↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Many medical image datasets suffer from noisy labels, significantly hindering the accuracy of deep learning models. Traditional methods for handling noisy labels often disregard the rich features provided by pre-trained vision foundation models (VFMs). This paper tackles this challenge by proposing CUFIT, a curriculum fine-tuning strategy that combines the benefits of linear probing (which is robust to noise) with the power of adapter fine-tuning. Existing clean sample selection methods suffer from the limitation of training from scratch; CUFIT, on the other hand, intelligently utilizes the knowledge embedded in VFMs.
CUFIT employs a three-module training process: Linear Probing Module, Intermediate Adapter Module, and Last Adapter Module. This curriculum design systematically filters noisy samples, progressively improving the quality of training data for the adapters. The results show CUFIT outperforms previous methods across multiple benchmark datasets by a significant margin (up to 5.8% improvement at a 40% noise rate). This suggests that CUFIT effectively addresses the problem of noisy labels in medical image classification while achieving state-of-the-art performance.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image classification and machine learning. It directly addresses the challenge of noisy labels, a pervasive issue impacting model accuracy. By introducing CUFIT, it offers a novel, effective approach, using pre-trained models, significantly enhancing the robustness and performance of medical image analysis. This opens up new avenues for improving the reliability of AI-based medical diagnoses and expanding the application of vision foundation models in challenging data scenarios. The findings also have implications for other fields dealing with noisy datasets and curriculum learning techniques.
Visual Insights#
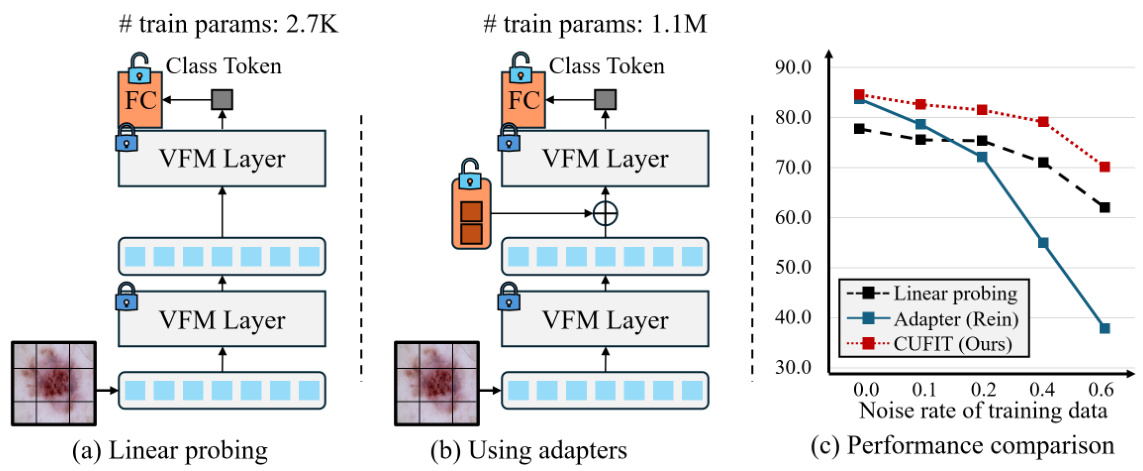
This figure illustrates the concepts of linear probing and adapter usage in the context of vision foundation models (VFMs) for medical image classification with noisy labels. Panel (a) shows linear probing where only a fully connected layer is trained, leaving the VFM weights frozen. Panel (b) shows adapter usage, where trainable adapters are added to the VFM, while the VFM weights remain frozen. Panel (c) compares the performance of linear probing and adapter usage against the proposed CUFIT method on a simulated noisy dataset, highlighting the robustness of linear probing to noise and the superior performance of adapters in clean data scenarios. CUFIT aims to combine the benefits of both approaches.

This table presents the average test accuracy achieved by different methods (Full-training, Linear probing, Rein, Co-teaching, JoCor, CoDis, and CUFIT) on four medical image datasets (HAM10000, APTOS-2019, BloodMnist, and OrgancMnist) under various simulated noise levels (0.1, 0.2, 0.4, and 0.6). The results highlight the performance of CUFIT in comparison to other methods, showcasing its robustness to noisy labels.
