↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Annotating 3D medical images for deep learning is expensive and time-consuming. Active learning (AL), which strategically selects which images need annotation, has shown promise but existing AL methods aren’t optimized for medical image characteristics. Specifically, volume-based AL is inefficient, and the potential benefits of AL combined with weak supervision remain unexplored.
This research proposes a new active learning method. It uses deep metric learning with Coreset to select the most informative slices to annotate for 3D segmentation. By leveraging the natural groupings within medical images (e.g., slices from the same patient), the method learns a metric that highlights relevant differences for training. Experiments show that this new method significantly outperforms existing methods across four datasets, achieving superior results, even with limited annotations. This is particularly valuable in medical imaging, where annotation budgets are often tight.
Key Takeaways#
Why does it matter?#
This paper is important because it addresses the critical issue of high annotation costs in 3D medical image segmentation, a major bottleneck in deep learning applications. By introducing a novel approach that combines deep metric learning with Coreset for slice-based active learning, it offers a more efficient and cost-effective solution that outperforms existing methods. This opens avenues for applying deep learning to medical image analysis where data annotation is expensive and time-consuming.
Visual Insights#

This figure illustrates the active learning pipeline used in the paper. It shows how unlabeled samples are processed using a contrastive learning-based encoder to create embeddings in a metric space. These embeddings are then used with Coreset to select the most informative samples for labeling, which are then added to the labeled dataset. This iterative process continues until a desired level of annotation is reached.
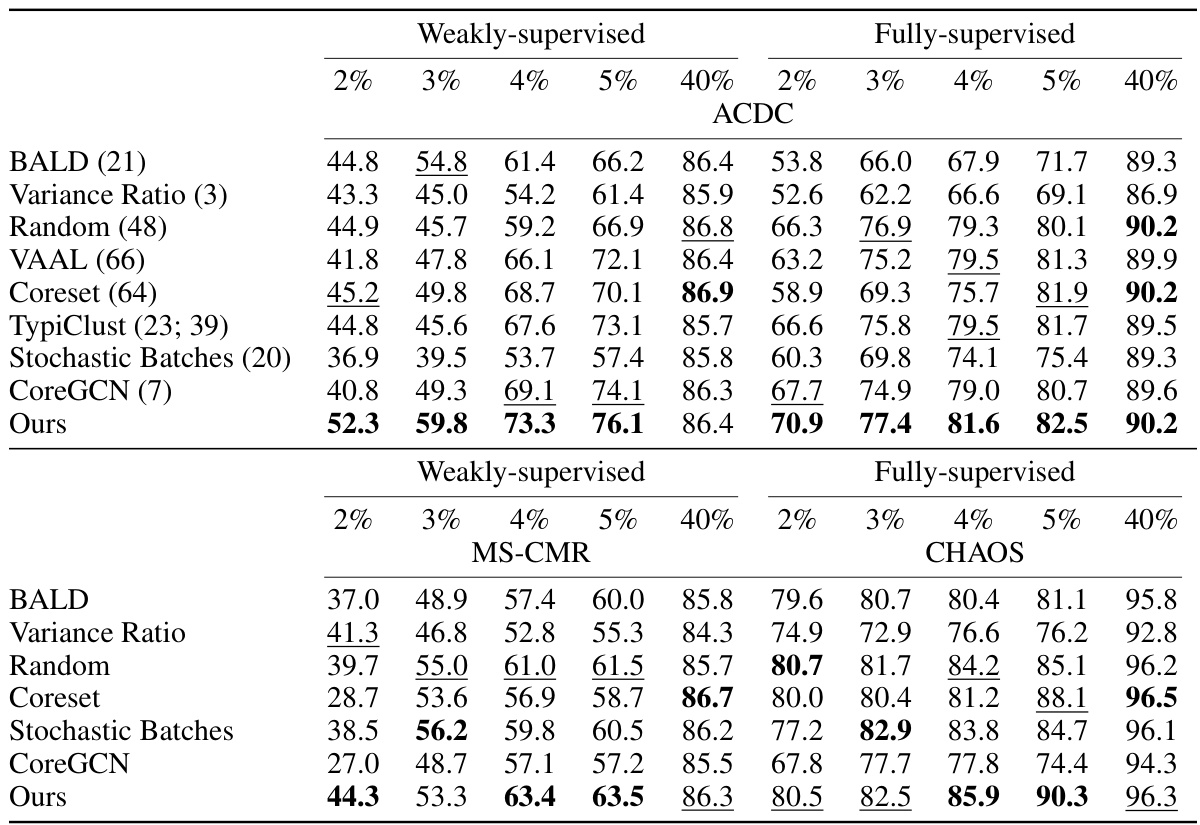
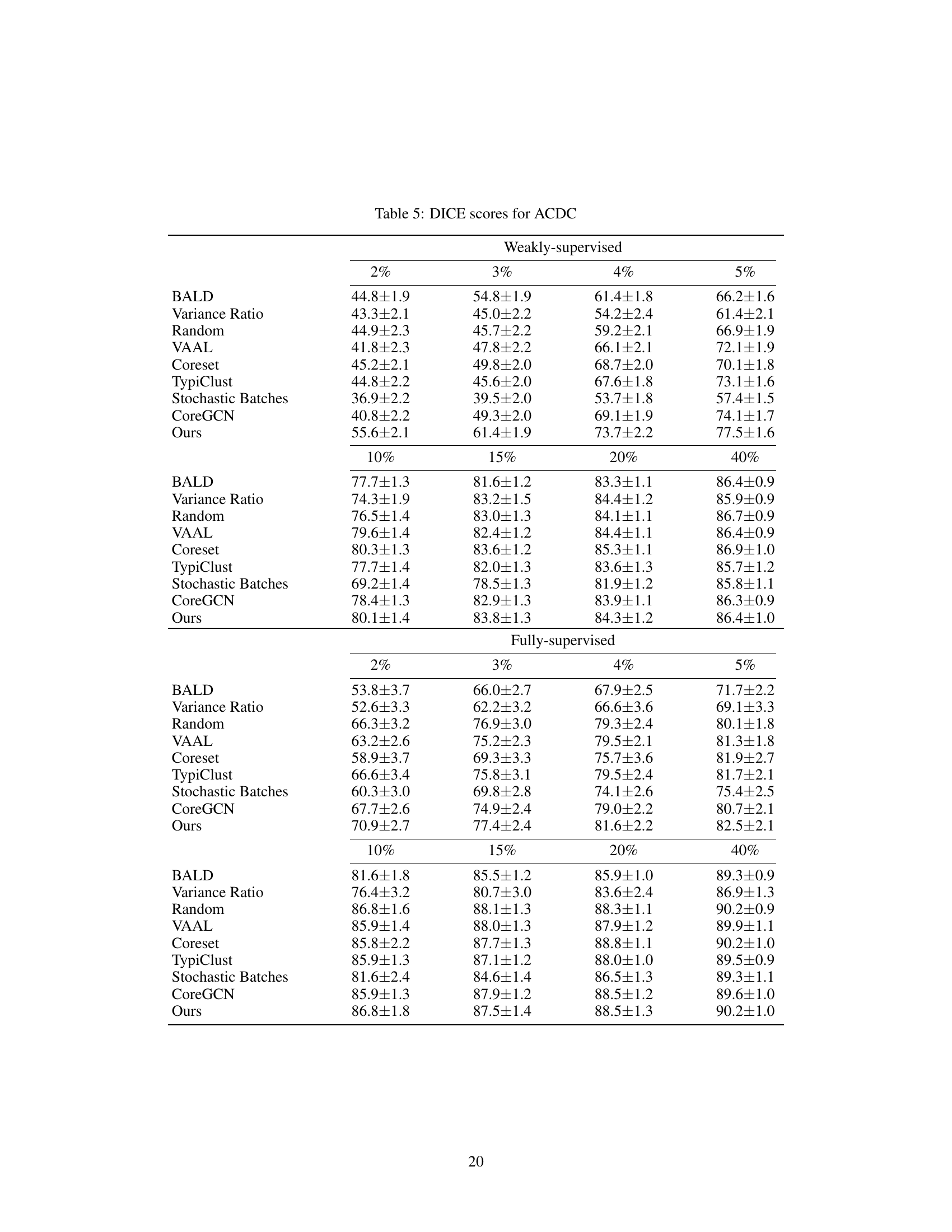
This table presents the DICE scores achieved by different active learning methods on three datasets: ACDC, MS-CMR, and CHAOS. The results are broken down by the percentage of annotation used (2%, 3%, 4%, 5%, and 40%) and whether weak or full supervision was used. The table allows comparison of the performance of the proposed method against several baselines across various annotation budgets and supervision levels.
In-depth insights#
Deep Metric Coreset#
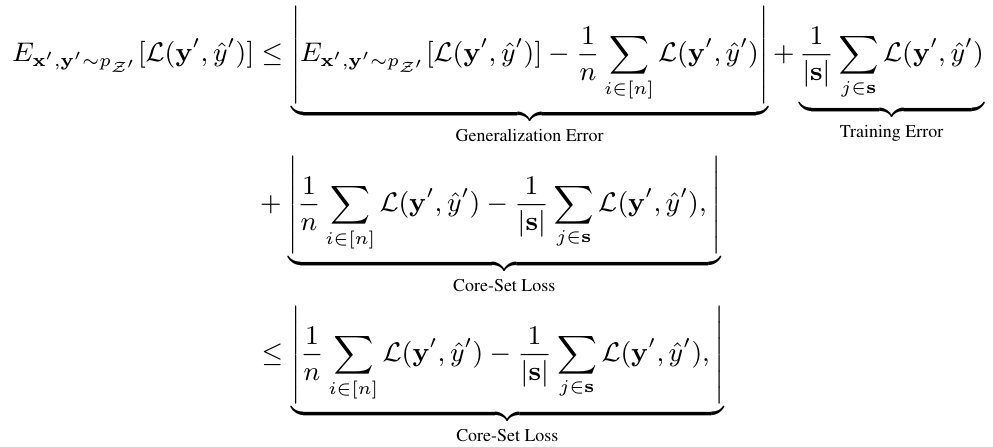
A hypothetical ‘Deep Metric Coreset’ method would combine the strengths of deep metric learning and coreset construction for efficient and effective data summarization. Deep metric learning would learn a data embedding where semantically similar data points are closer together, improving coreset selection. Coreset construction would aim to select a small subset of representative data points. By integrating deep metric learning, the coreset selection could prioritize samples that maximize