↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Multiple Instance Learning (MIL) excels in histopathology but lacks reliable explanation methods. Existing methods often fail with large datasets or complex instance interactions, hindering understanding and trust in diagnostic AI. This limits knowledge discovery and model debugging, crucial for reliable clinical applications.
The paper introduces xMIL, a refined MIL framework, and xMIL-LRP, a novel explanation method using Layer-wise Relevance Propagation (LRP). xMIL-LRP addresses the limitations of existing methods by handling instance interactions and large datasets effectively. Extensive experiments demonstrate superior performance compared to existing methods across various histopathology datasets. This improved explainability facilitates better model understanding, knowledge extraction, and ultimately, enhanced AI-powered diagnostics in histopathology.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in histopathology and explainable AI. It addresses the limitations of current MIL explanation methods, offering a novel framework (xMIL) and technique (xMIL-LRP) that produce more faithful and informative explanations. This directly impacts knowledge discovery, model debugging, and improves trust in AI-driven diagnostics. The open-source code further facilitates broader adoption and future research.
Visual Insights#

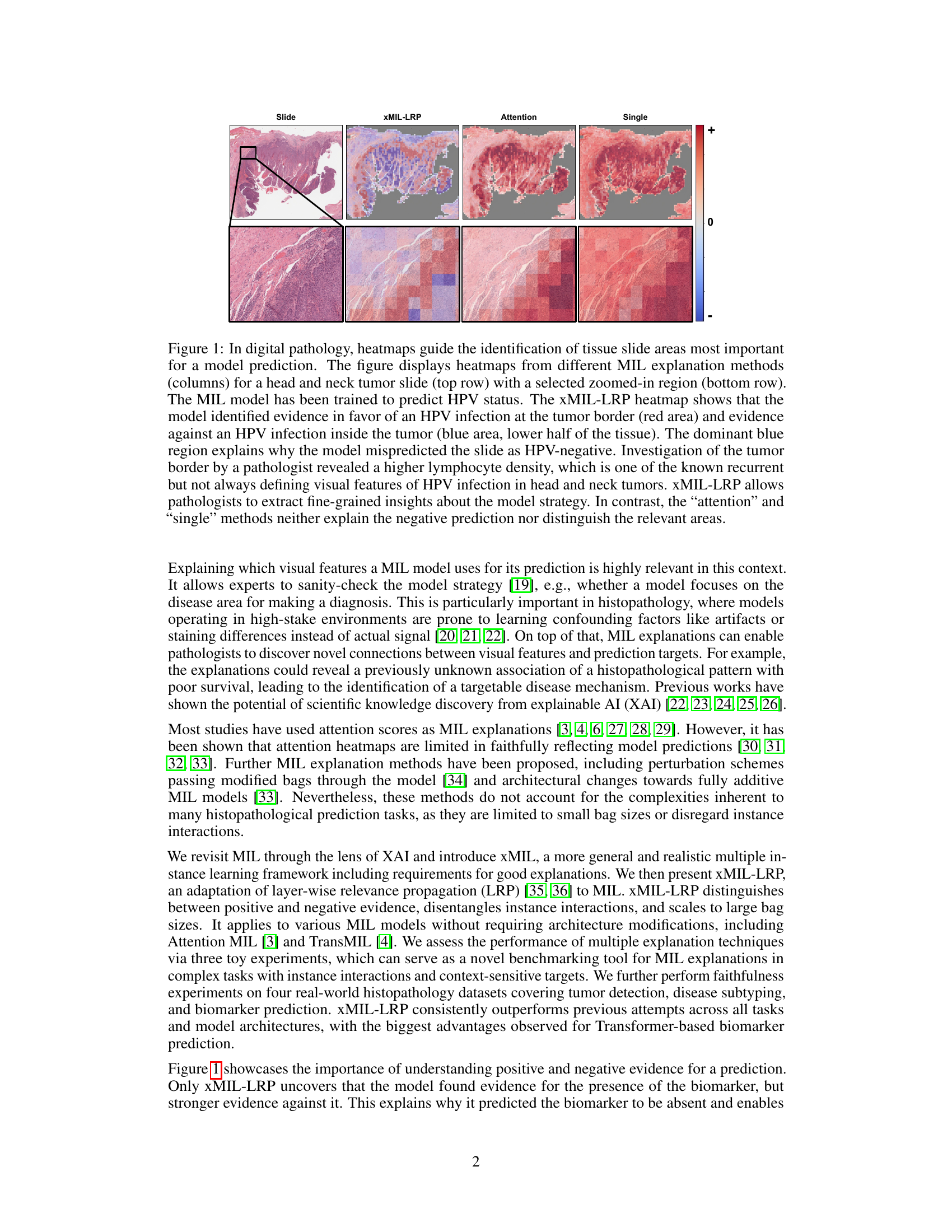
This figure shows heatmaps generated by different MIL explanation methods (xMIL-LRP, Attention, Single) for a head and neck tumor slide. The goal is to predict the HPV status. The xMIL-LRP heatmap highlights the areas most relevant to the model’s prediction, showing both positive (red) and negative (blue) evidence. The other methods fail to provide such detailed insights. The zoomed-in region helps illustrate the differences in the heatmaps more clearly.