In-depth insights#
Noisy Label Handling#
Handling noisy labels is crucial for reliable machine learning, especially in medical image analysis where obtaining perfectly labeled data is challenging. This paper tackles this issue by introducing a curriculum fine-tuning approach, CUFIT, that leverages pre-trained vision foundation models (VFMs). The strategy cleverly exploits the robustness of linear probing on VFMs to initial noisy data, gradually introducing adapter modules for improved performance. This curriculum-based training ensures that the model learns from increasingly clean samples, enhancing the overall accuracy while mitigating the impact of noisy labels. The core strength of this method lies in its effective integration of VFMs and a parameter-efficient fine-tuning strategy that minimizes overfitting to noisy data. The empirical results highlight CUFIT’s superior performance over traditional methods, demonstrating the potential of this approach for medical image analysis and potentially other domains suffering from label noise issues.
VFM Curriculum#
A ‘VFM Curriculum’ in a research paper would likely detail a structured learning approach for Vision Foundation Models (VFMs). This likely involves a staged training process, perhaps starting with linear probing of the pre-trained VFM, to leverage its inherent robustness to noisy data. Subsequently, the curriculum might introduce adapter modules for fine-tuning, beginning with simpler tasks or cleaner data subsets. This would gradually increase the complexity or noise level, allowing the model to progressively adapt without catastrophic forgetting or overfitting to noisy labels. The goal is to exploit VFMs’ rich feature representations while mitigating the negative impact of label noise. Curriculum learning could involve methods such as self-training or co-teaching to iteratively refine data selection and model training. The paper would likely emphasize performance improvements over baseline approaches through empirical evaluations on relevant medical image datasets. Robustness to noisy labels would be a crucial aspect, as demonstrated by improvements in metrics like precision, recall and overall classification accuracy.
Adapter Fine-tuning#
Adapter fine-tuning, in the context of large vision foundation models (VFMs), offers a compelling approach to parameter-efficient adaptation. Instead of retraining the entire VFM, which is computationally expensive and risks catastrophic forgetting, adapter modules are inserted into the VFM’s architecture. These adapters contain a small number of trainable parameters, allowing for targeted adjustments to the model’s behavior without affecting the pre-trained weights. This strategy is particularly valuable when dealing with noisy or limited data, as it mitigates the risk of overfitting and allows for a more robust and generalizable model. Curriculum learning could further enhance the effectiveness of adapter fine-tuning by progressively introducing increasingly complex data or tasks, starting with simpler, potentially cleaner data samples selected using a linear probing approach to ensure the adapters learn efficiently and effectively, while avoiding memorization of noise. Careful selection of adapter architecture and placement within the VFM is critical to the success of this method, impacting both computational efficiency and the model’s ability to generalize.
Medical Image Benchmarks#
A robust evaluation of medical image analysis methods necessitates diverse and challenging benchmarks. Dataset selection is critical, considering factors like image quality, annotation accuracy, and class balance. The choice between publicly available datasets (e.g., publicly available chest x-ray images) and privately held data significantly influences generalizability. Performance metrics must align with clinical relevance (e.g., sensitivity and specificity are crucial in medical diagnosis). Quantitative metrics provide objective comparisons, while qualitative analysis offers insights into model strengths and weaknesses. Finally, the reproducibility of benchmark results should be prioritized via detailed methodology descriptions. Addressing these multifaceted elements is crucial for establishing reliable and meaningful comparisons of medical image analysis techniques.
CUFIT Framework#
The CUFIT framework offers a novel approach to medical image classification by leveraging the strengths of Vision Foundation Models (VFMs). It cleverly addresses the challenge of noisy labels, a common problem in medical datasets, through a curriculum learning paradigm. This paradigm consists of three modules: a linear probing module (LPM) for initial robust classification, an intermediate adapter module (IAM) for refining predictions using a subset of clean samples identified by the LPM, and a final adapter module (LAM) for further refinement. This staged approach gradually refines the model’s understanding of the data, while minimizing the negative impact of noisy labels. The framework demonstrates superior performance to existing methods across various medical image benchmarks. The use of VFMs and the curriculum learning approach enables CUFIT to benefit from pre-trained features, enhancing its efficiency and effectiveness. The core contribution lies in the strategic integration of linear probing’s robustness and adapter fine-tuning’s flexibility, overcoming limitations of traditional clean sample selection methods that begin training from scratch.