diversity and informativeness, resulting in a smaller coreset that better captures the full dataset. This would be particularly useful for large datasets where traditional coreset methods might fail to achieve an adequate representation. The method could improve active learning by selecting the most informative samples to label. However, challenges remain in optimizing the coreset size and ensuring that the deep metric learning doesn’t introduce bias or computational overhead.
Slice-based AL#
Slice-based active learning (AL) offers a compelling approach to reduce the annotation burden in 3D medical image segmentation. Traditional volume-based AL methods can be computationally expensive and time-consuming. Slice-based AL addresses this by focusing on selecting individual slices for annotation rather than entire volumes. This targeted approach significantly reduces annotation costs and time, making it more practical for real-world applications. The core idea is to leverage inherent data groupings within medical images (e.g., slices from the same patient or volume tend to exhibit similar characteristics) to intelligently guide the selection process, ensuring diversity and maximizing information gain from each labeled slice. Effective slice-based AL methods require careful consideration of the sampling strategy to avoid biasing the model towards specific regions or characteristics. A thoughtful algorithm should balance exploration and exploitation, considering both uncertainty and diversity within the chosen metric space, to ensure optimal performance with minimal annotation effort. The key advantage is the significant cost reduction compared to volume-based AL, making it a more sustainable approach for 3D medical image segmentation projects. The effectiveness of slice-based AL relies on a robust metric that accurately captures the similarity or dissimilarity between slices.
Weak Supervision#
Weak supervision, in the context of 3D medical image segmentation, offers a practical solution to the significant annotation burden associated with deep learning models. Instead of relying on expensive, time-consuming, pixel-perfect annotations, weak supervision leverages simpler, readily available annotations like scribbles, bounding boxes, or even points. This approach significantly reduces the cost and effort of data labeling, making deep learning more accessible in medical settings. The combination of weak supervision with active learning is particularly powerful, as it allows the model to iteratively request only the most informative annotations from human experts, further optimizing the annotation process. The effectiveness of this strategy is highlighted by the paper’s results, demonstrating that a model trained with weak supervision under an active learning framework achieves performance comparable to, and sometimes surpassing, fully supervised models, particularly with limited annotation budgets. This underscores the importance of exploring weak supervision techniques in domains with limited data and resources, such as medical imaging.
