↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
3D medical image segmentation is crucial for various clinical applications but existing deep learning models are computationally expensive and resource-intensive, hindering their deployment in real-world scenarios. This makes the development of efficient and accurate models paramount.
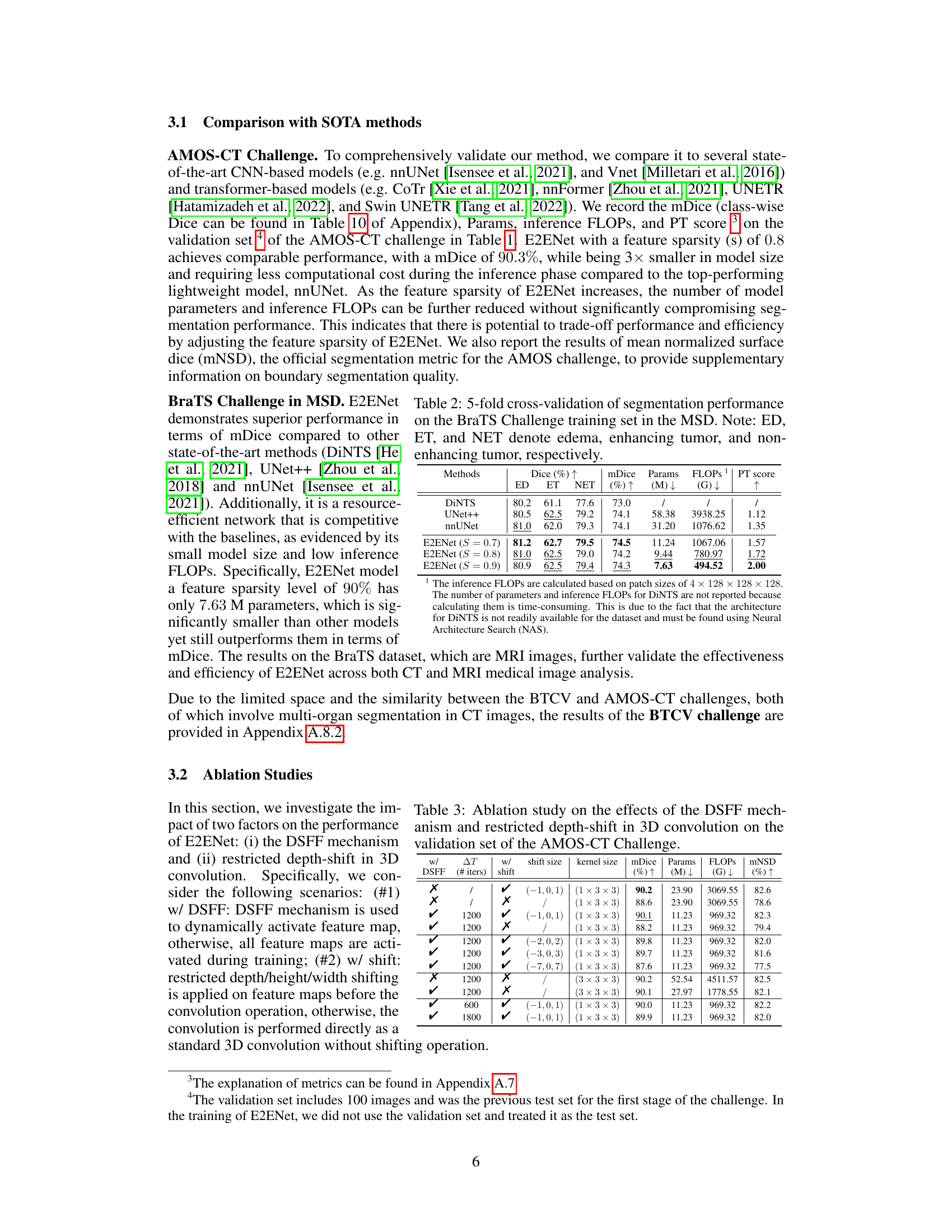
The proposed E2ENet model tackles this challenge by introducing two key innovations: a Dynamic Sparse Feature Fusion (DSFF) mechanism and a Restricted Depth-Shift strategy in 3D convolution. DSFF selectively integrates multi-scale features, improving efficiency without sacrificing accuracy. Restricted Depth-Shift reduces computational complexity while preserving 3D information. Evaluated on multiple datasets, E2ENet demonstrates a superior balance between accuracy and efficiency, achieving competitive accuracy while significantly reducing computational costs compared to state-of-the-art methods.
Key Takeaways#
Why does it matter?#
This paper is important because it presents E2ENet, a novel and efficient 3D medical image segmentation model that achieves a superior trade-off between accuracy and efficiency. It addresses the critical issue of computational cost in 3D medical image segmentation, making deep learning techniques more applicable to real-world resource-constrained settings. The dynamic sparse feature fusion and restricted depth-shift strategies are valuable contributions to the field, offering new avenues for improving efficiency and accuracy in similar tasks.
Visual Insights#
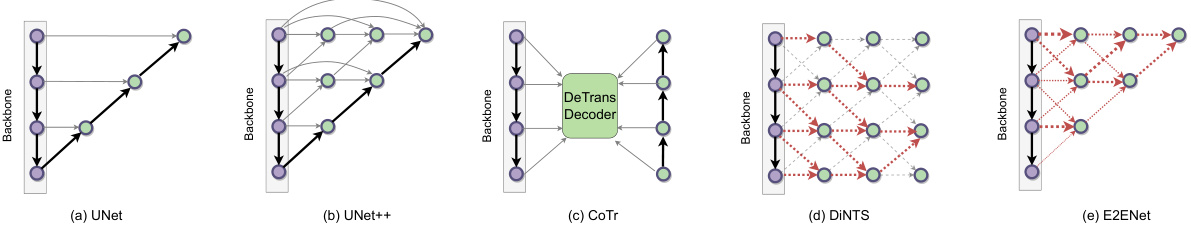
This figure compares different feature fusion schemes used in various architectures for medical image segmentation. It illustrates how different methods (UNet, UNet++, CoTr, DiNTS) combine multi-scale features extracted from a backbone network. The main focus is on E2ENet (e), which utilizes a Dynamic Sparse Feature Fusion (DSFF) mechanism, dynamically learning the most informative features to integrate and reducing redundancy. The thickness of red lines in E2ENet visually represents the sparsity of the feature fusion, with thicker lines indicating stronger connections and thinner lines indicating weaker ones.