More visual insights#
More on figures

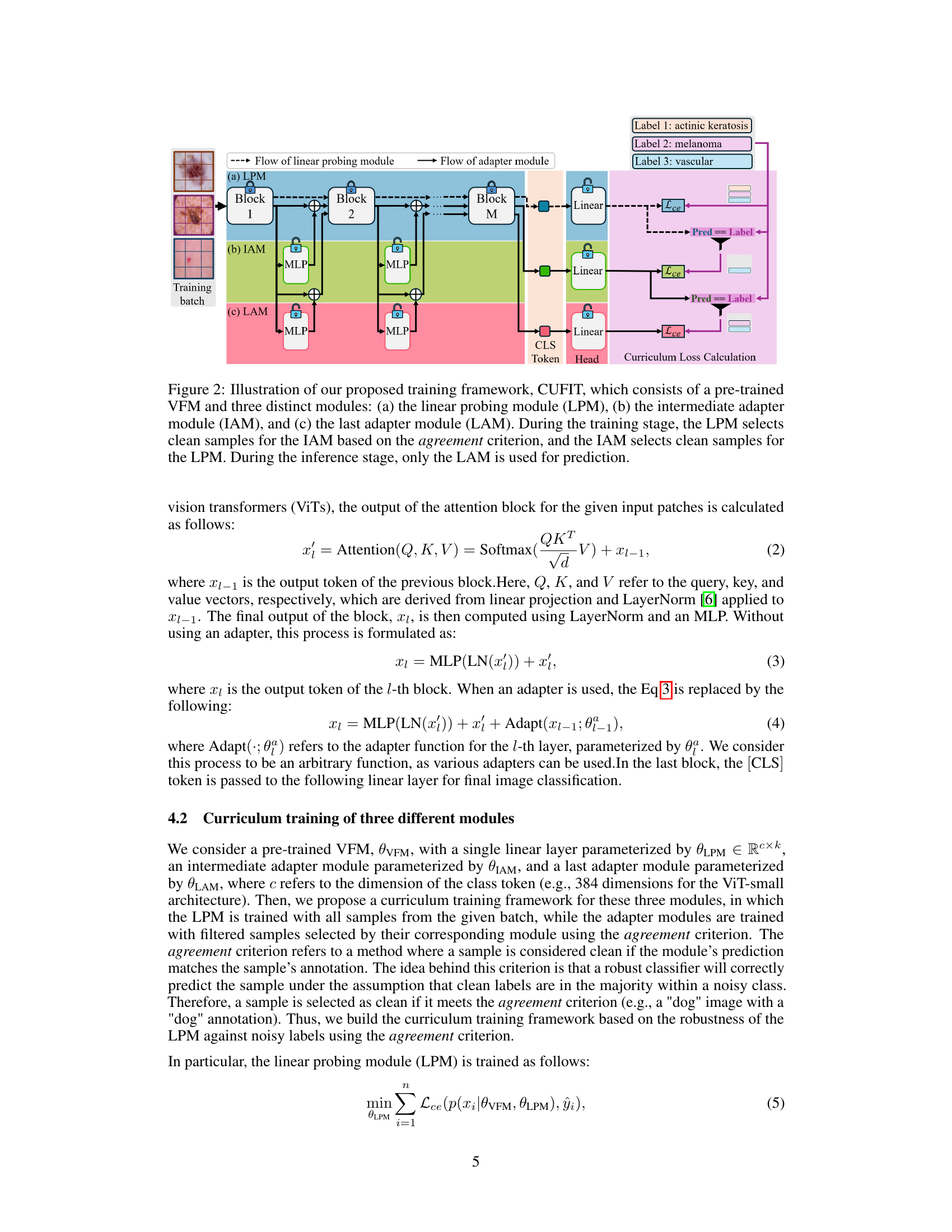
This figure illustrates the Curriculum Fine-tuning framework (CUFIT) proposed in the paper. CUFIT consists of three modules that are trained simultaneously: a linear probing module (LPM), an intermediate adapter module (IAM), and a last adapter module (LAM). The LPM uses all samples and serves as a filter for the IAM. The IAM, trained on samples selected by the LPM based on a criterion, further filters the samples for the LAM. Finally, only the LAM is used for inference to make predictions. This curriculum approach helps mitigate the negative effects of noisy labels by progressively refining the sample selection and adapter training.

