↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many clinical prediction tasks benefit from integrating electronic health records (EHR) and chest X-rays (CXR). However, these data are often asynchronous, with CXR images taken infrequently. Using the last available CXR can lead to inaccurate predictions. This paper tackles this issue by focusing on improving the timeliness of the data.
The proposed method, DDL-CXR, dynamically generates an up-to-date CXR representation using a latent diffusion model. This model is conditioned on previous CXR images and EHR time series to capture anatomical structures and disease progression. The generated representation is then combined with the EHR and historical CXR for improved prediction. Experiments demonstrate that this approach significantly outperforms traditional methods, highlighting the effectiveness of generating dynamic, patient-specific CXR for improving the accuracy and timeliness of clinical predictions.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in clinical AI and healthcare. It directly addresses the asynchronicity problem in multimodal data fusion, a significant hurdle in many clinical prediction tasks. The proposed method, DDL-CXR, offers a novel approach to handle asynchronous data, improving the accuracy of clinical predictions. This work opens new avenues for research into dynamic data generation, contrastive learning, and multimodal fusion, relevant to various healthcare applications. Its findings have immediate implications for improving patient care and developing more reliable clinical AI systems.
Visual Insights#

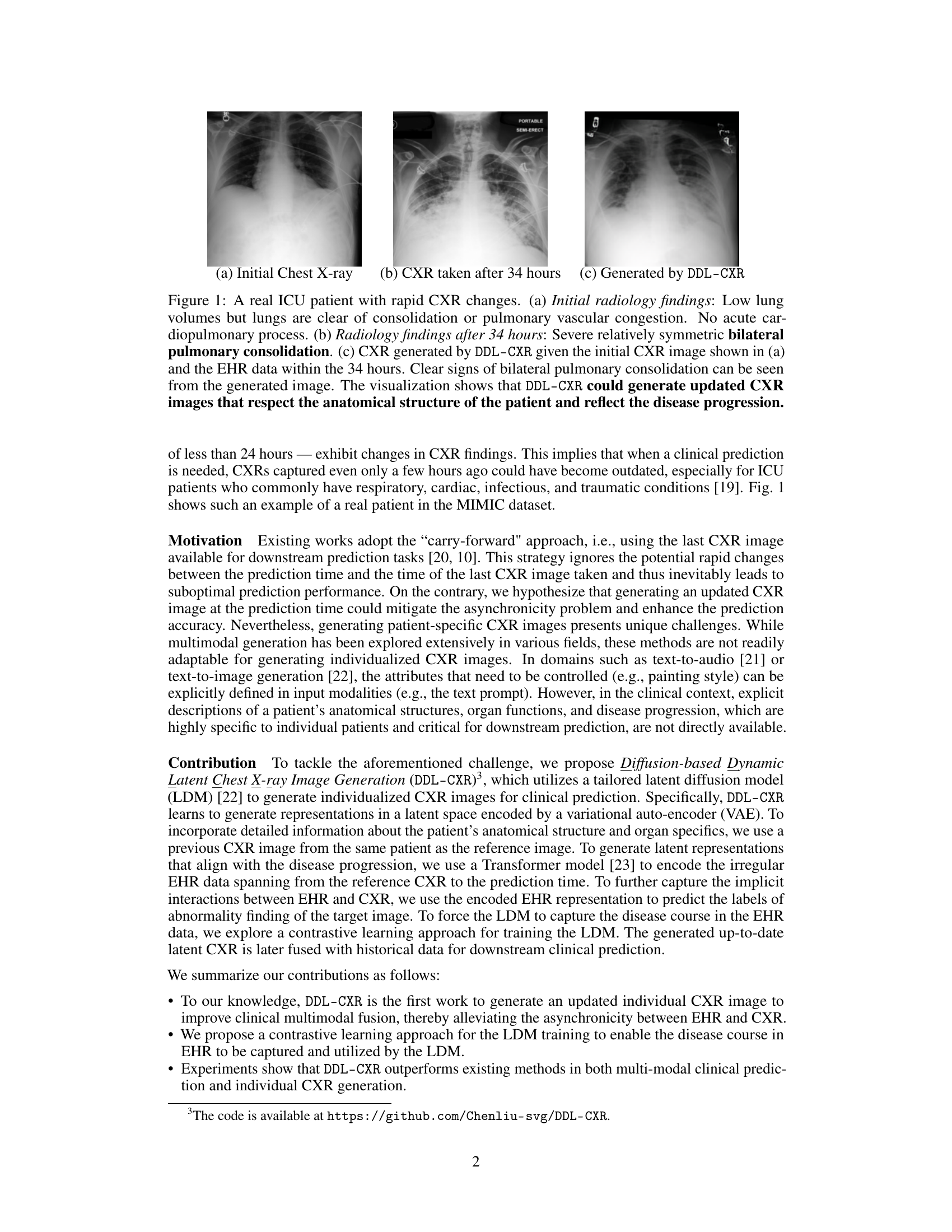
This figure shows a real example of an ICU patient’s chest X-ray (CXR) images taken at different time points, demonstrating rapid disease progression. The initial CXR (a) shows clear lungs, while the CXR taken 34 hours later (b) reveals severe bilateral pulmonary consolidation. The DDL-CXR model, given the initial CXR and the patient’s electronic health record (EHR) data from the 34-hour period, successfully generates a CXR image (c) that accurately reflects the disease progression shown in (b). This highlights the model’s ability to generate up-to-date CXR images, addressing the asynchronous nature of multimodal clinical data.
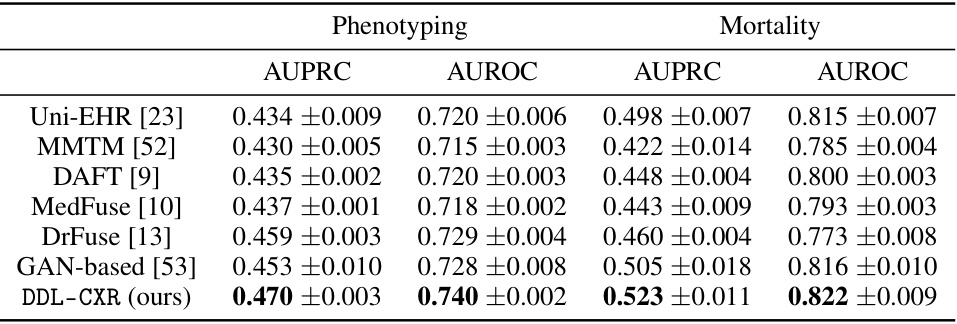
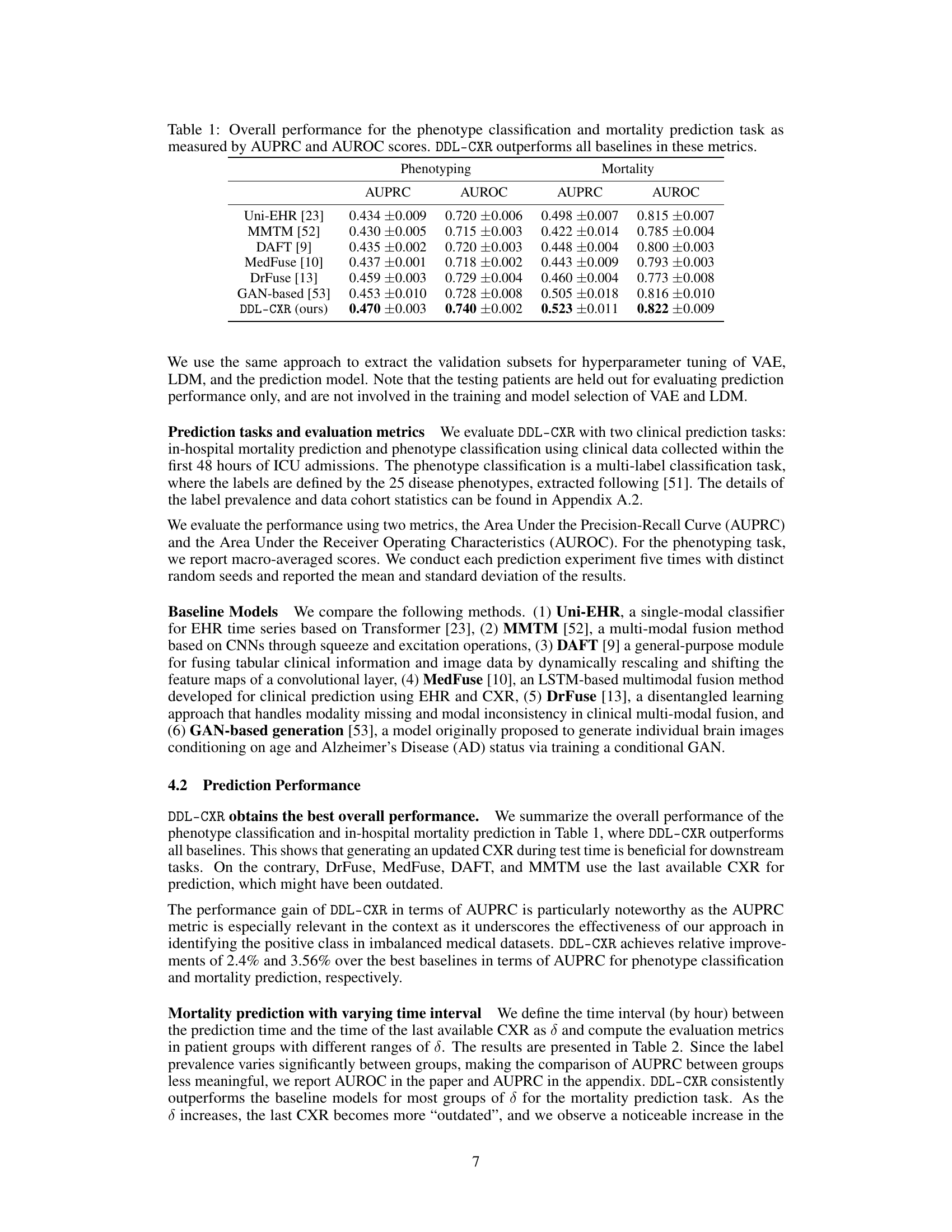
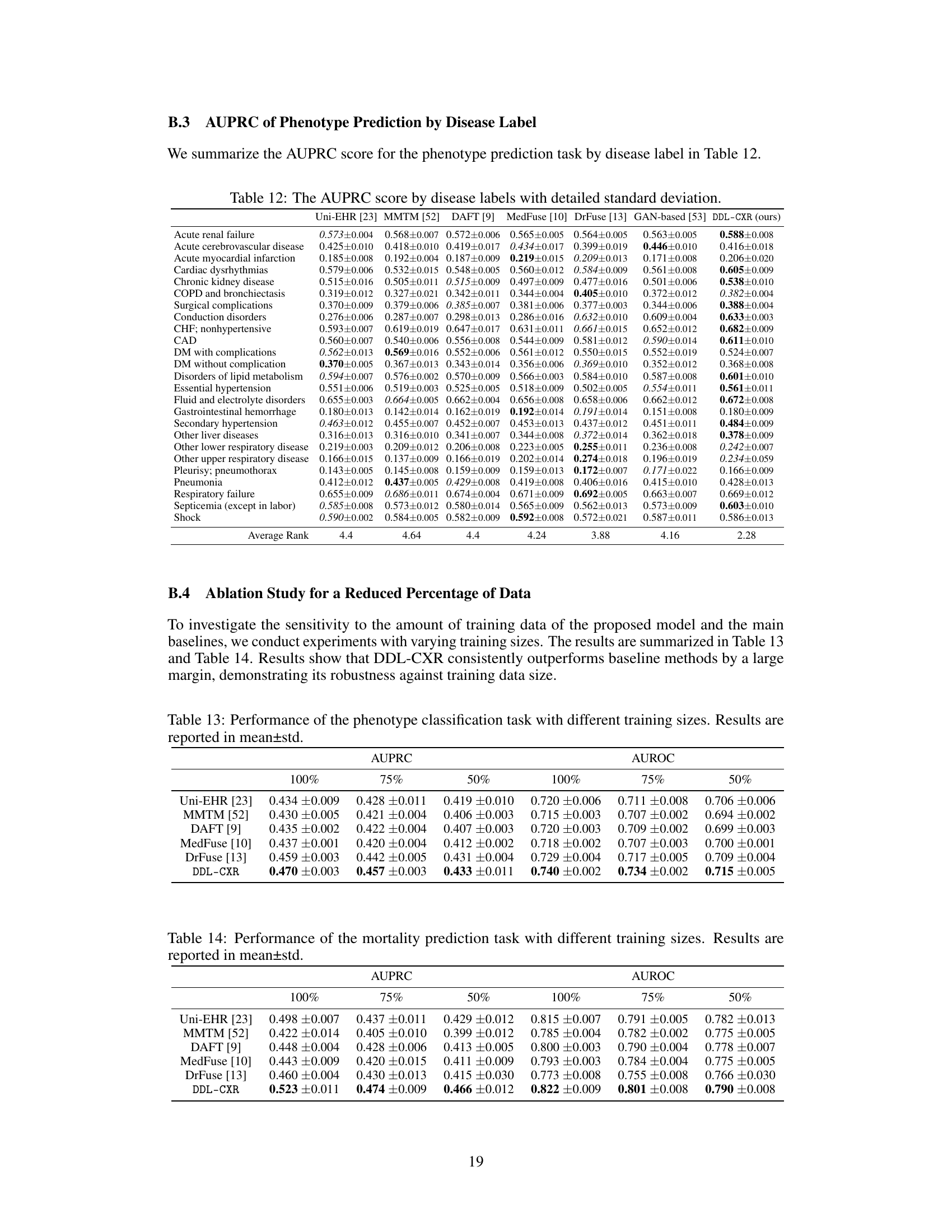
This table presents the AUPRC and AUROC scores for two clinical prediction tasks: phenotype classification and mortality prediction. The performance of DDL-CXR is compared against several baseline methods (Uni-EHR, MMTM, DAFT, MedFuse, DrFuse, and GAN-based). The results demonstrate that DDL-CXR achieves superior performance across all metrics, indicating its effectiveness in addressing the asynchronicity challenge in multimodal clinical data fusion.
In-depth insights#
Async Multimodal Fusion#
Asynchronous multimodal fusion presents a significant challenge in medical applications due to the inherent time discrepancies between data modalities. Electronic health records (EHRs) provide continuous longitudinal data, whereas medical images like chest X-rays (CXRs) are acquired sporadically. This asynchronicity leads to outdated information at the time of clinical prediction, negatively impacting accuracy. Effective fusion strategies must address temporal misalignment. One approach involves generating synthetic, up-to-date CXR images using latent diffusion models conditioned on the available EHR data and past CXRs. This dynamically generates a current representation, enabling more accurate multimodal fusion. However, challenges remain in accurately capturing complex disease progression and anatomical details within the synthetic image generation. Further research should explore more sophisticated methods for temporal modeling and improved model evaluation metrics to address this challenging problem of asynchronous multimodal fusion in clinical applications.