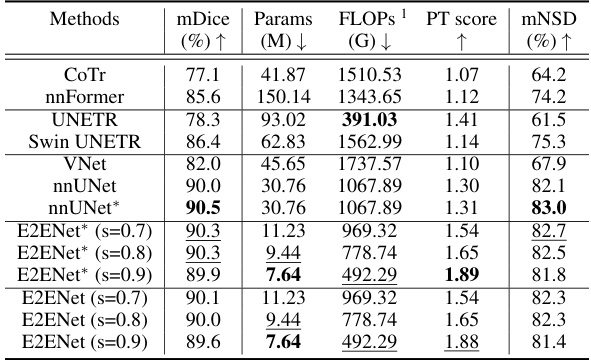
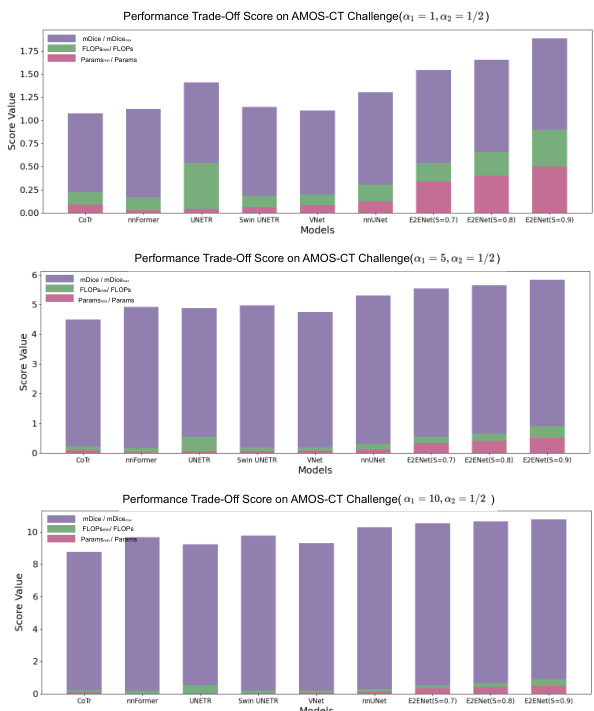
This table presents a comparison of the segmentation performance of different methods on the AMOS-CT dataset’s validation set. Metrics include mDice (mean Dice Similarity Coefficient), the number of model parameters (Params), floating point operations (FLOPs) during inference, a performance trade-off score (PT score) that balances accuracy and efficiency, and mNSD (mean Normalized Surface Dice). The methods compared include several state-of-the-art (SOTA) models and the proposed E2ENet model with varying levels of feature sparsity. The * indicates results that included post-processing steps.
In-depth insights#
Sparse Feature Fusion#
Sparse feature fusion is a crucial technique in modern machine learning, particularly in areas like image processing and medical image analysis. The core idea revolves around selectively combining information from different feature maps to improve efficiency and accuracy. Unlike traditional dense methods that consider all features, sparse methods strategically identify and utilize only the most relevant features, thereby reducing computational cost and memory usage. Dynamic sparse feature fusion takes this a step further by learning which features to combine during training, adapting to the specific task at hand. This adaptive approach offers greater flexibility and efficiency, resulting in more compact models and faster inference times. However, challenges remain in determining optimal sparsity levels and ensuring that crucial information is not inadvertently discarded. Future research should focus on developing more sophisticated selection mechanisms and investigating the impact of sparsity on model robustness and generalization ability.
3D Convolutions#
3D convolutions offer the capability to capture spatial relationships within volumetric data, a crucial advantage in processing 3D medical images. However, they come with a significant increase in computational cost compared to 2D convolutions. Efficient strategies are therefore essential to harness the power of 3D convolutions without compromising performance. One such technique is restricted depth-shift, which leverages the 3D spatial information while maintaining computational complexity comparable to 2D methods. This approach involves carefully shifting channels along the depth dimension before applying a smaller 3D kernel (e.g., 1x3x3). This reduces computational cost without sacrificing the ability to extract 3D features, offering a compelling trade-off between accuracy and efficiency. The effectiveness of this approach might depend on the specific application, dataset, and model architecture. Further research may explore more sophisticated sparse convolution techniques that dynamically adapt to the complexity and sparsity of the data to maximize computational efficiency and accuracy. Ultimately, the choice between 2D and 3D convolutions (and the specific implementation of 3D convolutions) often involves a careful balancing act, weighing the need for rich feature representations against the constraints of available computational resources.
Accuracy-Efficiency#
The overarching goal in many computer vision tasks, especially those involving resource-constrained environments like medical image segmentation, is to achieve a balance between accuracy and efficiency. A highly accurate model is desirable but often comes with a high computational cost, making deployment challenging. Conversely, a highly efficient model may sacrifice accuracy to achieve speed. The ‘Accuracy-Efficiency’ trade-off necessitates careful consideration of model architecture, training strategies, and hardware limitations. Approaches that leverage techniques such as model pruning, quantization, and knowledge distillation aim to reduce the model’s size and computational complexity without significant performance degradation. Furthermore, specialized hardware and optimized algorithms play a crucial role in enhancing efficiency, especially for computationally intensive tasks like 3D medical image segmentation. Therefore, the success of any model hinges on the design and implementation choices that successfully navigate the inherent trade-off between accuracy and efficiency, thereby maximizing both precision and feasibility in a given application context.