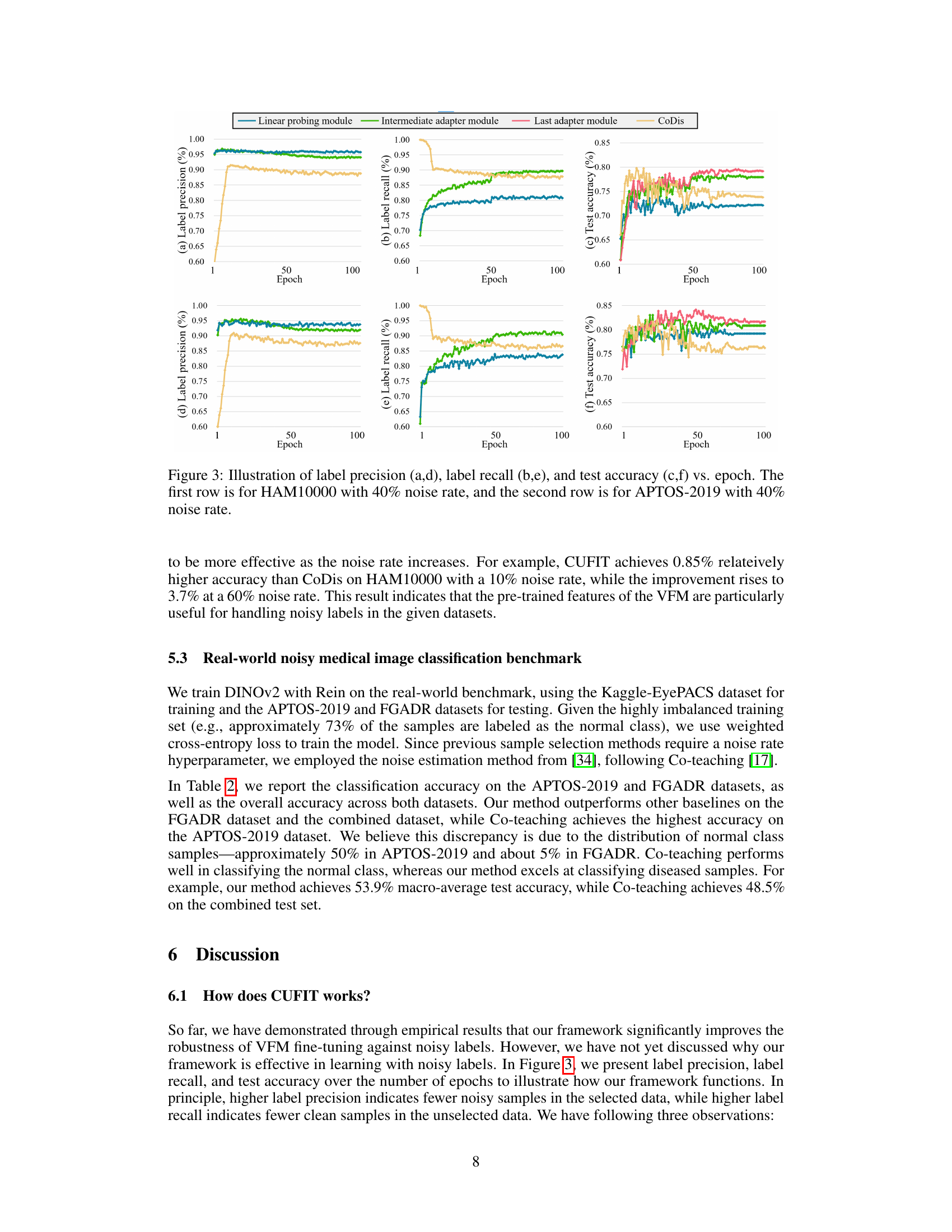
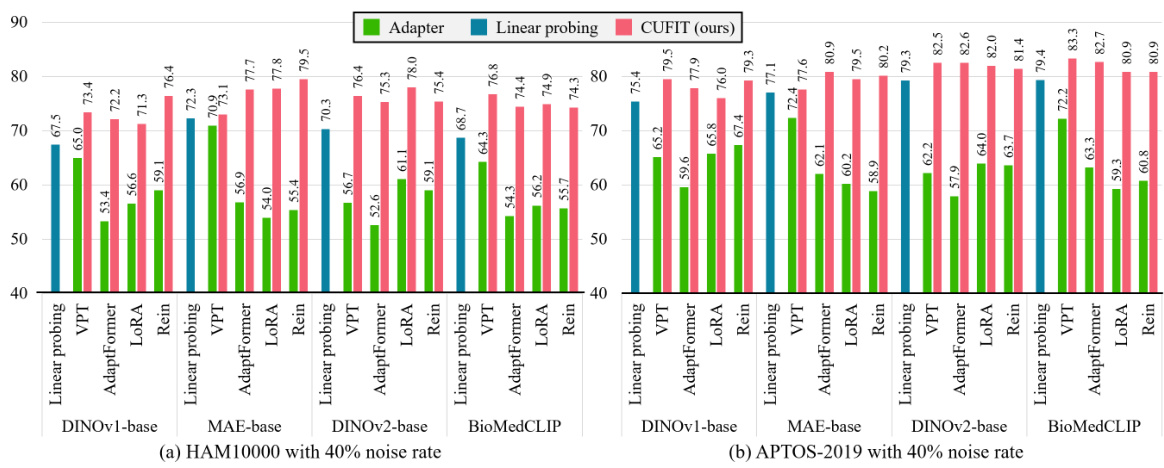
This figure compares the label precision, label recall, and test accuracy of the linear probing module, intermediate adapter module, last adapter module, and CoDis method across 100 epochs for two datasets, HAM10000 and APTOS-2019. Both datasets have a 40% noise rate. The plots show the performance of each module over time. The results highlight the effectiveness of the proposed curriculum training framework.
