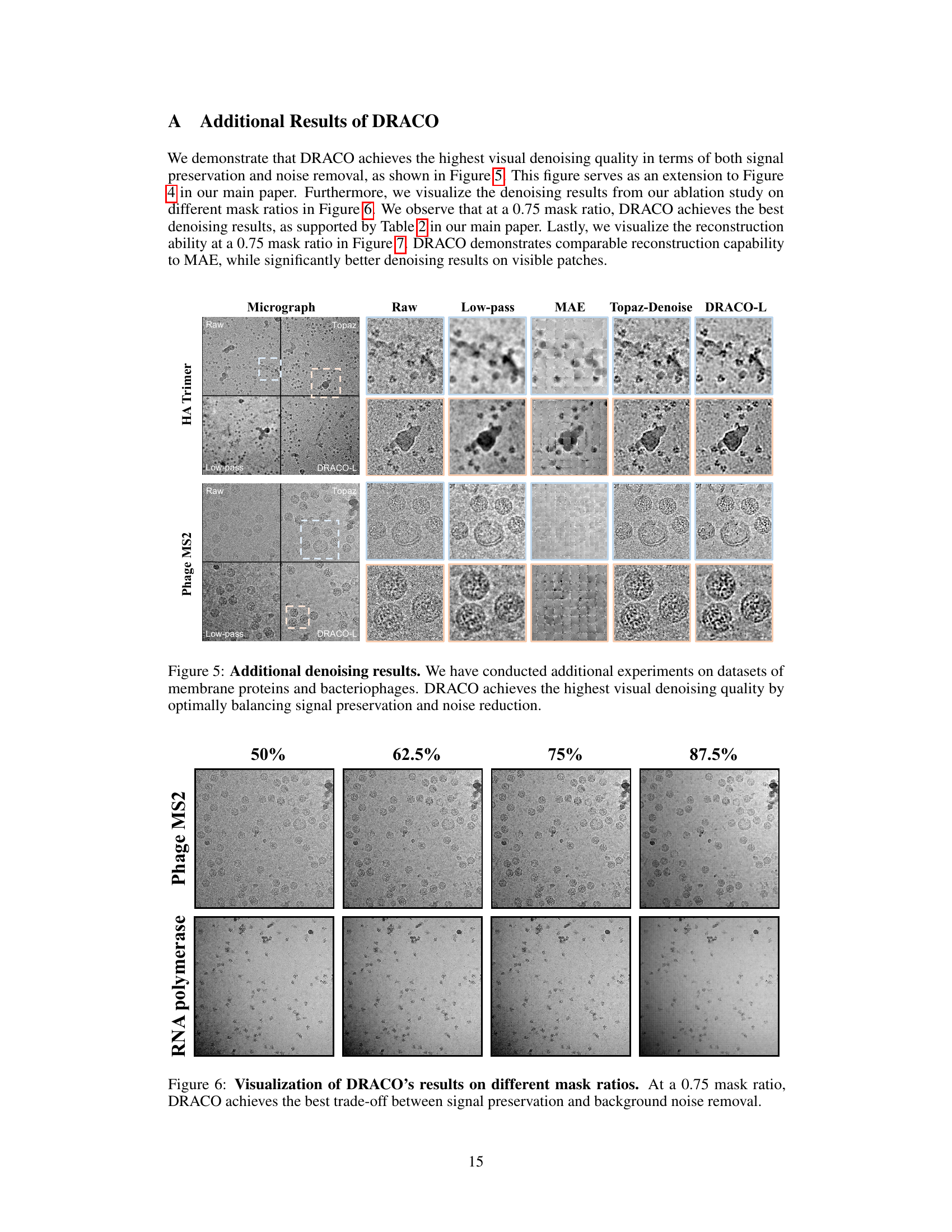
This table compares the performance of different models on a micrograph curation task, which involves classifying micrographs as either high-quality or low-quality. The models compared include Miffi (which uses its own general model), ResNet18 (trained from scratch), MAE (a masked autoencoder), and two versions of DRACO (with different ViT architectures). The table shows that DRACO outperforms all other methods across all four evaluation metrics: accuracy, precision, recall, and F1-score, demonstrating its superior ability to distinguish between high-quality and low-quality micrographs.
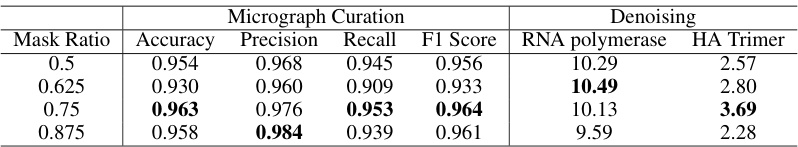
This table presents the ablation study on different mask ratios in DRACO model for micrograph curation and denoising tasks. It shows the performance (Accuracy, Precision, Recall, F1 Score, SNR) of the DRACO model with mask ratios of 0.5, 0.625, 0.75, and 0.875. The results indicate that a mask ratio of 0.75 yields the best performance across both tasks.
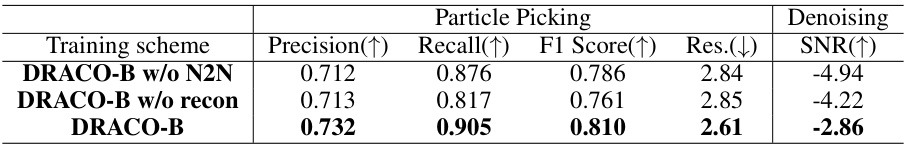
This table presents an ablation study on the DRACO model, specifically evaluating the impact of the Noise2Noise (N2N) loss and the reconstruction loss on particle picking and denoising performance using the 70S ribosome dataset. Three different training schemes are compared: DRACO-B without N2N loss, DRACO-B without reconstruction loss, and the full DRACO-B model. The results indicate the contribution of each loss to the overall performance, suggesting that both losses are essential for optimal results.
Full paper#