Latent CXR Generation#
The concept of ‘Latent CXR Generation’ in this research paper is crucial for addressing the asynchronicity problem inherent in multimodal clinical data fusion. The core idea is to dynamically generate an up-to-date latent representation of a patient’s chest X-ray (CXR) image, conditional on both previous CXR images and the patient’s Electronic Health Record (EHR) time series. This approach cleverly leverages the strengths of both data modalities; the previous CXR captures the patient’s anatomical structure, while the EHR time series provides information about disease progression. Using a latent diffusion model allows the generation of a realistic and patient-specific CXR, avoiding the limitations of simply carrying forward outdated CXR images. This dynamic generation process effectively bridges the temporal gap between image acquisition and clinical prediction, leading to more accurate and timely clinical predictions. The method is innovative as it generates patient-specific CXR images rather than relying solely on pre-existing data, thus mitigating the inherent challenges of asynchronicity in multimodal data fusion.
Contrastive Learning#
Contrastive learning, in the context of multimodal clinical data fusion, is a powerful technique to learn better representations by leveraging the inherent relationships between different data modalities. The approach involves simultaneously learning from similar and dissimilar data points. In the given research, the method likely focuses on contrasting EHR time series representing different disease progression stages with corresponding chest X-ray images. By contrasting these data pairs, the model aims to disentangle the information from anatomical structures and disease progressions, improving the accuracy of CXR generation and enhancing downstream clinical predictions. The success hinges on effectively defining similarity and dissimilarity metrics for these heterogeneous data modalities, which requires careful consideration of temporal aspects and inherent noise within clinical data. A well-designed contrastive loss function is crucial to guide the model’s learning process and improve the quality of generated latent CXRs. Overall, it’s a sophisticated technique to leverage the nuances of multimodal clinical data by emphasizing comparative learning.
MIMIC-IV Results#
An analysis of MIMIC-IV results would require access to the full paper. However, considering the paper’s focus on addressing asynchronous multimodal data in clinical prediction using chest X-rays (CXR) and electronic health records (EHR), a thoughtful analysis of MIMIC-IV results would likely center on evaluating the model’s performance. Key metrics would likely include AUROC and AUPRC for both mortality prediction and phenotype classification tasks. The results would show whether dynamically generating up-to-date CXR representations, conditioned on past CXRs and EHR data, significantly improves predictive performance compared to methods that simply use the last available CXR. A strong analysis would investigate the impact of varying time intervals between the prediction time and the last available CXR image. This would highlight the effectiveness of addressing asynchronicity. Finally, comparing the results across different time intervals would provide crucial insight into the model’s ability to handle dynamic disease progression, indicating the robustness and practical implications of the proposed approach.