Generalizability#
The concept of generalizability in a research paper assesses the model’s ability to perform well on unseen data or different tasks beyond the specific data it was trained on. A highly generalizable model is robust and adaptable, performing well across various conditions. In the context of a medical image segmentation paper, generalizability might explore how well the model performs on different imaging modalities (e.g., MRI vs. CT scans), different patient demographics, or different disease states. Evaluating generalizability typically involves testing the model on datasets separate from the training data, which are diverse in terms of image acquisition parameters, patient characteristics, or organ variations. Analyzing the performance metrics across these diverse test sets provides crucial insights into the model’s reliability and robustness in real-world clinical applications. A model’s generalizability is a key indicator of its practical value. It demonstrates the model’s ability to generalize knowledge learned from a specific training dataset to make accurate predictions on completely new and unseen data, ultimately highlighting its wider applicability and significance.
Future Work#
Future research directions stemming from this efficient 3D medical image segmentation method (E2ENet) could explore several promising avenues. Investigating a learnable shift offset within the restricted depth-shift convolution could potentially enhance its performance further. Exploring the integration of E2ENet’s dynamic sparse feature fusion (DSFF) mechanism and restricted depth-shift strategies into other 3D segmentation models presents an opportunity to evaluate their generalizability and potential for improved efficiency. Additionally, the impact of hardware acceleration tailored for sparse neural networks should be investigated to fully realize the speed advantages of the sparse architecture. Benchmarking E2ENet on a wider array of medical image datasets will assess the model’s robustness and generalizability across diverse clinical applications and imaging modalities. Finally, examining the effect of varying network depths and widths and the impact on accuracy-efficiency trade-offs would provide further insights into optimal network design for different resource constraints.
More visual insights#
More on figures

This figure illustrates the architecture of the E2ENet model. It shows the Efficient backbone extracting features at multiple levels, which are then fed into a Feature Pyramid. These features are gradually aggregated across multiple stages using a Dynamic Sparse Feature Fusion (DSFF) mechanism. In each stage, the features are fused using a fusion operation that includes LeakyReLU and InstanceNorm, then passed through a Shift CNN. Finally, the fused features are processed by an Output Module to generate the final output feature map. The figure visually depicts how bottom-up, top-down, and forward connections are integrated for multi-scale feature fusion in the E2ENet network.
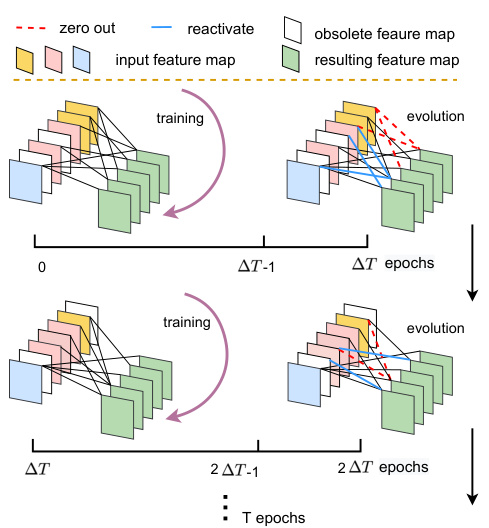
This figure illustrates the Dynamic Sparse Feature Fusion (DSFF) mechanism. The DSFF mechanism starts with sparse connections between feature maps. Over a period of ΔT training epochs, the network dynamically adjusts these connections. Connections with low importance (measured by the L1 norm of their kernels) are removed (zeroed out, shown in red), while the same number of inactive connections are randomly reactivated (shown in blue). This process repeats every ΔT epochs, maintaining a constant sparsity level (S) throughout training. The figure visually represents this iterative process of sparsity adjustment and connection evolution.

This figure illustrates the restricted depth-shift operation used in E2ENet’s 3D convolutions. The input feature maps are divided into three parts along the channel dimension. Each part is then shifted by -1, 0, or 1 unit along the depth dimension. Finally, 3D convolutions with a 1x3x3 kernel are applied to these shifted feature maps to capture 3D spatial relationships while maintaining the efficiency of 2D convolutions.