Medical Image AL#
Active learning (AL) in medical image analysis is crucial due to the high cost and time involved in manual annotation of large datasets. The core challenge is to strategically select the most informative samples for annotation, maximizing model performance with minimal human effort. Slice-based AL offers a potential cost reduction compared to volume-based approaches, focusing on individual slices rather than the entire 3D volume. The integration of deep metric learning further enhances AL by learning a distance metric in the feature space that prioritizes diversity and relevance, thereby improving the selection of samples for annotation. This approach is particularly beneficial when data inherently has groupings (e.g., slices from the same patient). The use of weakly supervised methods can further reduce annotation burden, and combining this with AL shows strong promise. However, challenges remain in balancing computational efficiency with model accuracy and generalizability across diverse medical imaging modalities. Future work might focus on robust group-based contrastive learning strategies and exploration of how to optimize AL under various weak supervision conditions.
Future of AL-Seg#
The future of AL-Seg (Active Learning in 3D Segmentation) is promising, driven by the need to reduce the substantial annotation burden in medical imaging. Further research should focus on more sophisticated metric learning techniques that can better capture the complex relationships between image features and the desired segmentation outcomes. Developing methods for handling diverse data modalities (e.g., combining MRI and CT scans) would greatly enhance the generalizability of AL-Seg. Exploring novel acquisition functions which move beyond simple uncertainty estimates towards more robust and informative sampling strategies is crucial. Efficiently integrating weak supervision with AL-Seg will significantly impact cost-effectiveness, requiring advanced methods to handle noisy or incomplete labels effectively. Finally, rigorous evaluation methodologies are needed to quantify the gains from AL-Seg in various clinical settings, and address the inherent biases in medical datasets.
More visual insights#
More on figures

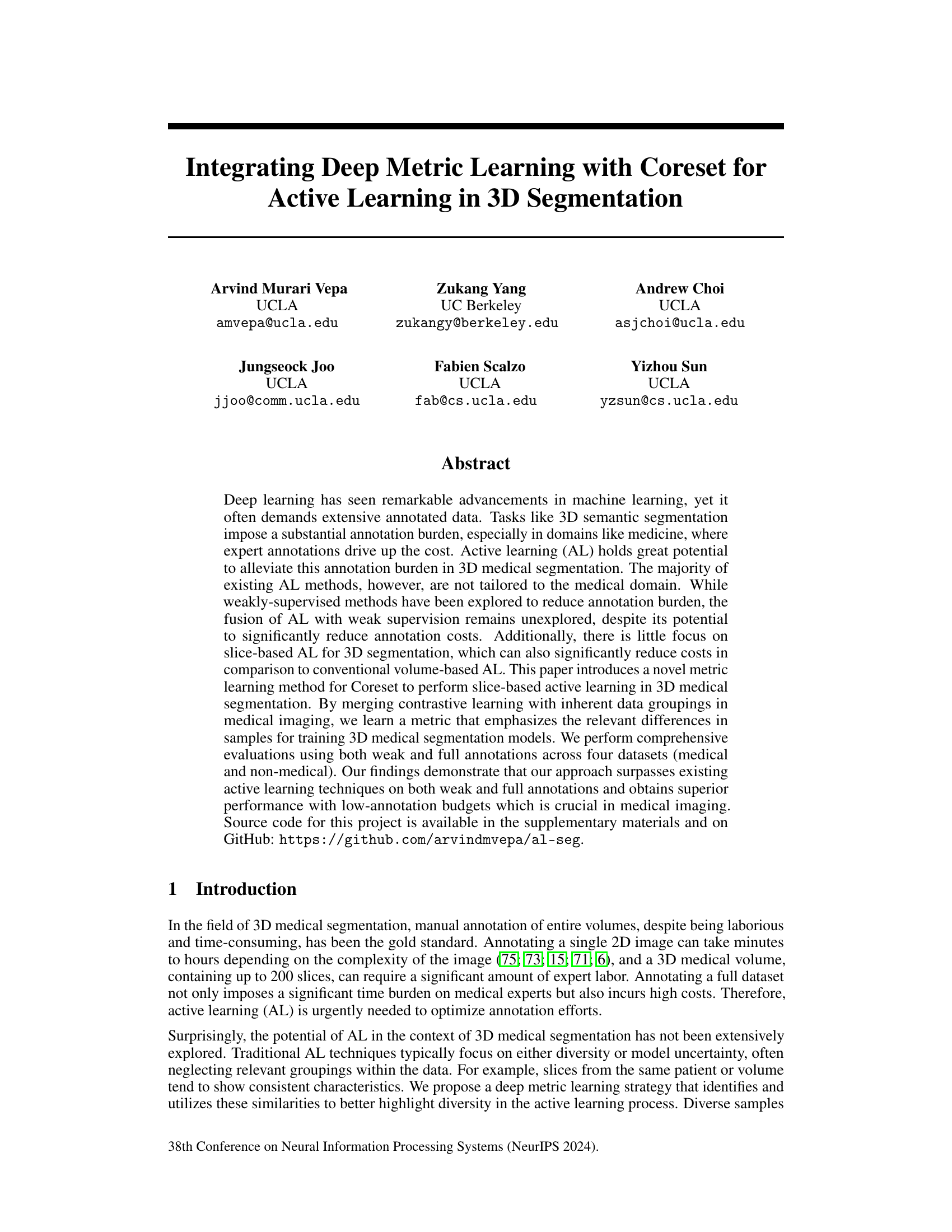
This figure shows a comparison of the model performance (Dice score) against the annotation time (percentage of fully labeled data) for different active learning methods. It compares weakly supervised 2D slices, fully supervised 2D slices using the proposed method, and random sampling of fully supervised 3D volumes on the ACDC dataset. The result shows that the proposed method achieves higher Dice score using weakly supervised 2D slices with significantly less annotation time than random sampling using 3D volumes.