Future Research#
Future research directions stemming from this work could explore several promising avenues. Improving the model’s ability to handle highly irregular or sparse EHR data is crucial, as real-world data often exhibits significant inconsistencies. This might involve exploring advanced imputation techniques or developing more robust temporal modeling methods. Another key area is enhancing the model’s generalizability across diverse patient populations and healthcare settings. The current study uses a specific dataset; further validation on other datasets is needed to demonstrate wider applicability and robustness. Investigating the explainability of the generated CXR images would significantly improve clinical trust and acceptance. Methods for visualizing and interpreting the model’s decision-making process are needed, potentially through techniques like attention mechanisms or saliency maps. Finally, exploring the integration of additional modalities, such as pulmonary function tests or blood biomarkers, could potentially further enhance prediction accuracy and provide a more holistic clinical view. Investigating the cost-effectiveness of implementing this technology in various clinical settings and workflows will be important for adoption.
More visual insights#
More on figures
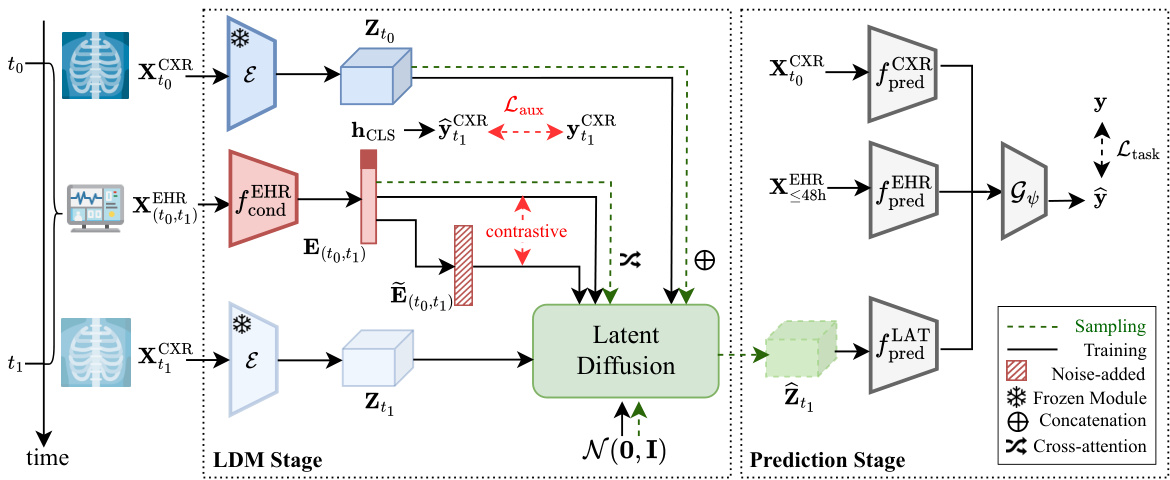
This figure illustrates the DDL-CXR framework, which consists of two main stages: the LDM (Latent Diffusion Model) stage and the prediction stage. The LDM stage dynamically generates an updated latent representation of a chest X-ray (CXR) image using a previous CXR image and EHR (Electronic Health Records) data. The prediction stage then uses this generated latent CXR representation, along with the latest available CXR and the full EHR time series, to make clinical predictions. The figure highlights key components like the VAE (Variational Autoencoder), Transformer, contrastive loss, and auxiliary prediction task, which work together to handle the asynchronicity of multimodal data and improve prediction accuracy.
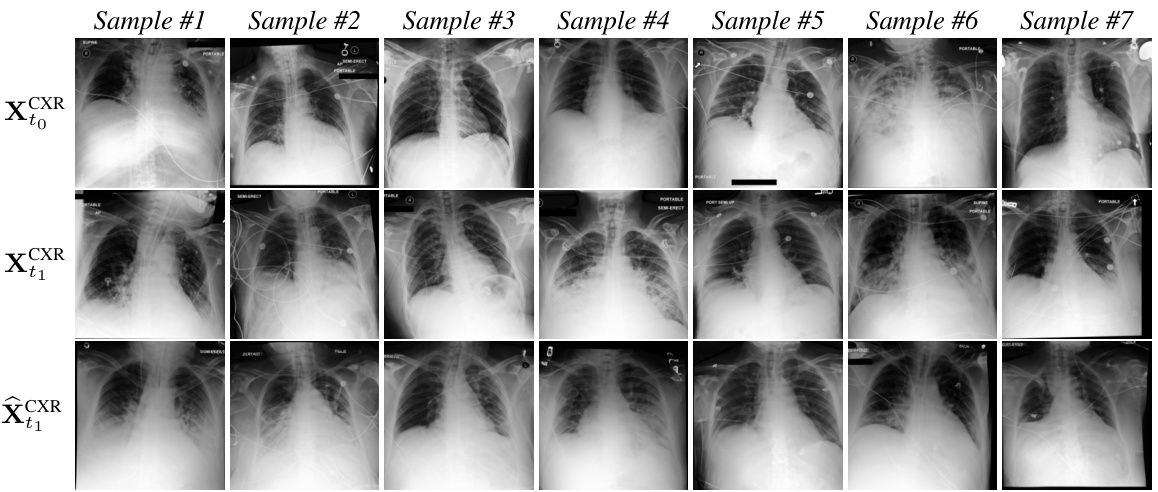
This figure shows examples of chest X-ray images generated by the DDL-CXR model. The top row displays the initial (reference) X-ray images, the middle row shows the actual X-ray images taken later, and the bottom row presents the images generated by the model. The results demonstrate that the model is able to generate images that reflect both the anatomical structure of the original image and the disease progression indicated in the EHR data.
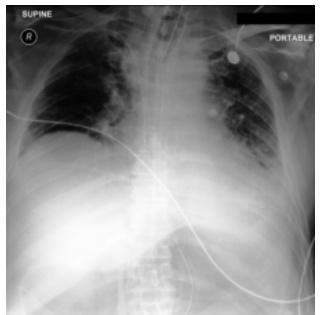
This figure shows a real example from the MIMIC-CXR dataset of a patient in the ICU with rapidly changing chest x-ray (CXR) findings. Subfigure (a) shows the initial CXR, indicating relatively normal lung volumes. Subfigure (b) depicts the CXR taken 34 hours later, revealing severe bilateral pulmonary consolidation. Subfigure (c) shows a CXR generated by the proposed DDL-CXR model, using only the initial CXR (a) and the EHR data from the intervening 34-hour period. The generated image accurately reflects the progression of the disease, demonstrating the model’s ability to generate up-to-date CXR representations.