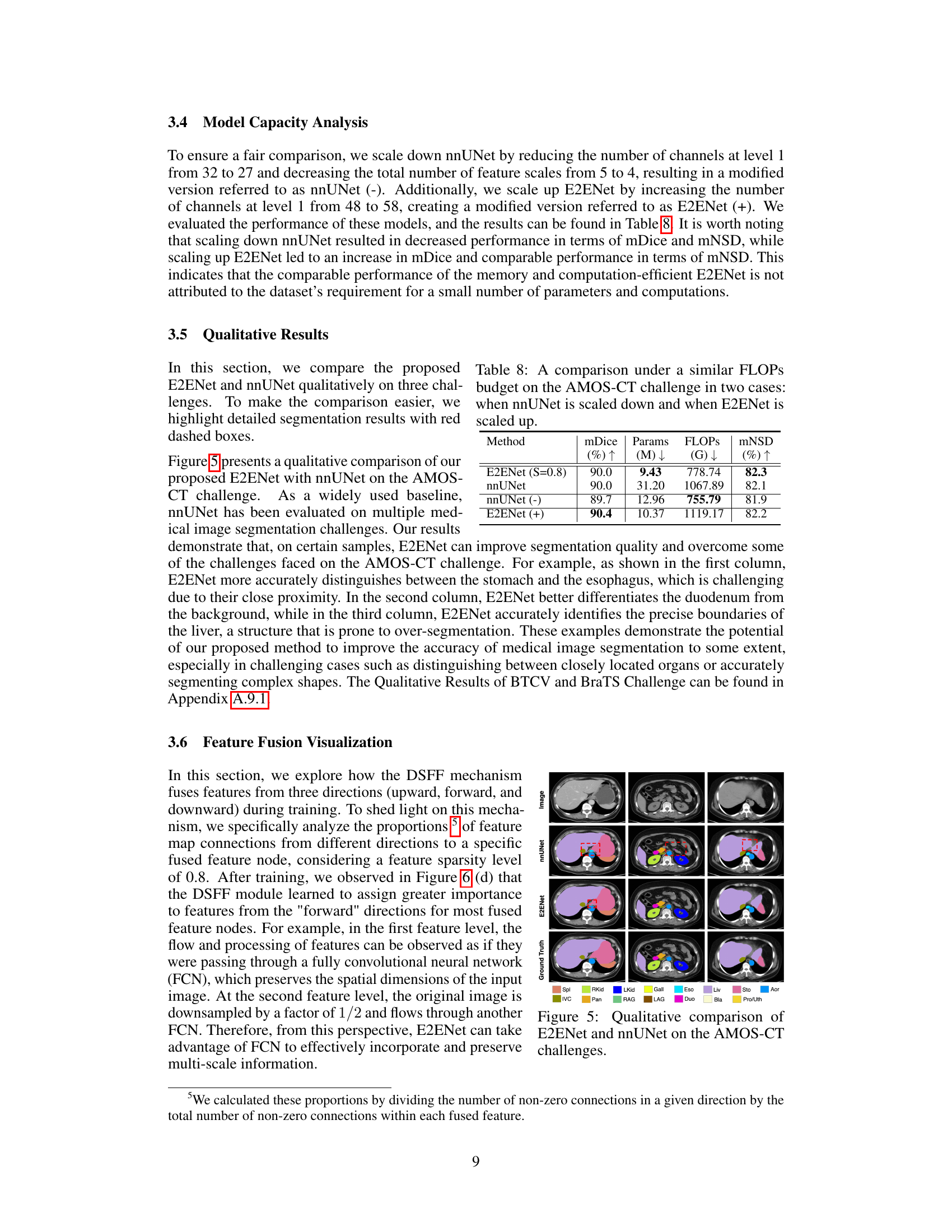
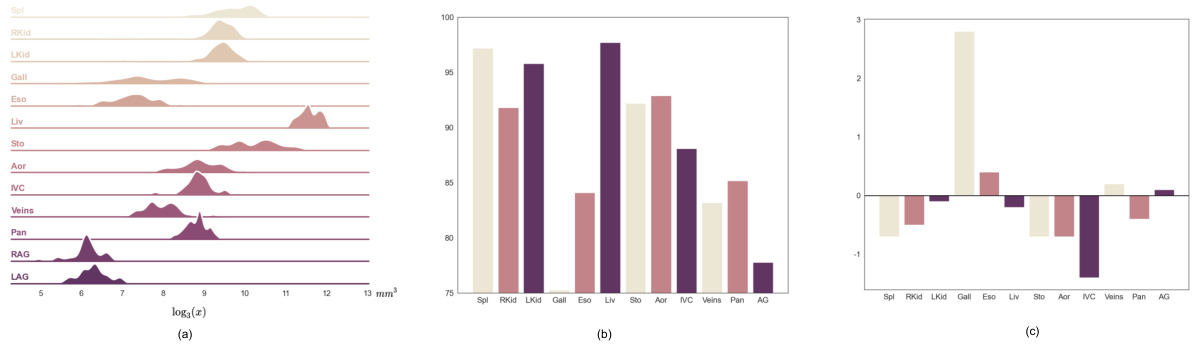
This figure shows a qualitative comparison of the segmentation results obtained using E2ENet and nnUNet on the AMOS-CT dataset. The comparison highlights the differences in the segmentation of various organs, such as the stomach, esophagus, and duodenum, showcasing E2ENet’s improved ability to distinguish between these closely located organs. The image includes visual representations of the ground truth, nnUNet segmentations, and E2ENet segmentations for better comparison.
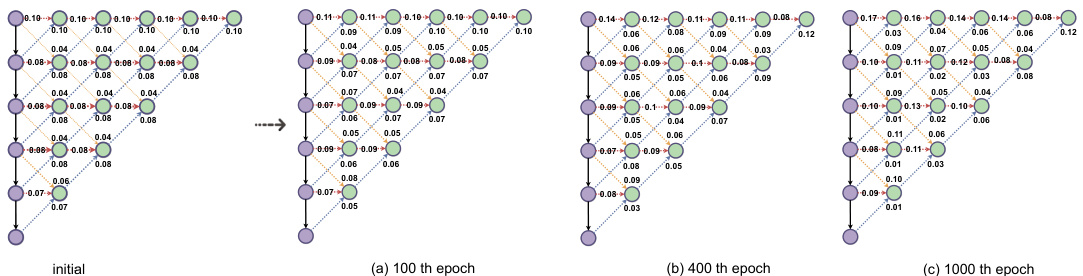
This figure visualizes how the Dynamic Sparse Feature Fusion (DSFF) mechanism learns to connect features from different levels during training. It shows the proportions of feature connections from ‘upward’, ‘downward’, and ‘forward’ directions at a feature sparsity level of 0.8. The proportions are shown at three different training epochs (100th, 400th, and 1000th). The visualization helps to understand how the DSFF mechanism adapts and learns to filter out less important connections, making the feature fusion more efficient during the training process.
This figure illustrates the Dynamic Sparse Feature Fusion (DSFF) mechanism. It shows how the fusion operation starts with sparse connections and then adapts during training. Over time, less important connections are removed, while a similar number of previously inactive connections are re-activated at random. This process maintains a constant level of sparsity (S) throughout training, improving efficiency by focusing on the most informative feature connections.
This figure illustrates the architecture of the Efficient to Efficient Network (E2ENet), a novel model designed for efficient 3D medical image segmentation. The model consists of a convolutional neural network (CNN) backbone responsible for extracting multiple levels of features from the input 3D image. These features are then hierarchically fused through several stages, involving a fusion operation that combines adjacent features to fully leverage the information across different scales. The fusion process incorporates multi-scale features, progressively aggregating information through the model’s stages, resulting in the final output.