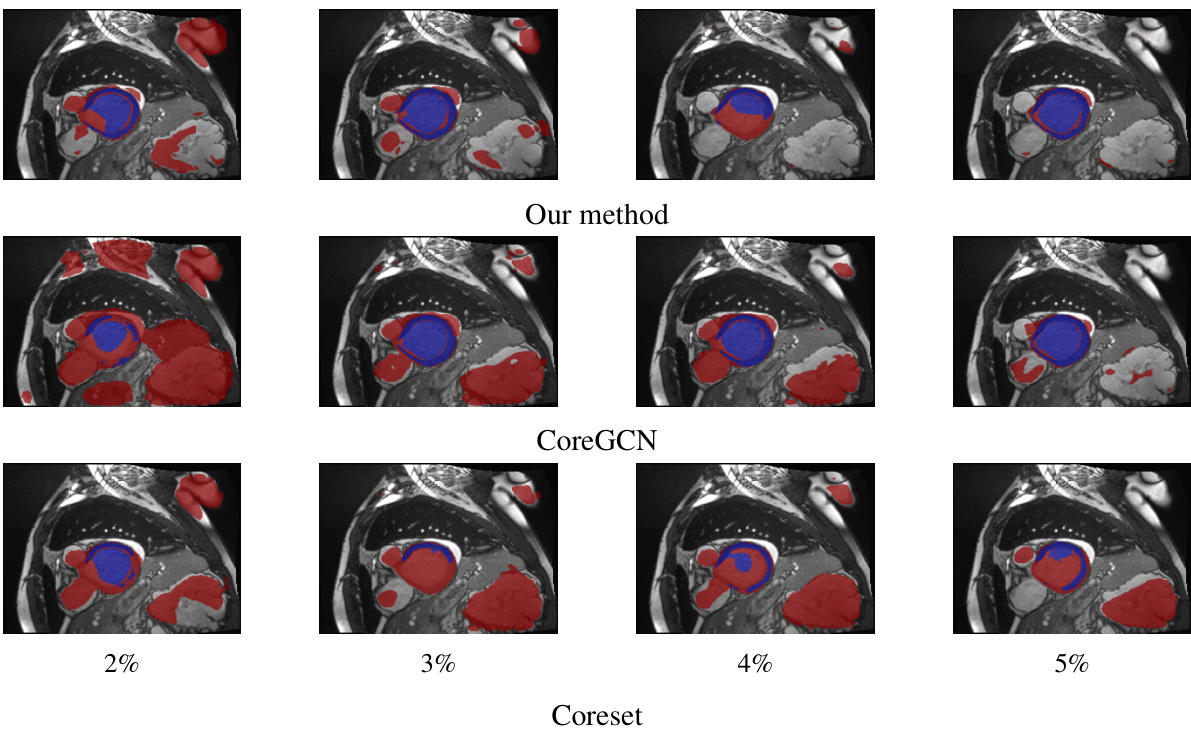
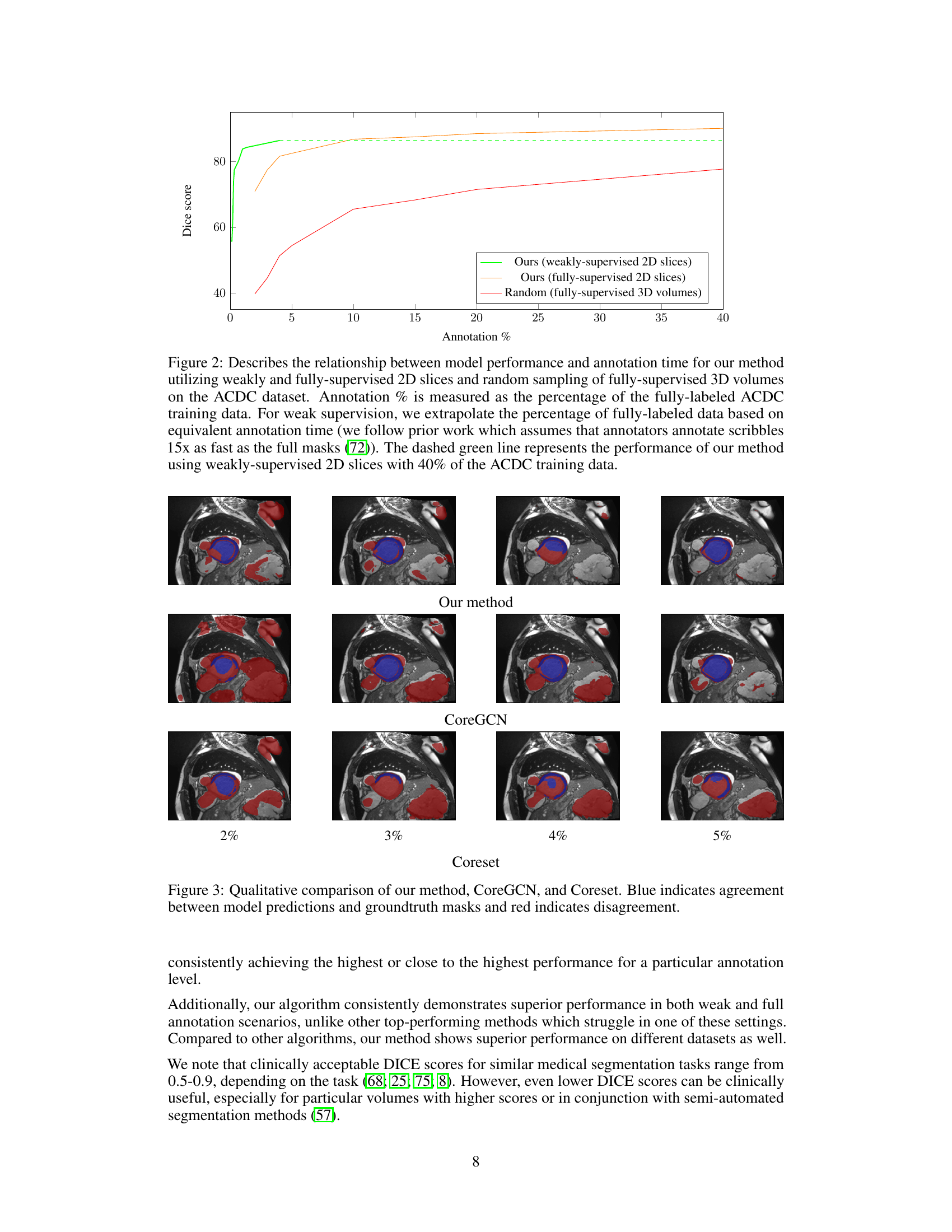
This figure shows a qualitative comparison of the segmentation results obtained by three different active learning methods: the proposed method, CoreGCN, and Coreset. For each method, segmentation results at annotation percentages of 2%, 3%, 4%, and 5% are shown. Blue coloration indicates areas where the model’s prediction and the ground truth mask agree, while red shows disagreement. The visual comparison allows for assessment of the qualitative differences in performance between methods at various annotation budgets.