This figure illustrates the core idea of the proposed method, CUFIT. It compares linear probing and adapter usage for handling noisy labels in medical image classification. (a) shows linear probing, where only the fully-connected layer is trained while the VFM’s weights are frozen. (b) shows adapter usage, where the weights of the adapters are trained to handle noisy labels. (c) shows the performance comparison with a simulated noisy dataset, illustrating that linear probing is more robust to noisy labels but has lower performance in the absence of noise. Conversely, adapter usage has lower robustness in the noisy setting but shows higher performance in the absence of noise. This motivates the need for curriculum fine-tuning.
More on tables
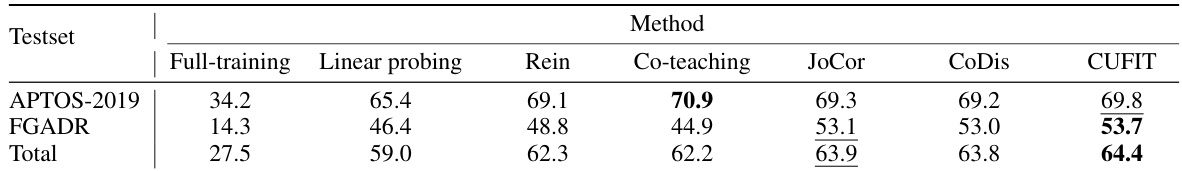
This table presents the average test accuracy achieved by different methods on two real-world noisy datasets (APTOS-2019 and FGADR) after training on the Kaggle-EyePACS dataset. The methods compared include various baseline approaches and CUFIT, the proposed method. The best and second-best results are highlighted for each dataset. The ‘Total’ row combines the results across both datasets.
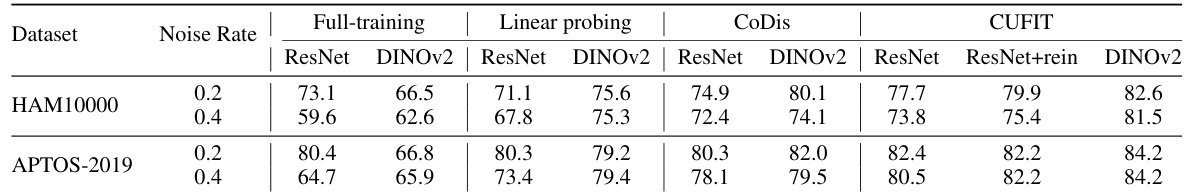
This table presents the average test accuracy achieved by different methods (Full-training, Linear probing, Rein, Co-teaching, JoCor, CoDis, and CUFIT) on four medical image datasets (HAM10000, APTOS-2019, BloodMnist, and OrgancMnist) with varying levels of simulated label noise (0.1, 0.2, 0.4, and 0.6). The results are averaged across the last ten epochs of training. The best and second-best results for each dataset and noise level are highlighted.