This figure shows a real example from the MIMIC-CXR dataset of an ICU patient whose chest X-ray (CXR) changed drastically within 34 hours. Subfigure (a) displays the initial CXR, showing normal lung volumes but no other issues. Subfigure (b) shows the CXR after 34 hours, revealing severe bilateral pulmonary consolidation. Subfigure (c) shows a CXR generated using the proposed DDL-CXR method, leveraging the initial CXR (a) and the patient’s electronic health record (EHR) data from the 34-hour period. The generated image successfully reflects the significant disease progression seen in the actual CXR.

This figure showcases a real case from the MIMIC-CXR dataset illustrating the asynchronicity problem in clinical multimodal data. It presents three chest X-rays (CXRs) from a single ICU patient over 34 hours. (a) shows the initial CXR with normal findings. (b) shows the CXR after 34 hours, revealing severe bilateral pulmonary consolidation. (c) displays a CXR generated by the proposed DDL-CXR model using the initial CXR and EHR data from the 34-hour period. The generated CXR accurately reflects the disease progression, highlighting the method’s ability to generate updated CXRs.

This figure showcases a real ICU patient’s chest X-ray (CXR) images over a 34-hour period, demonstrating rapid disease progression. Subfigure (a) shows the initial CXR, revealing clear lungs. Subfigure (b) presents the CXR after 34 hours, displaying severe bilateral pulmonary consolidation. Finally, subfigure (c) shows a CXR generated by the proposed DDL-CXR model using the initial CXR (a) and the patient’s electronic health record (EHR) data from the intervening 34 hours. The generated image accurately reflects the patient’s condition after 34 hours, highlighting the model’s ability to generate realistic and up-to-date CXRs.

This figure shows a real example of an ICU patient with rapid changes in their chest X-ray (CXR) findings over a 34-hour period. It highlights the asynchronicity problem in clinical multimodal data: the last available CXR might be outdated when clinical prediction is needed. Subfigure (a) shows the initial CXR, (b) shows the CXR taken 34 hours later revealing significant disease progression, and (c) demonstrates the ability of the proposed DDL-CXR model to generate a synthetic CXR image accurately reflecting the disease progression, conditioned on the initial CXR and the EHR data.
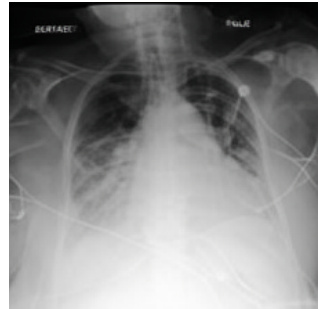
This figure showcases a real ICU patient’s chest X-ray (CXR) images over time, highlighting rapid disease progression. The first image (a) shows relatively clear lungs. The second image (b), taken 34 hours later, reveals severe bilateral pulmonary consolidation (lung filling). The third image (c) demonstrates the DDL-CXR model’s ability to generate a realistic, updated CXR based on the initial image (a) and the patient’s electronic health record (EHR) data over the 34-hour period. The generated image correctly reflects the disease progression, indicating the model’s potential to overcome asynchronicity in multimodal data fusion.
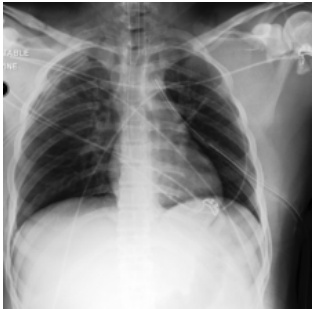
This figure shows a real case of an ICU patient whose chest X-ray (CXR) changed dramatically within 34 hours. The initial CXR (a) shows clear lungs, while the follow-up CXR (b) shows severe bilateral consolidation. The authors’ model, DDL-CXR, successfully generated a CXR image (c) that accurately reflects the disease progression observed in the patient’s EHR data, demonstrating the model’s ability to dynamically generate up-to-date CXR representations.

This figure shows a real case of an ICU patient whose chest X-ray (CXR) changed drastically within 34 hours. The initial CXR (a) shows clear lungs, but a follow-up CXR (b) 34 hours later reveals severe bilateral pulmonary consolidation. The authors’ proposed method, DDL-CXR, generated a CXR image (c) using the initial CXR and Electronic Health Record (EHR) data from the 34-hour period. Notably, the generated image accurately reflects the disease progression, demonstrating the potential of DDL-CXR to predict future CXR images based on past data.
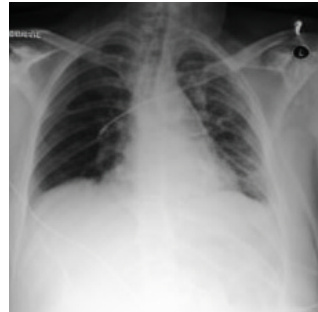
This figure shows a real case of an ICU patient whose lung condition deteriorated rapidly within 34 hours. The leftmost image (a) shows the initial chest X-ray, which was clear. The middle image (b) shows the chest X-ray taken 34 hours later, which shows severe bilateral pulmonary consolidation. The rightmost image (c) is a chest X-ray generated by the proposed model (DDL-CXR), which takes the initial chest X-ray and the EHR data during the 34-hour period as input. Image (c) accurately reflects the patient’s worsened lung condition after 34 hours, demonstrating the model’s ability to generate an up-to-date CXR that accurately reflects disease progression.

This figure shows a real case of an ICU patient with rapidly changing chest X-ray (CXR) findings over 34 hours. It compares the initial CXR (a), the CXR taken after 34 hours showing significant worsening (b), and a CXR generated by the proposed DDL-CXR method using only the initial image and the patient’s Electronic Health Record (EHR) data from that time period (c). The generated CXR successfully reflects the disease progression shown in the actual CXR taken 34 hours later.

This figure shows a real case of an ICU patient whose chest X-ray (CXR) changed dramatically within 34 hours. The initial CXR (a) shows clear lungs, while the CXR taken 34 hours later (b) shows severe bilateral pulmonary consolidation. The authors’ proposed method, DDL-CXR, generated a CXR image (c) based on the initial CXR and the patient’s electronic health record (EHR) data from that period. The generated image (c) successfully reflects the disease progression shown in the actual CXR (b), demonstrating the potential of DDL-CXR to predict future CXR images.