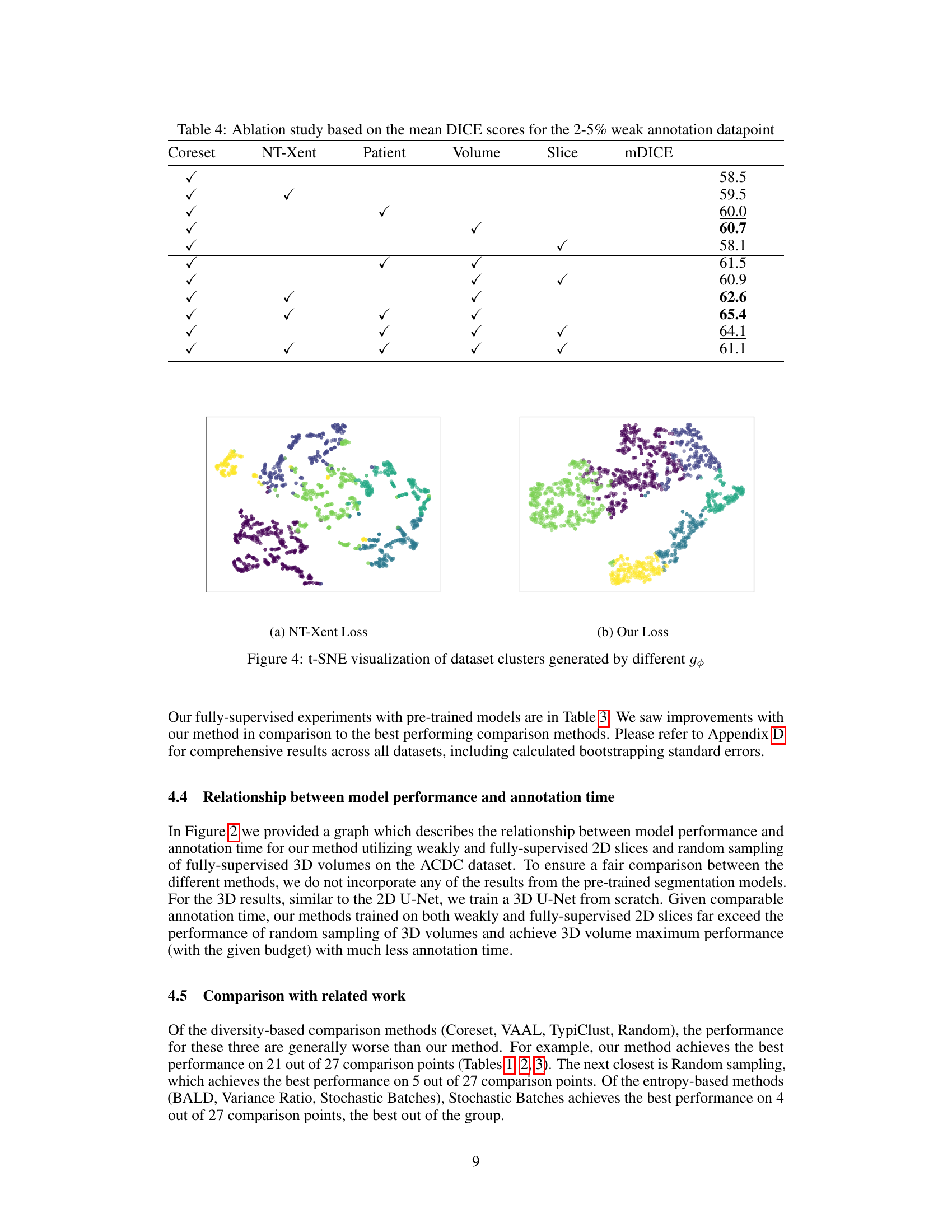
This figure visualizes the effect of different loss functions on the learned feature representations. The left panel shows the clusters generated using the NT-Xent loss, while the right panel shows the clusters generated using the proposed Group-based Contrastive Learning (GCL) loss. The t-SNE visualization helps to understand the quality of cluster separation and cohesion achieved by each loss function. The GCL loss shows better separation and cohesion, indicating that it learns more informative and relevant features for the Coreset.
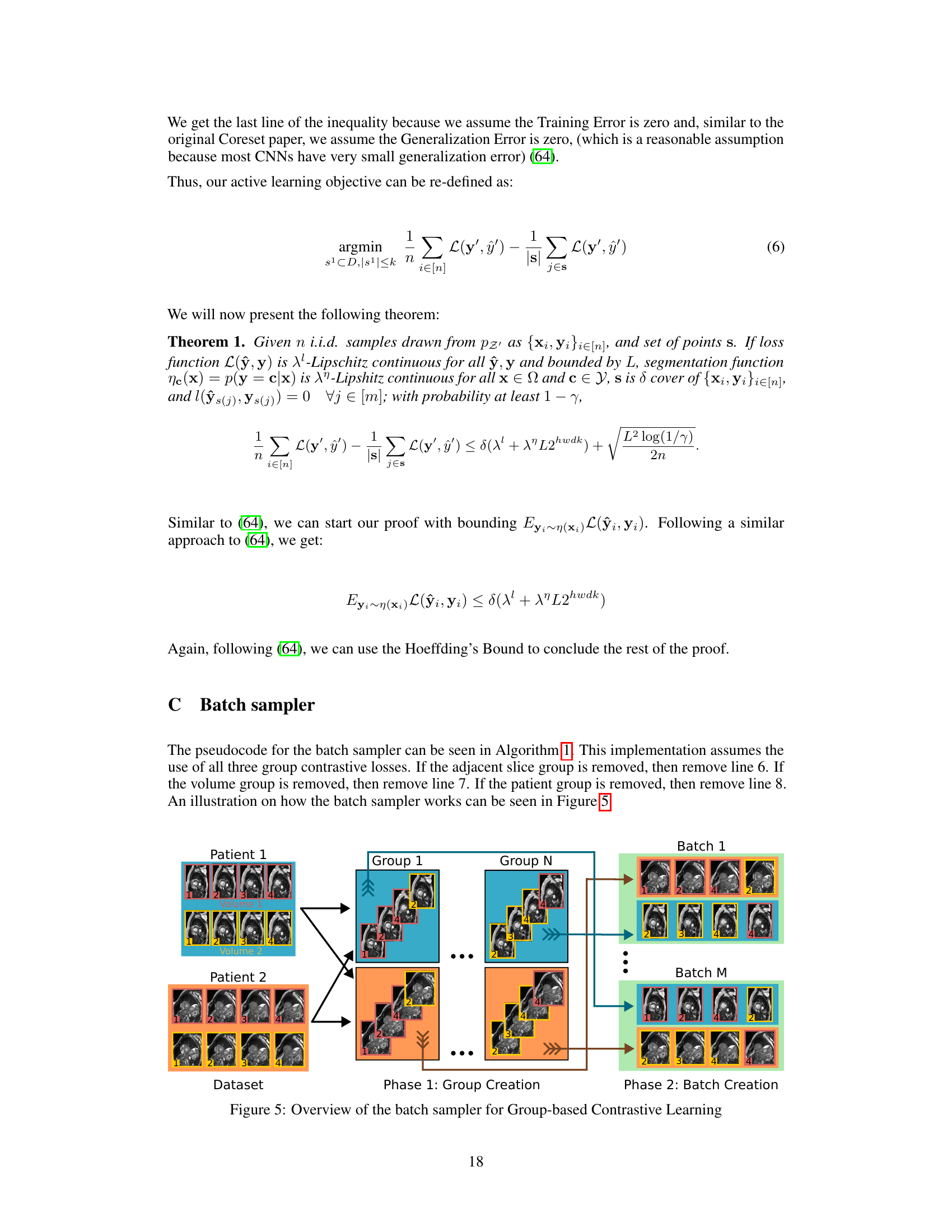
This figure shows the pipeline of the proposed active learning method. It starts with unlabeled samples and uses Coreset to select a subset of samples. These samples are then used for contrastive learning to obtain embeddings. The embeddings are used to calculate distances between samples in the metric space, which helps select the most informative samples. Finally, these newly labeled samples are added to the training set to improve the segmentation model.