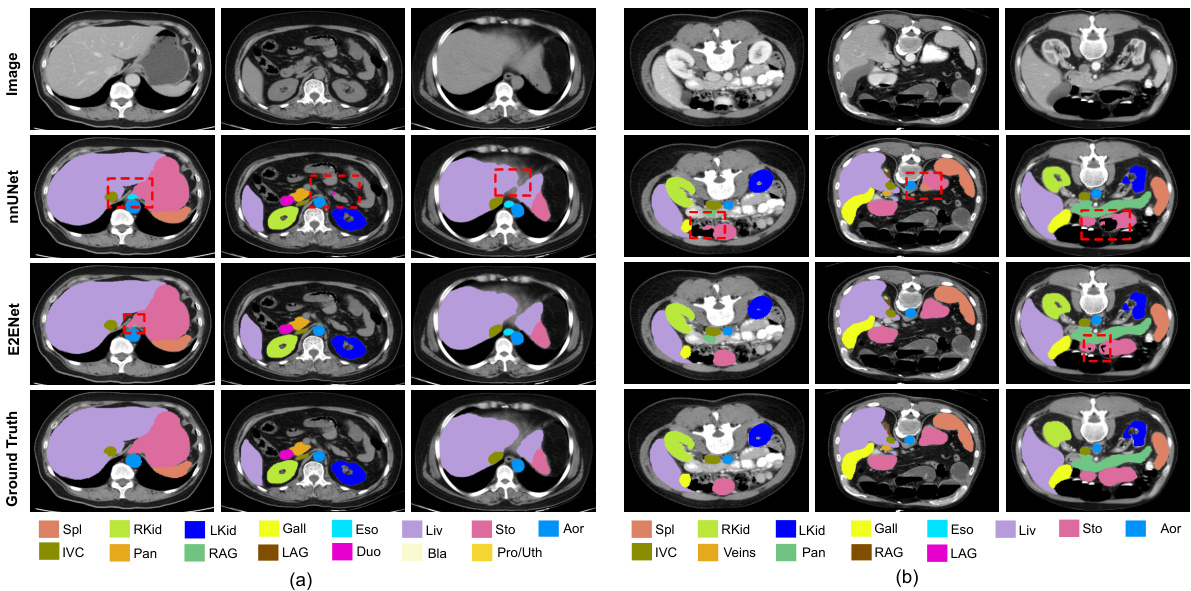
This figure shows a qualitative comparison of the segmentation results obtained using E2ENet and nnUNet on the AMOS-CT dataset. The image shows example slices from the dataset and highlights the differences in segmentation quality between the two models. Red boxes are used to highlight areas of particular interest where the differences in performance are more pronounced. The goal is to visually demonstrate the improved accuracy of E2ENet in segmenting challenging structures.
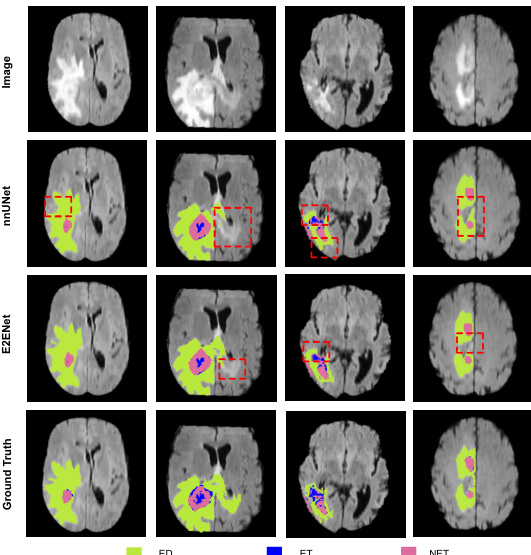
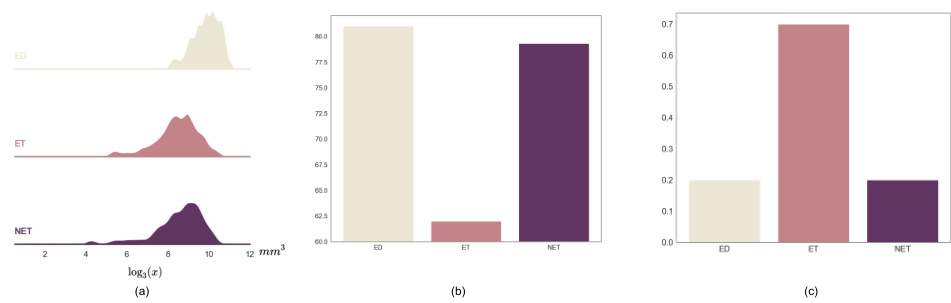
This figure shows a qualitative comparison of the segmentation results of the proposed E2ENet and the nnUNet baseline method on the BraTS challenge dataset. The figure displays four sample images from the BraTS dataset, along with their corresponding segmentation masks generated by both methods. The segmentation masks are color-coded to represent the different tumor regions of interest: edema (ED), enhancing tumor (ET), and non-enhancing tumor (NET). The red boxes highlight specific areas where the difference between the methods’ segmentations is most apparent. The comparison aims to demonstrate E2ENet’s ability to improve segmentation accuracy, particularly in challenging areas.