This figure showcases a real patient’s chest X-ray (CXR) images taken at different time points, illustrating rapid disease progression. The initial CXR shows clear lungs. After 34 hours, significant bilateral consolidation is observed. DDL-CXR, the proposed model, successfully generates a CXR that closely resembles the actual 34-hour image, demonstrating its ability to predict future CXR changes based on initial CXR and EHR data.
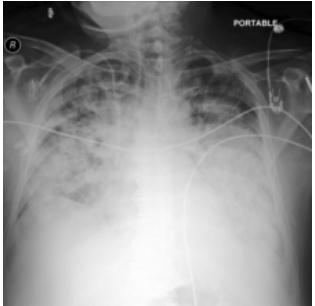
This figure shows a real ICU patient’s chest X-ray images taken at different times and a generated image. The first image shows a relatively clear lung. The second image shows severe bilateral pulmonary consolidation after 34 hours. The third image is generated by DDL-CXR using the first image and EHR data from the 34-hour period. The generated image shows the same consolidation as the second image, demonstrating that DDL-CXR is capable of generating updated images based on the progression of the disease.
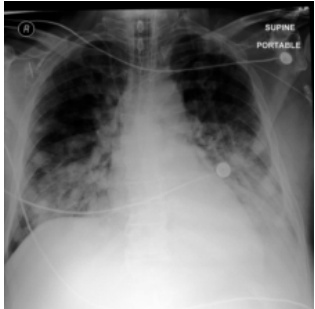
This figure shows a real case of an ICU patient with rapidly changing chest X-ray (CXR) findings over 34 hours. Image (a) shows the initial CXR, which was clear. Image (b) shows the CXR after 34 hours, revealing severe bilateral pulmonary consolidation. Image (c) displays a CXR generated by the proposed DDL-CXR model using the initial CXR (a) and the EHR data from the 34-hour period. The generated image successfully captures the bilateral consolidation, demonstrating the model’s ability to generate realistic and clinically relevant updated CXRs.
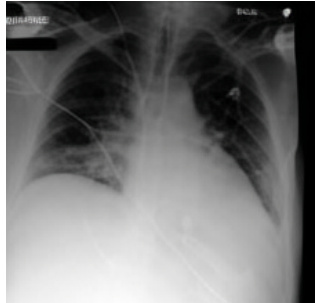
This figure shows a real ICU patient’s chest X-ray (CXR) images taken at different times, demonstrating rapid disease progression. Image (a) shows the initial CXR with clear lungs. Image (b) shows the CXR after 34 hours, revealing severe bilateral pulmonary consolidation. Image (c) shows a CXR generated by the proposed DDL-CXR model using the initial CXR and EHR data, accurately reflecting the progression to bilateral consolidation, showcasing the model’s ability to generate realistic and up-to-date CXRs.
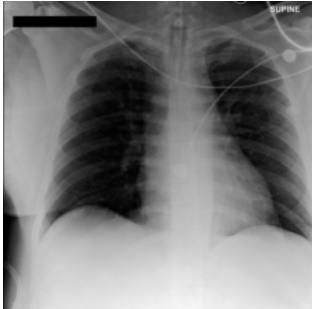
This figure shows a real case of an ICU patient with rapidly changing chest X-ray (CXR) findings over 34 hours. The initial CXR (a) shows clear lungs, while a subsequent CXR (b) reveals severe bilateral pulmonary consolidation. The model DDL-CXR, given the initial CXR and the patient’s Electronic Health Record (EHR) data from the intervening period, successfully generates a CXR (c) that accurately reflects the new disease progression, showing bilateral consolidation.
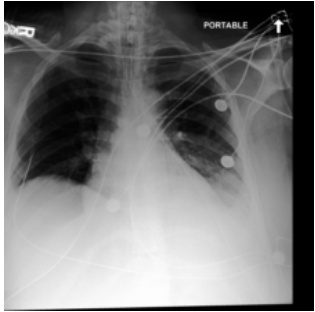
This figure shows a real case of an ICU patient with rapidly changing chest X-ray (CXR) findings within 34 hours. Subfigure (a) displays the initial CXR, showing clear lungs. Subfigure (b) shows the CXR taken 34 hours later, revealing severe bilateral pulmonary consolidation. Subfigure (c) presents a CXR generated by the proposed DDL-CXR model, using the initial CXR (a) and the patient’s electronic health record (EHR) data from the 34-hour period. The generated image accurately reflects the disease progression observed in the actual CXR (b), demonstrating the model’s ability to produce realistic and up-to-date CXR images.
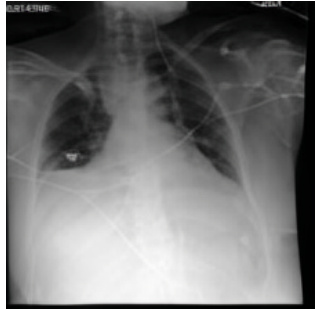
This figure shows a real example from the MIMIC-CXR dataset of an ICU patient’s chest X-ray (CXR) images over time. Subfigure (a) shows the initial CXR, which appears relatively normal. Subfigure (b) shows the CXR taken 34 hours later, revealing severe bilateral pulmonary consolidation (a significant worsening). Subfigure (c) presents a CXR generated by the proposed DDL-CXR method. This generated image uses the initial CXR (a) and the patient’s EHR data from the 34-hour period to accurately predict the later, significantly more severe condition shown in (b). This demonstrates the model’s ability to dynamically generate up-to-date CXR images reflective of a patient’s condition.
More on tables