This figure illustrates the active learning pipeline used in the paper. It shows how unlabeled samples are processed using contrastive learning to generate embeddings in a metric space. These embeddings are used with the Coreset algorithm to select the most informative samples for labeling. Newly labeled samples are then added to the training set, improving the model’s performance. The pipeline iteratively selects and labels samples until a desired budget or performance threshold is reached.
More on tables
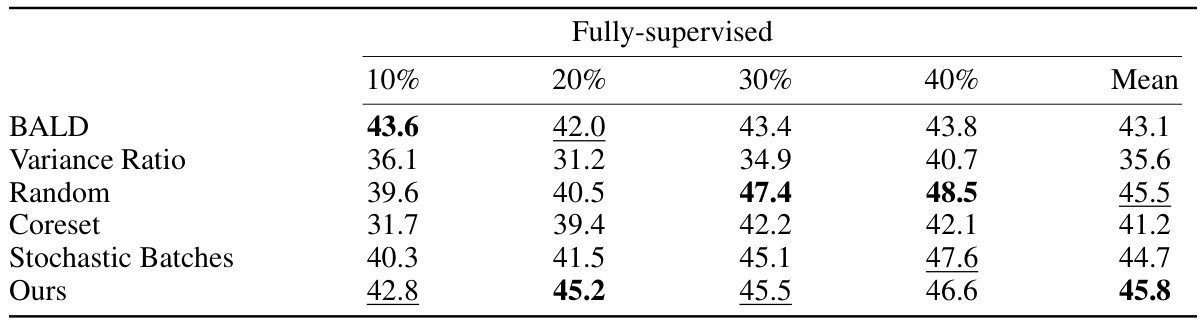
This table presents the DICE scores achieved by different active learning methods on the DAVIS dataset using fully supervised learning. The methods are compared across various annotation percentages (10%, 20%, 30%, 40%), showing the average DICE score obtained for each method at each annotation level. The table helps demonstrate the relative performance of each method in a fully-supervised setting with varying amounts of annotated data.

This table presents the mean Dice Similarity Coefficient (DSC) scores achieved by different active learning methods on three datasets (ACDC, CHAOS, and DAVIS) when using pre-trained segmentation models. The scores represent the average performance across all annotation levels, providing a comprehensive comparison of the methods’ performance with pretrained models. Higher DSC scores indicate better segmentation accuracy.
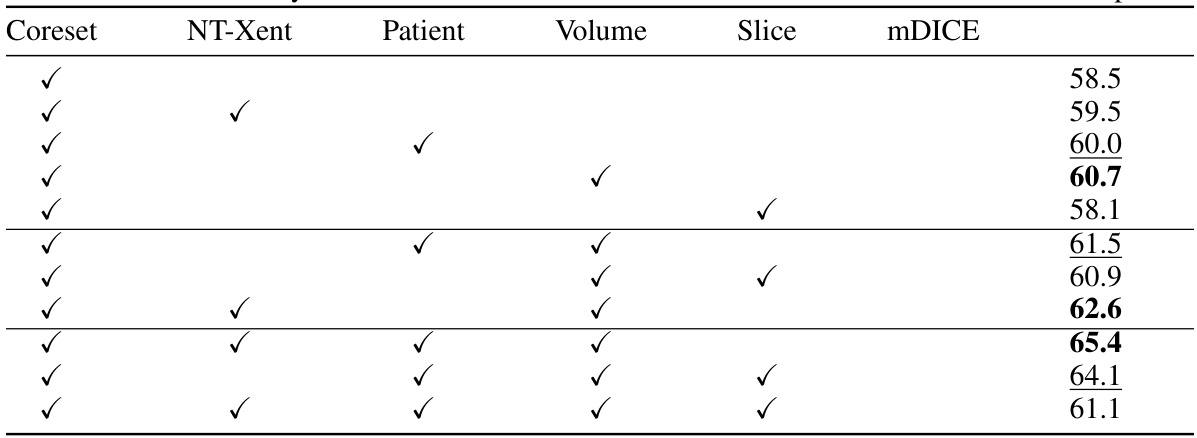
This table presents the results of an ablation study conducted to evaluate the impact of different combinations of contrastive losses on the model’s performance. The study focuses on the mean DICE scores obtained for a weak annotation datapoint (2-5%). The table shows how different combinations of NT-Xent loss (a contrastive learning method), patient group loss, volume group loss, and slice group loss affect the overall performance (mDICE). Each row represents a different model configuration indicated by checkmarks in the relevant columns. The mDICE score is a measure of the model’s segmentation accuracy.