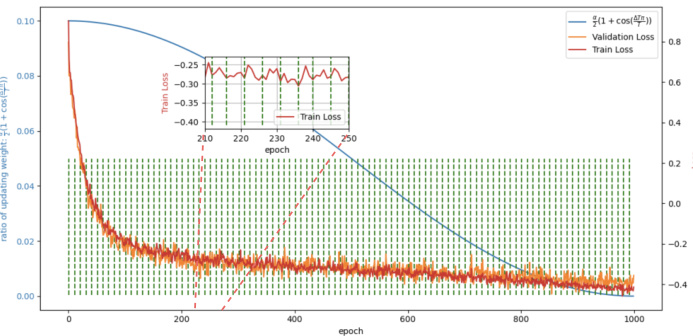
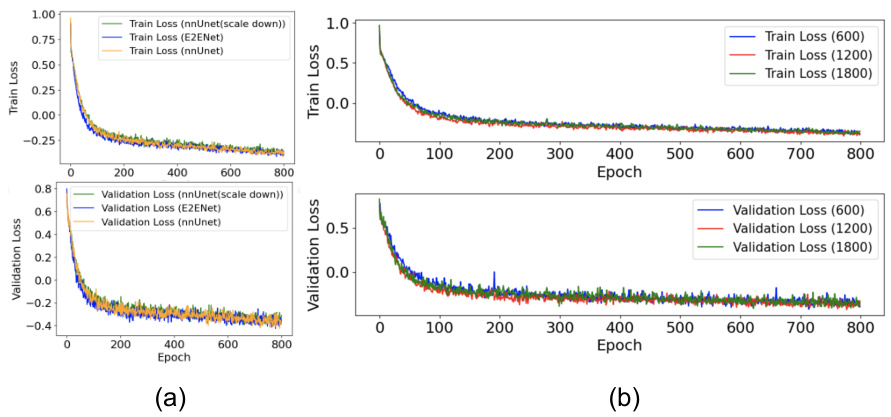
This figure shows the training and validation loss curves for the E2ENet model trained on the AMOS-CT dataset. The green dotted lines mark the epochs where the dynamic sparse feature fusion (DSFF) mechanism updates the network’s topology by activating or deactivating connections (weights). The blue line shows the ratio of deactivation to activation at each step. The plot illustrates how the training loss initially increases after topology updates, but the overall loss decreases over time, demonstrating the effectiveness of the DSFF mechanism for improving model efficiency.
This figure compares different feature fusion schemes in various architectures, including UNet, UNet++, CoTr, and DiNTS. It highlights the unique dynamic sparse feature fusion (DSFF) mechanism used in E2ENet, which allows for efficient and adaptive fusion of multi-scale features by dynamically learning the information flow paths. The thickness of the red lines in the E2ENet diagram represents the sparsity of the connections, demonstrating the model’s efficiency.

This figure compares different feature fusion schemes used in various neural network architectures for image segmentation. It highlights the differences between UNet, UNet++, CoTr, DiNTS, and the proposed E2ENet. The key difference is how E2ENet dynamically learns sparse connections between features, leading to efficient feature fusion, unlike the other methods that use dense connections or complex search algorithms. The purple nodes represent features from the backbone network, green represents the fused features, and the red lines represent the information flow, with line thickness in E2ENet indicating the strength of the learned connection.
This figure visualizes how the Dynamic Sparse Feature Fusion (DSFF) mechanism dynamically learns to fuse multi-scale features from three different directions (upward, forward, and downward) during training. The figure illustrates the proportions of feature map connections from these directions to a specific fused feature node at different training epochs (initial, 100th, 400th, and 1000th epoch) with a feature sparsity level of 0.8 on the AMOS-CT challenge. The proportions are presented visually, showing the learned importance of features from different directions for optimal fusion at each stage of the training process.
This figure compares different feature fusion schemes used in various architectures for image segmentation. It shows how different methods, including UNet, UNet++, CoTr, and DiNTS, handle the integration of multi-scale features extracted from the backbone network. The main focus is on E2ENet, highlighting its dynamic sparse feature fusion (DSFF) mechanism, which allows for adaptive learning of the feature fusion process, resulting in efficient feature integration.
More on tables
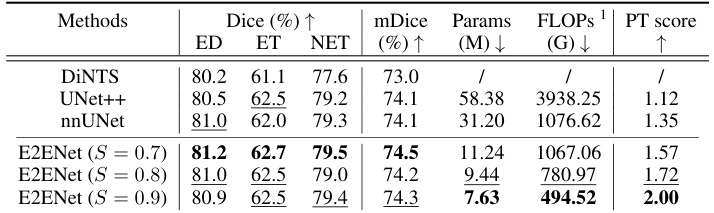
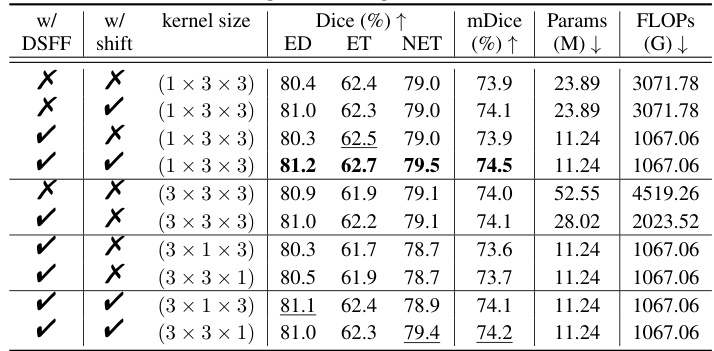
This table presents the results of a 5-fold cross-validation experiment on the BraTS (Brain Tumor Segmentation) Challenge training set within the Medical Segmentation Decathlon (MSD). It compares the performance of E2ENet at different sparsity levels (S) against other state-of-the-art methods such as DiNTS, UNet++, and nnUNet. The metrics reported are Dice scores for three tumor regions (edema, enhancing tumor, non-enhancing tumor), the mean Dice score (mDice), the number of model parameters (Params), the number of floating-point operations (FLOPs), and a performance trade-off score (PT score). The table shows that E2ENet achieves competitive performance while significantly reducing the number of parameters and FLOPs.