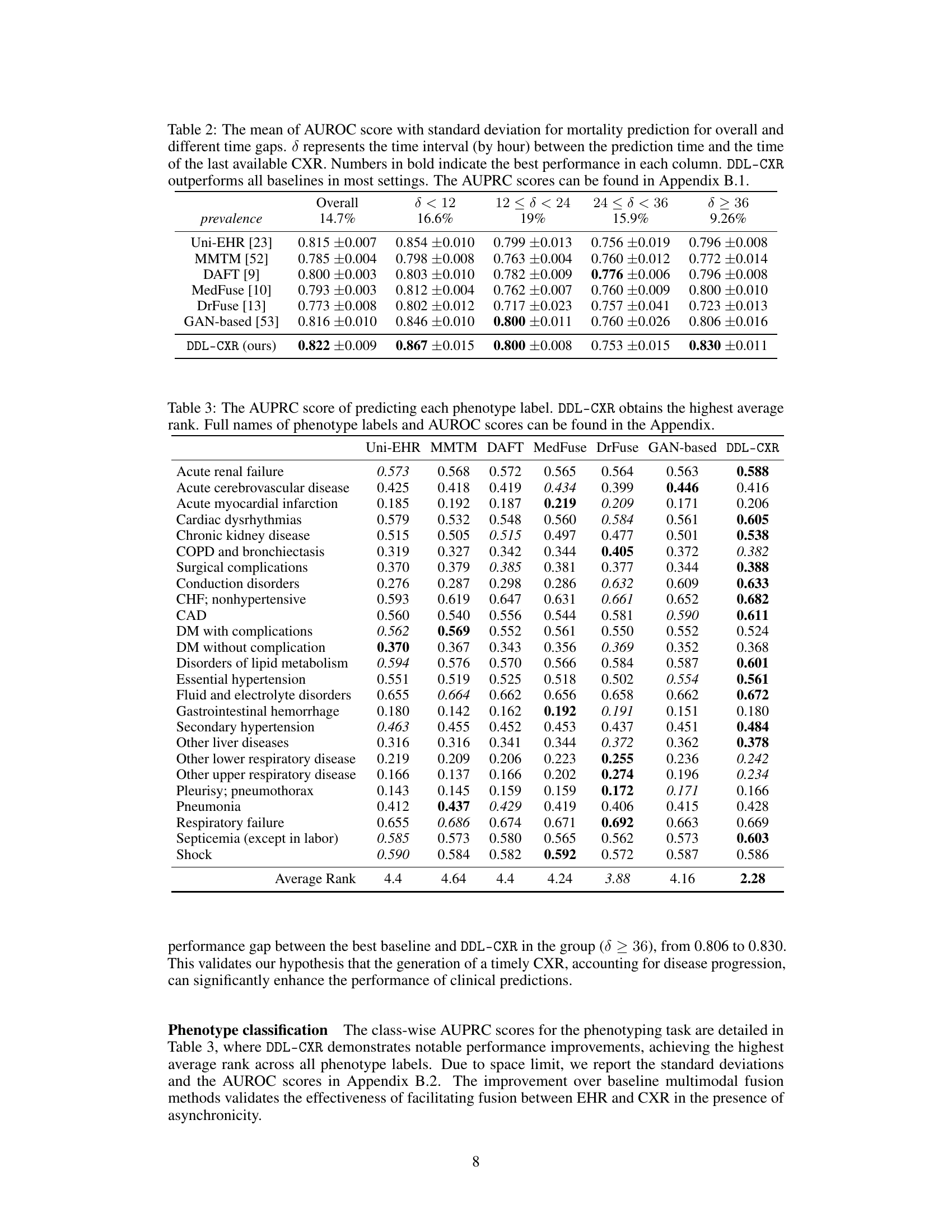
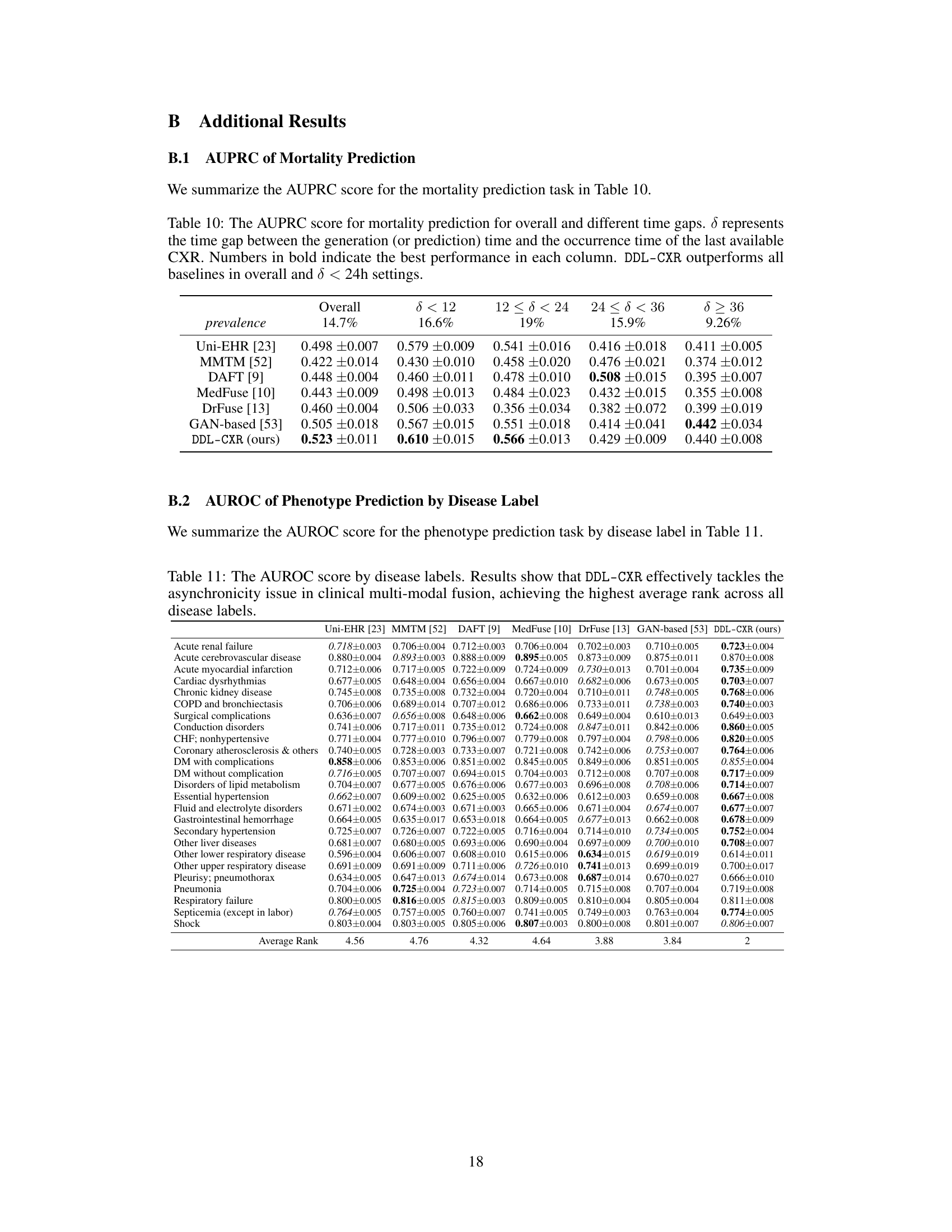
This table presents the Area Under the Receiver Operating Characteristic (AUROC) scores for mortality prediction using different methods. The results are broken down by the time interval (δ) between the prediction time and the last available chest X-ray image, showing how performance varies with the timeliness of the image data. The overall performance and performance within different time ranges (δ < 12, 12 ≤ δ < 24, 24 ≤ δ < 36, δ ≥ 36) are shown, highlighting the effectiveness of the proposed DDL-CXR method, especially when the CXR image is outdated.
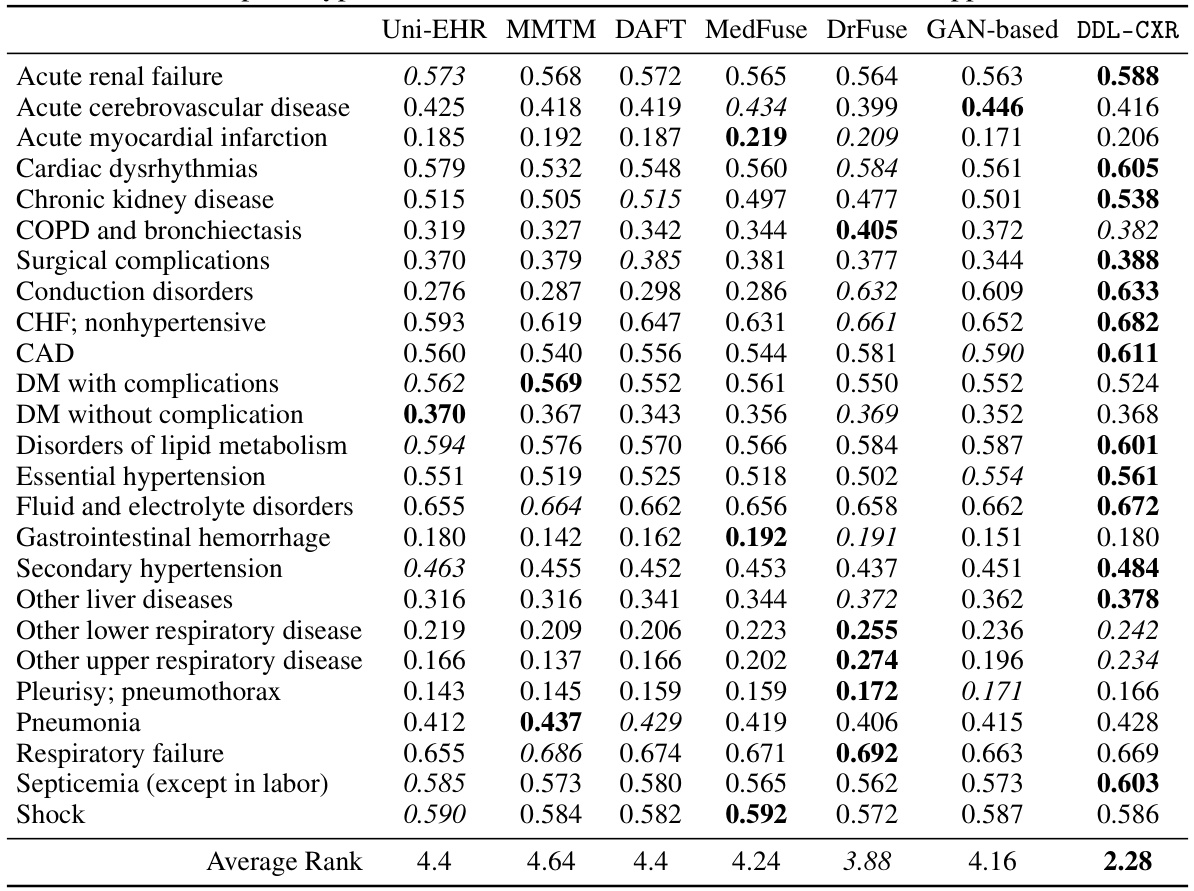
This table presents the performance of the proposed DDL-CXR model and several baseline models on two clinical prediction tasks: phenotype classification and mortality prediction. The performance is measured using two metrics: Area Under the Precision-Recall Curve (AUPRC) and Area Under the Receiver Operating Characteristic Curve (AUROC). The table shows that DDL-CXR achieves superior performance compared to the baselines across both tasks, demonstrating its effectiveness in addressing the asynchronicity issue in multimodal clinical data.