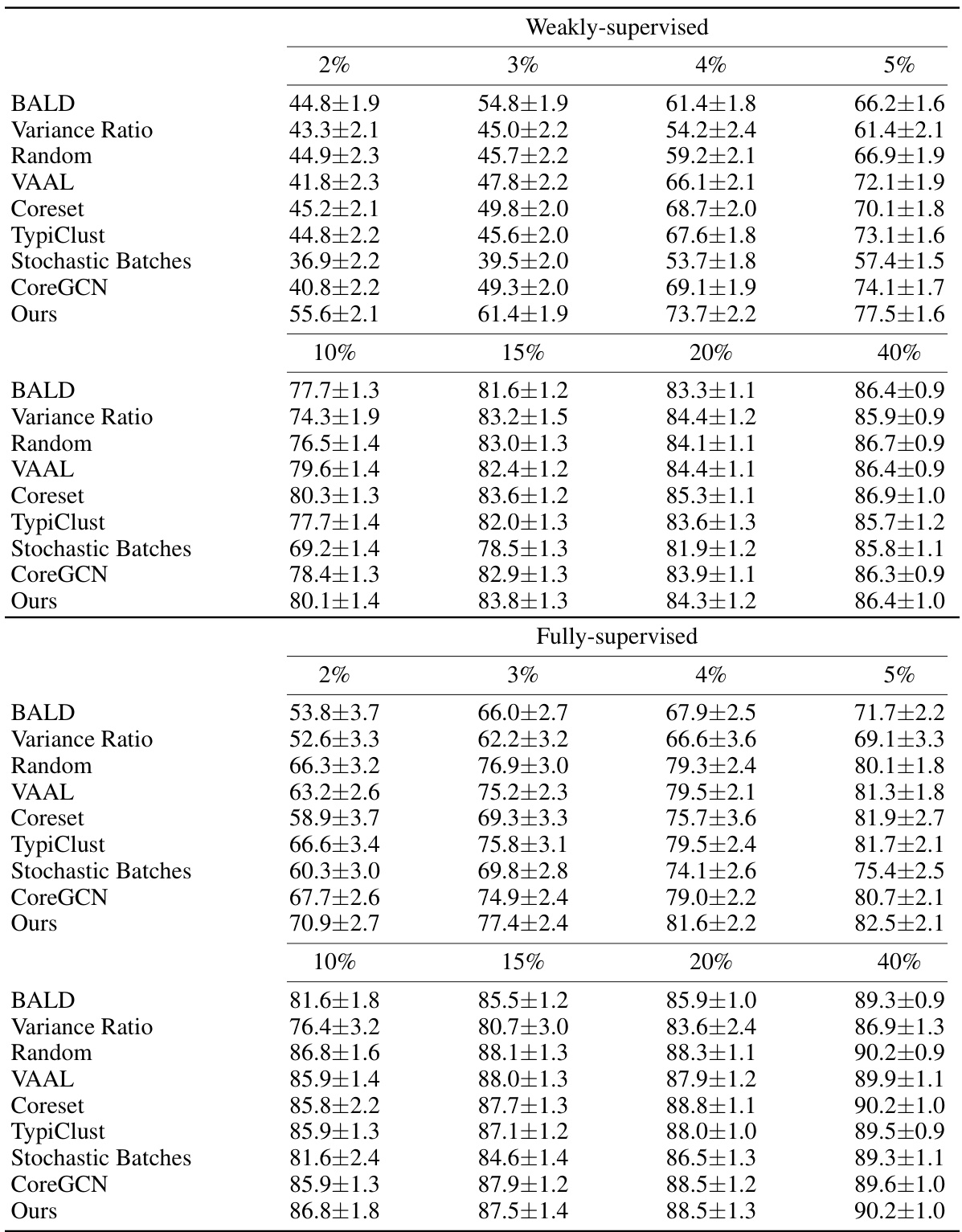
This table presents the DICE scores achieved by different active learning methods on three datasets: ACDC, MS-CMR, and CHAOS. Results are shown for both weakly-supervised and fully-supervised settings, across various annotation percentages (2%, 3%, 4%, 5%, and 40%). The table allows for a comparison of the performance of different algorithms in low annotation budget scenarios and at higher annotation levels.
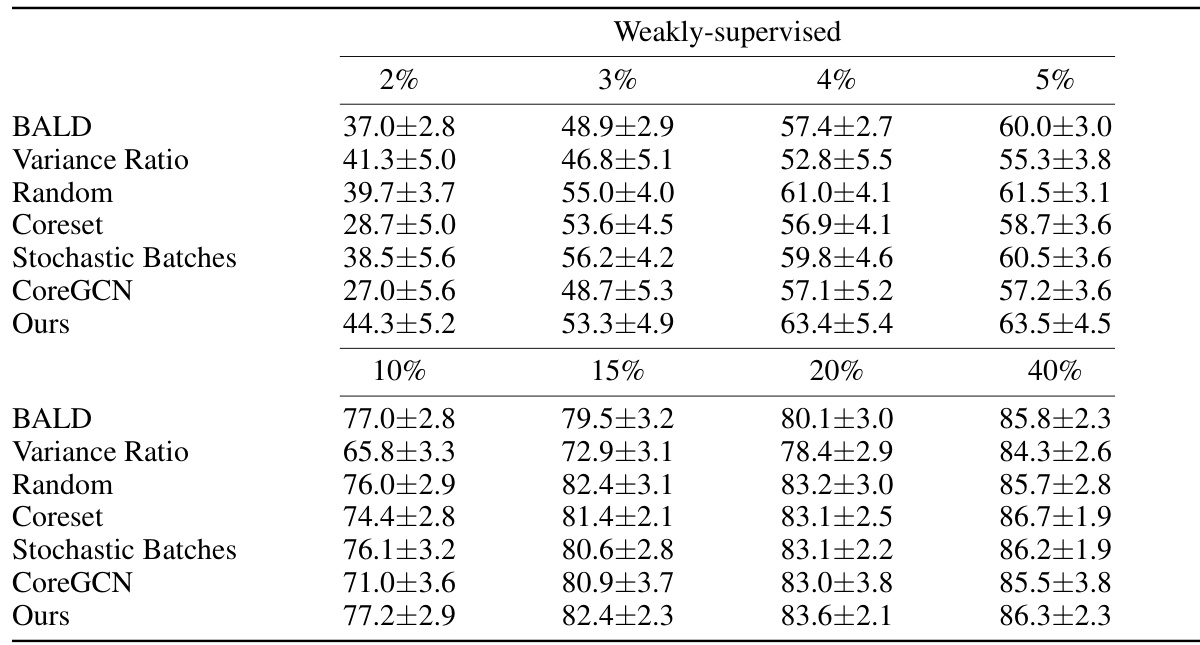
This table presents the Dice Similarity Coefficient (DSC) scores achieved by different active learning methods on three datasets: ACDC, MS-CMR, and CHAOS. The scores are shown for both weakly-supervised and fully-supervised settings, across various annotation percentages (2%, 3%, 4%, 5%, and 40%). The table allows for a comparison of the performance of different active learning strategies under different levels of annotation.