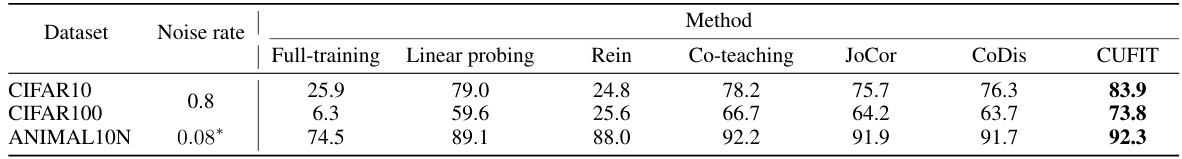
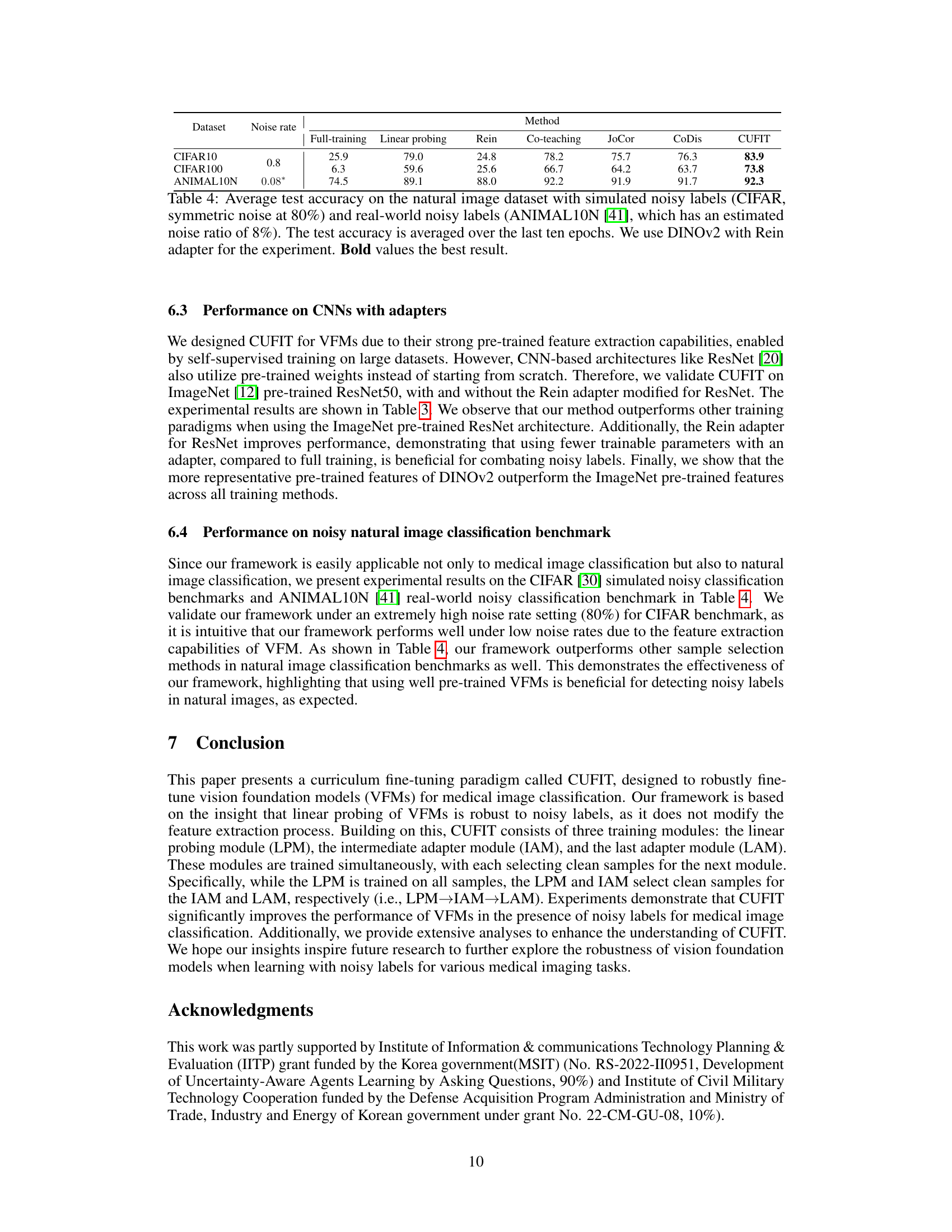
This table presents the average test accuracy achieved by different methods on three datasets: CIFAR10, CIFAR100, and ANIMAL10N. Each dataset has a different noise rate applied to its labels. The methods compared include full training, linear probing, Rein (adapter-based fine-tuning), Co-teaching, JoCor, CoDis, and CUFIT (the authors’ proposed method). The results show the performance of each method on each dataset, highlighting the superior performance of CUFIT, especially under high noise rates.
Full paper#