This table presents a quantitative evaluation of the quality of generated chest X-ray images using three metrics: Fréchet Inception Distance (FID), Fréchet Distance (FD), and Wasserstein Distance (WD). Lower scores indicate better quality. The table compares the performance of the proposed DDL-CXR model against a GAN-based baseline and also shows the impact of removing either the previous CXR image (Zt0) or the EHR data (E(t0,t1)) from the generation process.
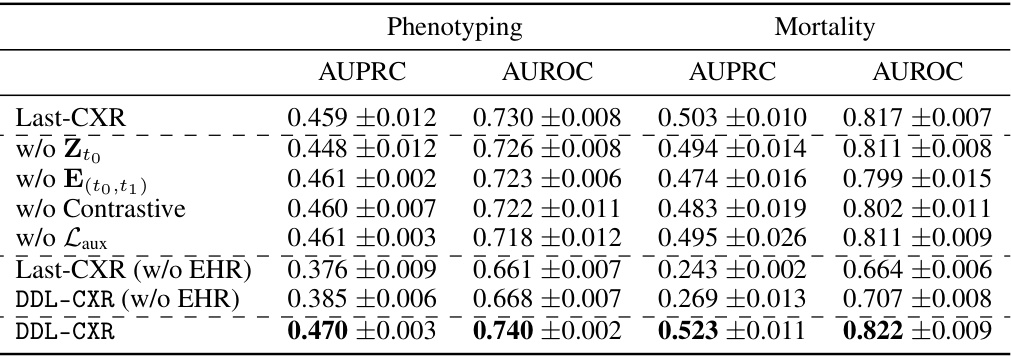
This ablation study investigates the impact of removing different components from the LDM stage on the performance of the model. The components investigated include the reference latent CXR (Zto), the encoded EHR representation (E(to,t1)), the contrastive learning component, and the auxiliary loss (Laux). The results demonstrate that removing any of these components leads to a decrease in the performance of both phenotype classification and mortality prediction tasks, highlighting the importance of each component in the model’s effectiveness. The table also shows results when EHR data is completely excluded in both the LDM stage and prediction stage. This emphasizes that the improvement from DDL-CXR in performance comes primarily from generating an updated CXR and leveraging the interaction between CXR and EHR.

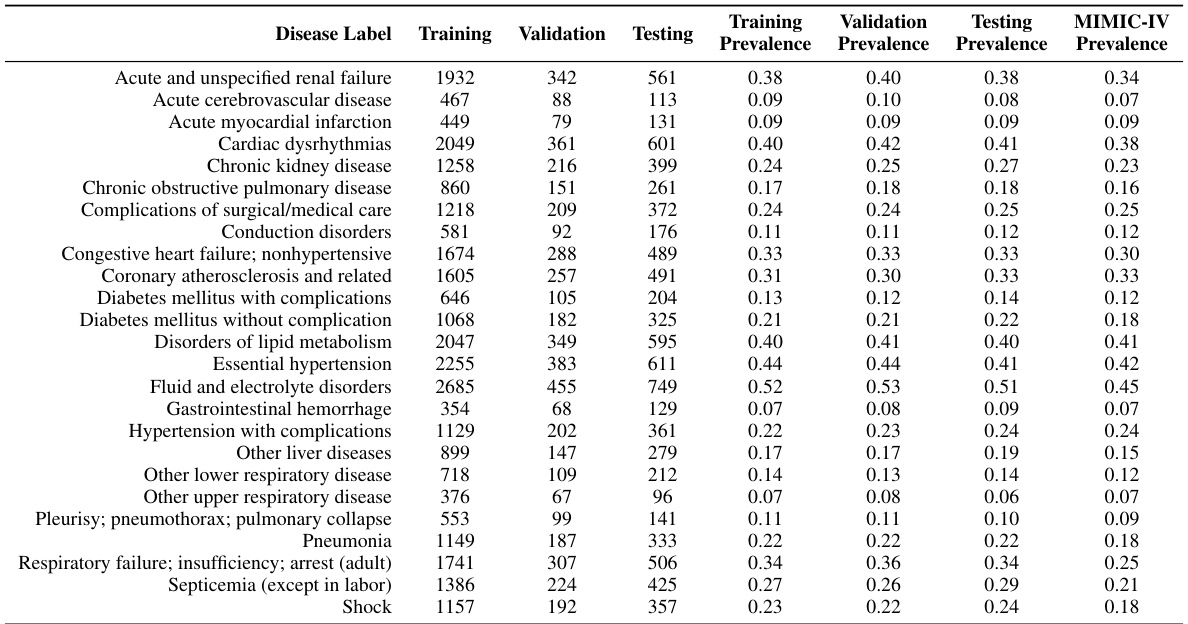
This table shows the number of samples used for training, validation, and testing in the two stages of the proposed model: the Latent Diffusion Model (LDM) stage and the prediction stage. The LDM stage uses image pairs for training, while the prediction stage uses complete EHR data and the generated latent CXR image. The split is done by patient identifiers to avoid overlaps.