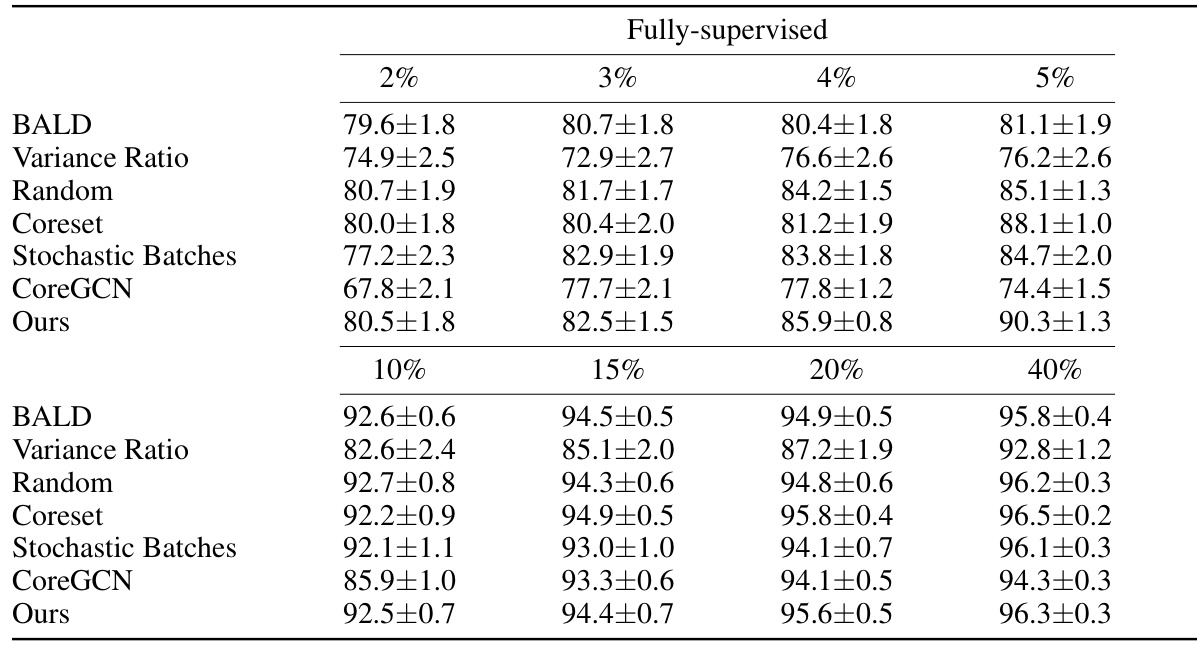
This table presents the DICE scores achieved by various active learning methods across three different datasets (ACDC, MS-CMR, and CHAOS) under both weakly and fully supervised settings. The results are shown for different annotation percentages (2%, 3%, 4%, 5%, and 40%), allowing comparison of performance under varying annotation budgets. This comparison helps assess the efficiency of each active learning technique in improving segmentation accuracy with limited annotations.
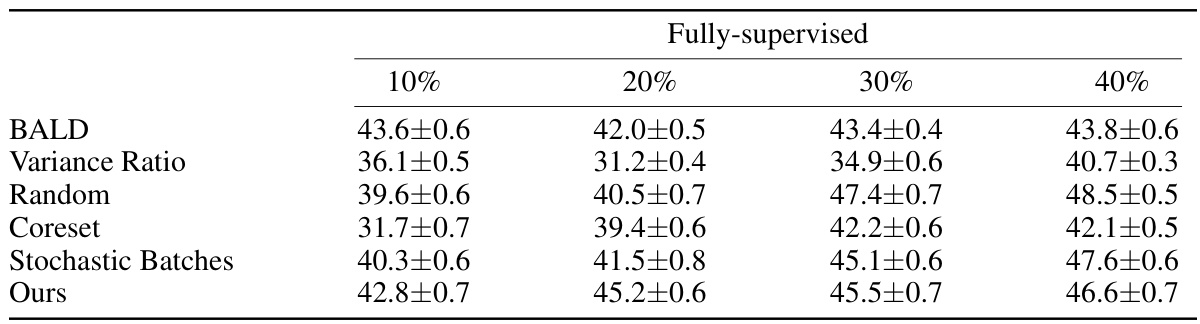
This table presents the Dice Similarity Coefficient (DSC) scores achieved by different active learning methods on the DAVIS dataset. The DSC is a common metric for evaluating the performance of segmentation models, representing the overlap between the predicted and ground truth segmentations. The table shows the results for both weakly and fully supervised settings and for various annotation percentages (10%, 20%, 30%, 40%). Higher DSC scores indicate better segmentation accuracy.

This table presents the DICE scores achieved by different active learning methods on the ACDC dataset when using pre-trained segmentation models. The results are shown for various annotation percentages (1%, 2%, 3%, 4%, and 5%), comparing the performance of Random sampling, Stochastic Batches, Coreset, and the authors’ proposed method.

This table presents the performance of different active learning methods on the CHAOS dataset using pretrained segmentation models. The DICE scores are shown for different annotation percentages (1%, 2%, 3%, 4%, and 5%), comparing the performance of Random sampling, Stochastic Batches, Coreset, and the proposed method.
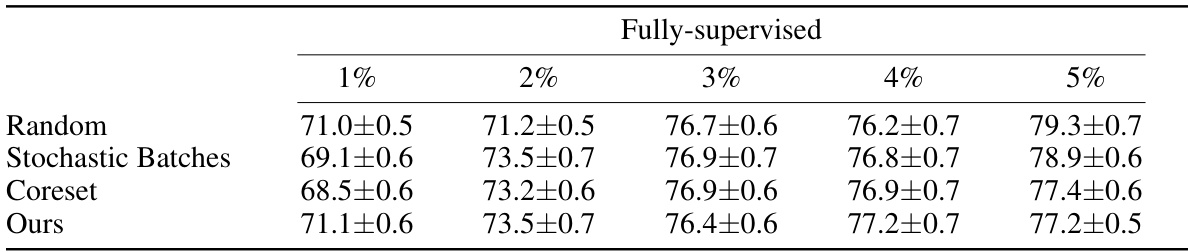
This table presents the Dice Similarity Coefficient (DSC) scores achieved by different active learning methods on the ACDC dataset using pretrained segmentation models. The results are broken down by the percentage of fully-supervised annotations used (1%, 2%, 3%, 4%, and 5%). The methods compared include Random sampling, Stochastic Batches, Coreset, and the authors’ proposed method. Higher DSC scores indicate better segmentation performance.
Full paper#