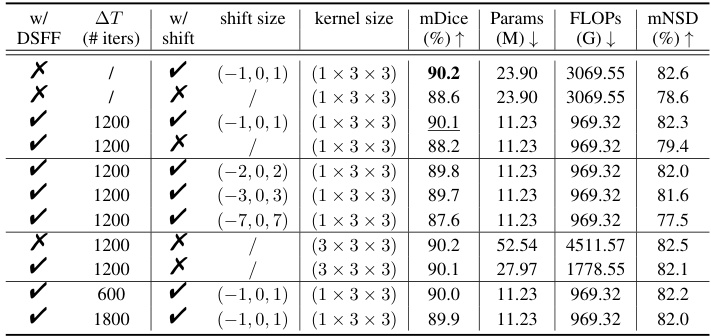
This table presents the ablation study results on the AMOS-CT dataset validation set to evaluate the impact of the Dynamic Sparse Feature Fusion (DSFF) mechanism and the restricted depth-shift in 3D convolution on the model’s performance. It compares different model configurations, including with and without DSFF, with and without depth-shift, and with different depth-shift sizes and kernel sizes. The results are measured using mDice, the number of parameters, FLOPs (floating-point operations), and mNSD (mean normalized surface dice).
This table presents the ablation study results on the validation set of the AMOS-CT challenge. It shows the impact of two key components of E2ENet: the Dynamic Sparse Feature Fusion (DSFF) mechanism and the restricted depth-shift in 3D convolution. The table compares the performance (mDice, Params, FLOPs, mNSD) under different configurations: with/without DSFF, with/without depth-shift, and with different kernel sizes. This helps understand the contribution of each component to the overall performance and efficiency of the model.

This table compares the performance of different segmentation models on the AMOS-CT dataset, including the proposed E2ENet model. It shows the mDice score (a metric for evaluating segmentation accuracy), the number of model parameters (Params), the number of floating point operations (FLOPs) during inference, and the PT score (a composite metric considering accuracy and efficiency). The results highlight that E2ENet achieves competitive performance while using significantly fewer parameters and FLOPs.
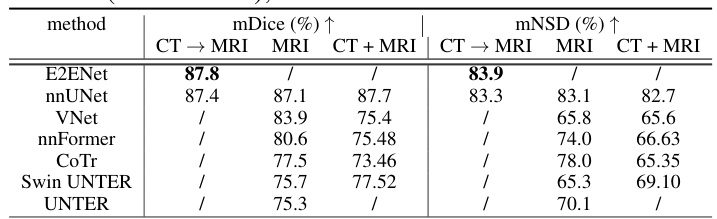
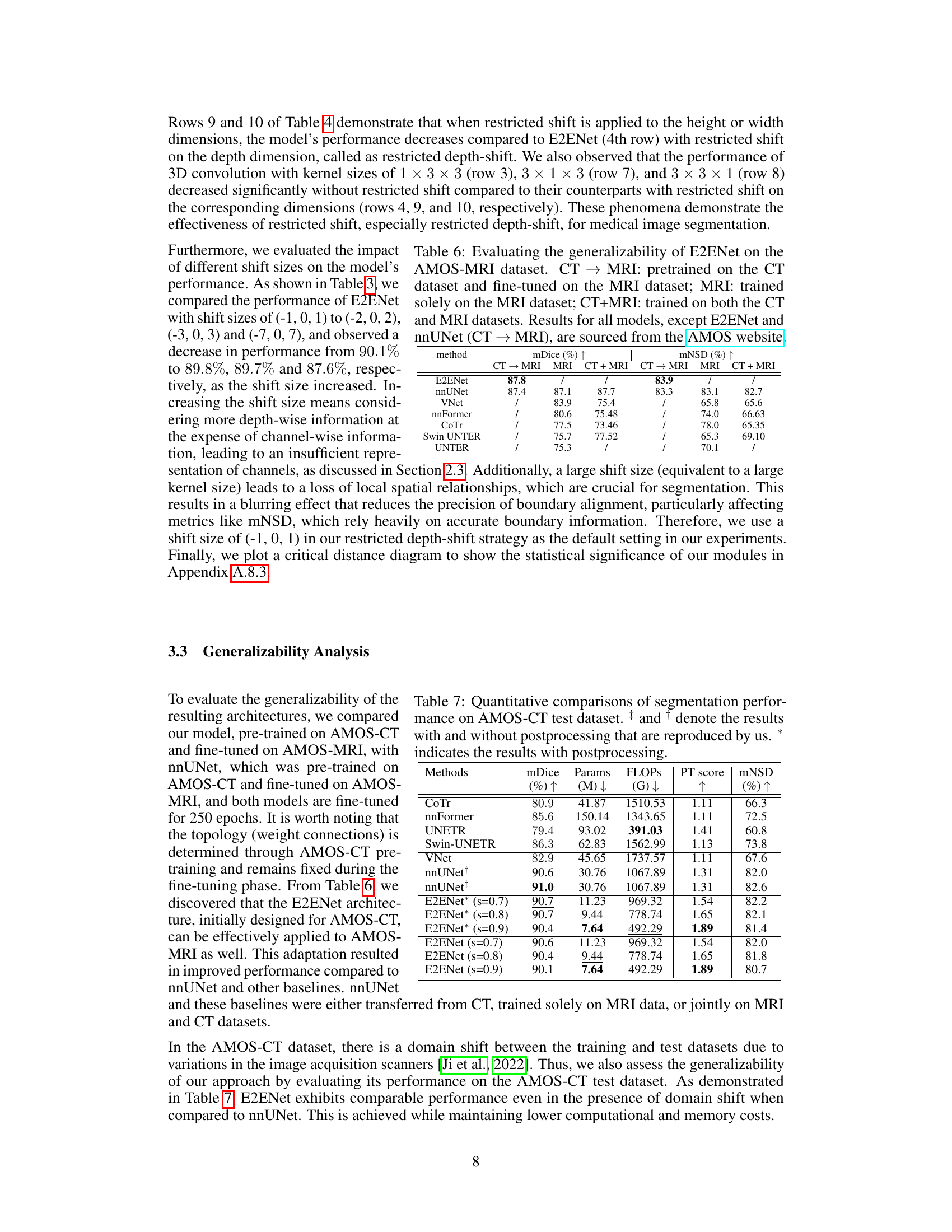
This table presents a comparison of the segmentation performance of different models on the AMOS-MRI dataset. Three training strategies are compared: using a model pre-trained on the AMOS-CT dataset and then fine-tuned on the AMOS-MRI dataset; training a model solely on the AMOS-MRI dataset; and training a model on both the AMOS-CT and AMOS-MRI datasets. The results demonstrate the generalizability of E2ENet and its performance compared to other state-of-the-art methods.