This table shows the distribution of positive and negative cases in the training, validation, and test sets used for the in-hospital mortality prediction task. The ratio of negative to positive cases is also provided for each set, showing a relatively consistent imbalance across all sets.
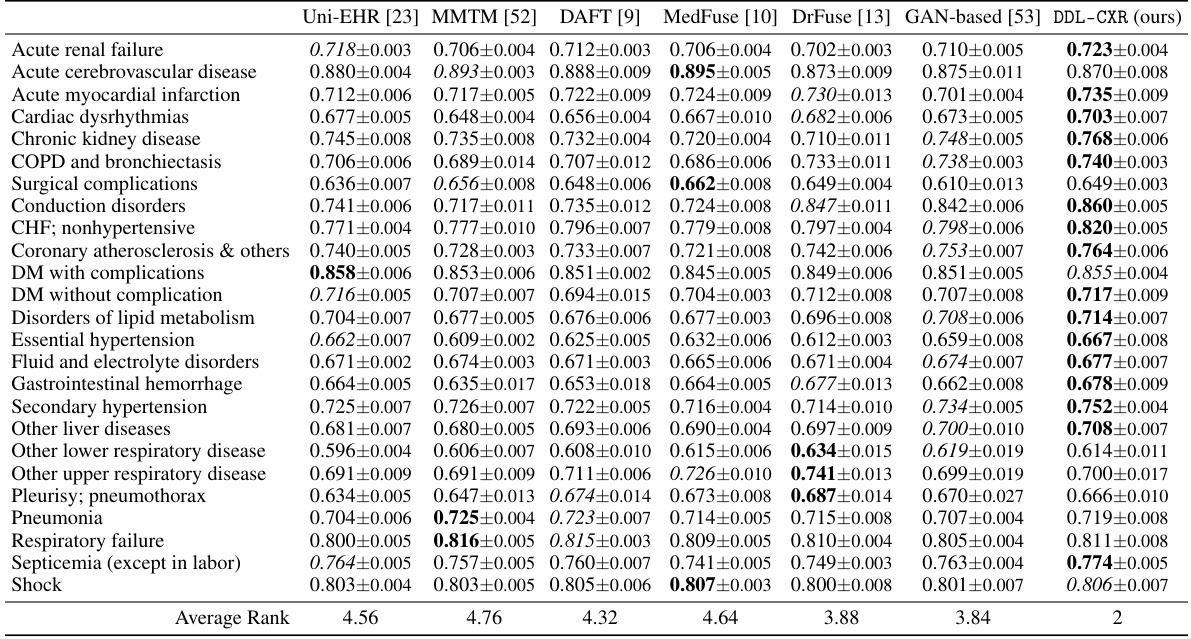
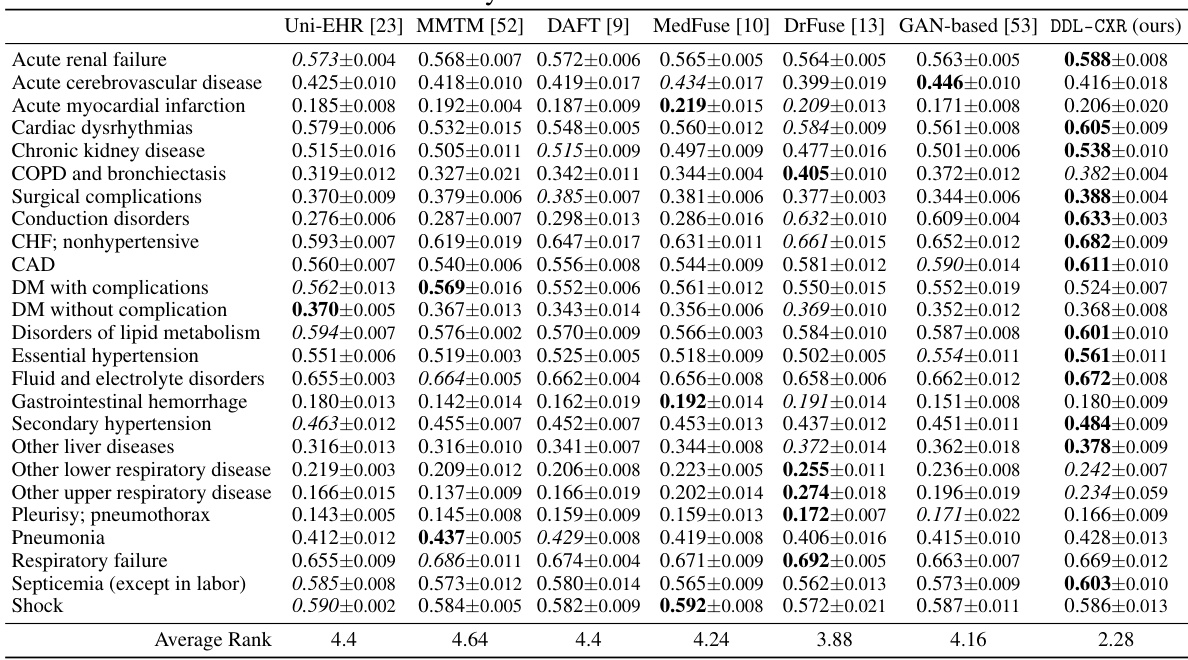
This table presents the AUPRC (Area Under the Precision-Recall Curve) scores for predicting each of the 25 disease phenotypes. The AUPRC is a metric that assesses the performance of a classification model, particularly useful when dealing with imbalanced datasets (like medical datasets). The table compares the performance of DDL-CXR against several other baseline methods for each phenotype, demonstrating DDL-CXR’s superiority across the board by achieving the highest average rank. More detailed results, including AUROC scores, are available in the Appendix.

This table presents the performance comparison of DDL-CXR against several baseline methods for two clinical prediction tasks: phenotype classification and mortality prediction. The Area Under the Precision-Recall Curve (AUPRC) and Area Under the Receiver Operating Characteristic (AUROC) scores are reported for each task and each method, demonstrating DDL-CXR’s superior performance.
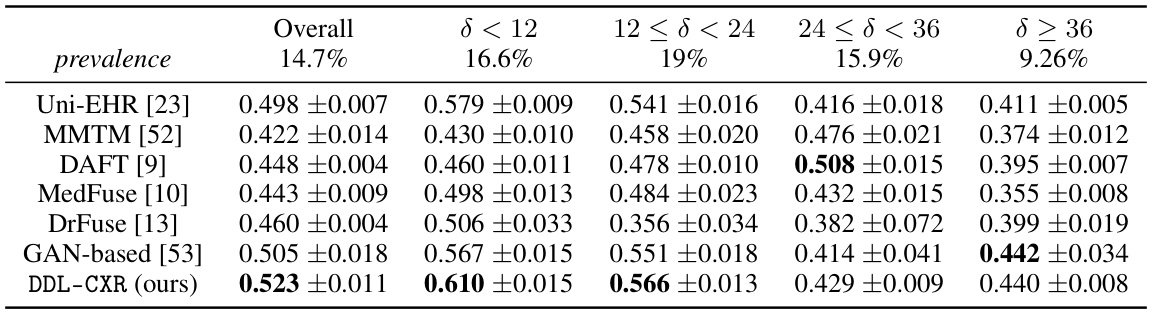
This table presents the Area Under the Receiver Operating Characteristic (AUROC) scores for mortality prediction using different methods. It breaks down the performance based on the time interval (δ) between the prediction time and the last available chest X-ray (CXR) image. The table shows that DDL-CXR consistently outperforms other methods across different time intervals, highlighting its ability to handle the asynchronicity issue in multimodal data. Appendix B.1 provides additional results using the Area Under the Precision-Recall Curve (AUPRC).
This table presents the performance comparison of DDL-CXR against several baseline models for two clinical prediction tasks: phenotype classification and mortality prediction. The performance is evaluated using two metrics: Area Under the Precision-Recall Curve (AUPRC) and Area Under the Receiver Operating Characteristics (AUROC). The results show that DDL-CXR consistently achieves better AUPRC and AUROC scores across both tasks, indicating its superior performance compared to the existing methods.
This table presents the performance comparison of DDL-CXR against several baseline methods on two clinical prediction tasks: phenotype classification and mortality prediction. The performance is evaluated using two metrics: Area Under the Precision-Recall Curve (AUPRC) and Area Under the Receiver Operating Characteristic curve (AUROC). The results demonstrate that the proposed DDL-CXR model consistently outperforms the baseline methods in terms of both AUPRC and AUROC scores.

This table presents the results of phenotype classification and mortality prediction using different methods. The performance is measured using AUPRC (Area Under the Precision-Recall Curve) and AUROC (Area Under the Receiver Operating Characteristic) scores. The table shows that the proposed method, DDL-CXR, outperforms all other baseline methods in terms of both AUPRC and AUROC for both tasks.

This table presents the performance of different models on phenotype classification and mortality prediction tasks using AUPRC and AUROC scores. The results show that the proposed model, DDL-CXR, outperforms existing methods across both tasks, demonstrating its effectiveness in clinical prediction.
Full paper#