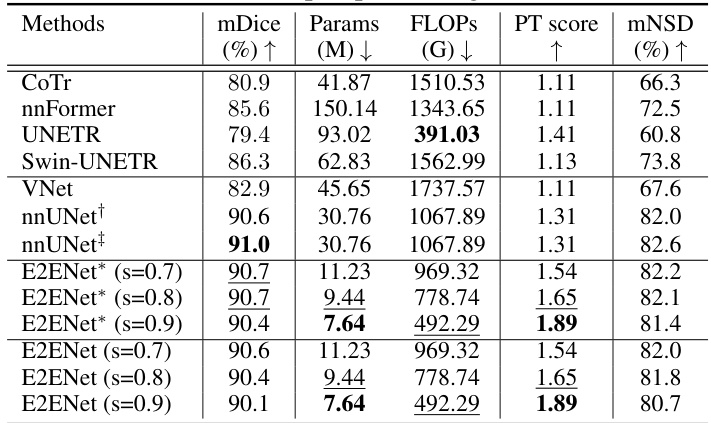
This table compares the performance of E2ENet with other state-of-the-art methods on the AMOS-CT dataset. The metrics used for comparison include mDice (mean Dice similarity coefficient), the number of model parameters (Params), floating point operations during inference (FLOPs), the Performance Trade-off score (PT score), and mean normalized surface dice (mNSD). The table shows that E2ENet achieves competitive performance (mDice) with significantly fewer parameters and FLOPs, demonstrating its efficiency.

This table compares the performance of E2ENet and nnUNet models under similar FLOPs constraints. To ensure a fair comparison, a smaller version of nnUNet (nnUNet(-)) and a larger version of E2ENet (E2ENet(+)) were created by adjusting the number of channels and feature scales, respectively. The table shows that even with a reduction in parameters and FLOPs, E2ENet maintains comparable performance to nnUNet, demonstrating its efficiency in memory and computation.
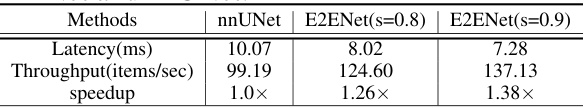
This table presents a comparison of the inference speed (latency and throughput) of E2ENet and nnUNet using an Intel Xeon Platinum 8360Y CPU with 18 cores. Patches of images sized 32x32x32 were used as input for the comparison. The results demonstrate that E2ENet shows significant speedups compared to nnUNet.
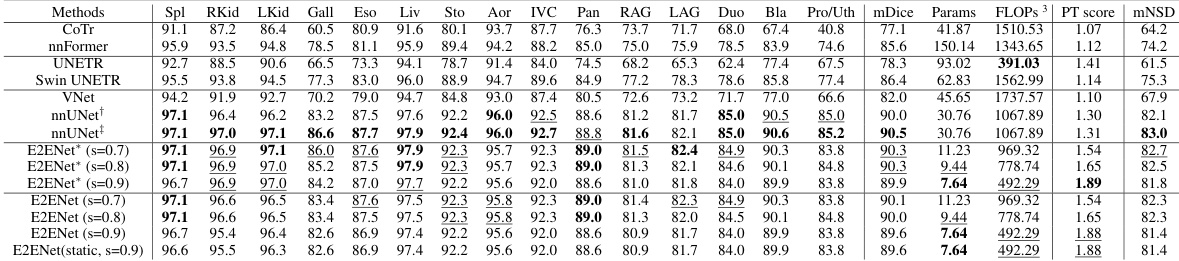
This table presents a quantitative comparison of the segmentation performance of various state-of-the-art (SOTA) methods on the AMOS-CT dataset’s validation set. The metrics used for comparison include mDice (mean Dice similarity coefficient), Params (model parameters in millions), FLOPs (floating point operations in billions), PT score (performance trade-off score, which combines accuracy and efficiency), and mNSD (mean normalized surface dice). The asterisk (*) indicates that post-processing was applied to the results. The table allows readers to compare the accuracy and efficiency of different methods, particularly highlighting E2ENet’s performance in relation to others.
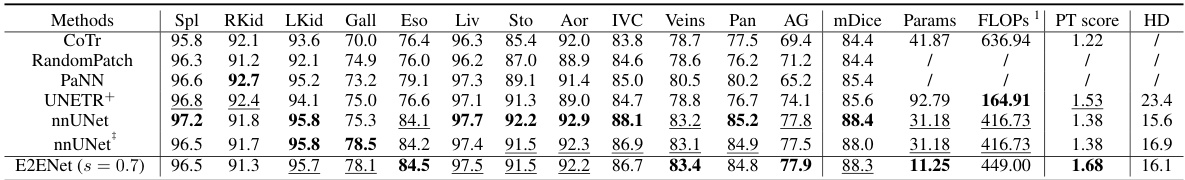
This table compares the performance of E2ENet against several baselines on the BTCV challenge dataset. It shows class-wise Dice scores for various organs, as well as overall metrics like mDice, the number of parameters, FLOPS, a performance trade-off score, and Hausdorff distance. Note that some results are taken from the standard leaderboard and others from a free leaderboard, and that one method (UNETR+) was trained without external data, unlike others.
Full paper#