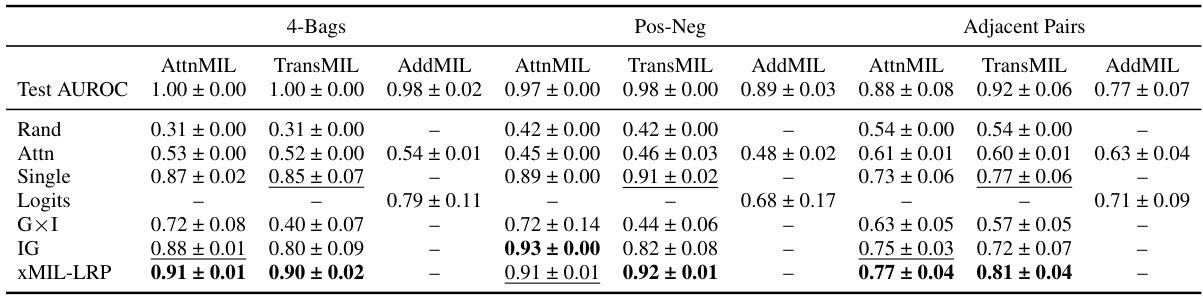
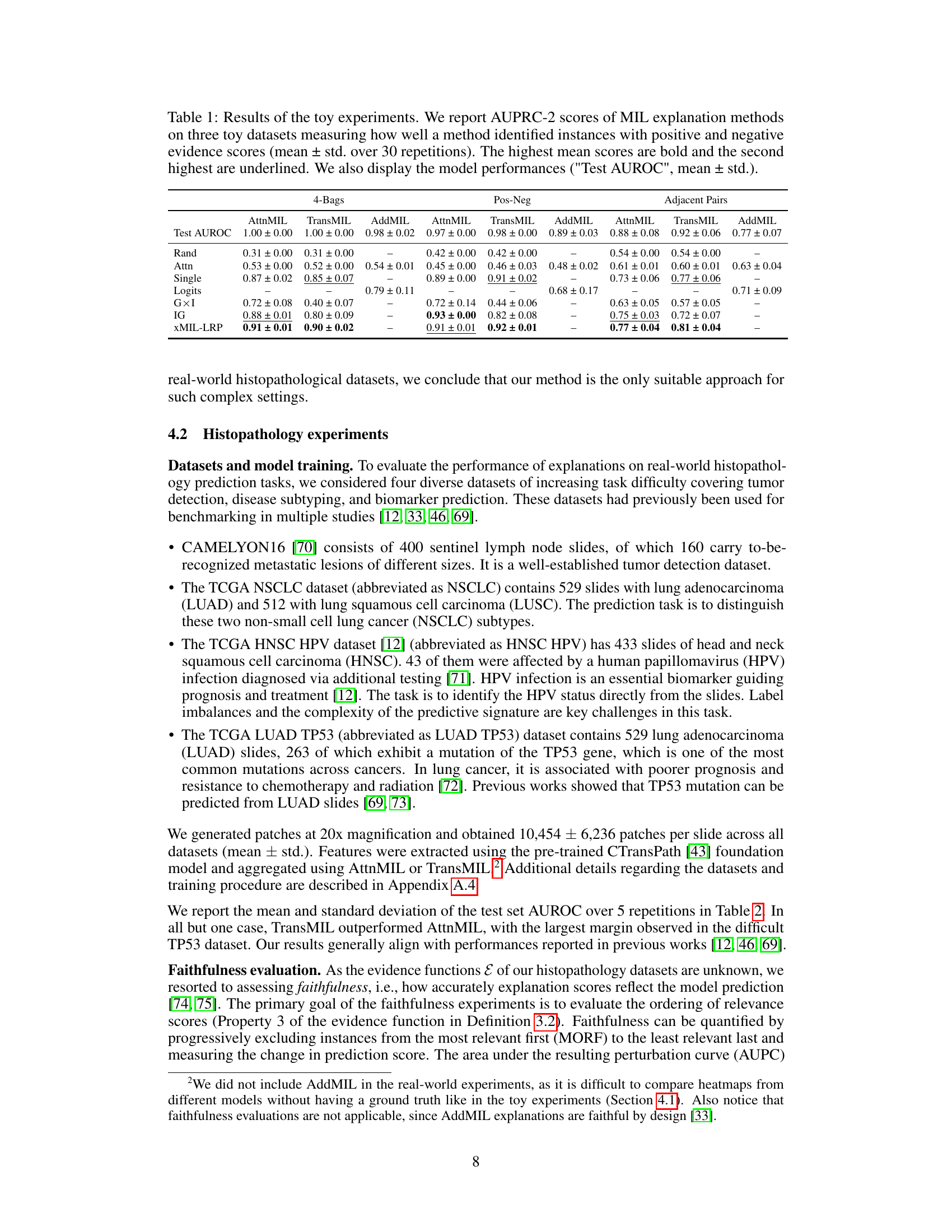
This table presents the results of three toy experiments designed to evaluate the performance of different MIL explanation methods in identifying instances with positive, negative, and neutral evidence. The experiments use MNIST images and evaluate three different MIL tasks (4-Bags, Pos-Neg, and Adjacent Pairs) measuring context sensitivity and positive/negative evidence handling. The table shows the Area Under the Precision-Recall Curve (AUPRC) scores for each method, highlighting the best-performing methods for each task. Additionally, model performance (Test AUROC) is included to provide context.
In-depth insights#
xMIL: A Novel Framework#
The proposed xMIL framework offers a significant advancement in explainable multiple instance learning (MIL), particularly within the context of histopathology. Its core innovation lies in shifting the focus from relying on assumptions about instance labels to directly estimating an evidence function. This function quantifies the impact of each instance on the overall bag prediction, thus enabling a more nuanced and faithful interpretation of model predictions. xMIL addresses inherent limitations of traditional MIL approaches by accounting for context sensitivity, positive and negative evidence, and complex instance interactions. This framework is particularly valuable for complex histopathological analyses where interactions among features are crucial for reliable predictions. By integrating concepts from explainable AI, xMIL-LRP, an adaptation of layer-wise relevance propagation (LRP) to MIL, provides a practical method for estimating the evidence function, producing improved explanations and surpassing existing methods in faithfulness experiments. This allows pathologists to gain richer insights into model behavior and decision-making processes, contributing to model debugging and the discovery of novel knowledge in histopathology.
LRP for MIL Explanations#
Applying Layer-wise Relevance Propagation (LRP) to Multiple Instance Learning (MIL) explanations presents a powerful approach for enhancing the interpretability of MIL models, particularly in complex domains like histopathology. LRP’s ability to decompose the model’s prediction into instance-level relevance scores offers a significant advantage over traditional attention-based methods. These methods often fail to capture the nuances of instance interactions and struggle to distinguish between positive and negative evidence within a bag of instances. xMIL-LRP, a refined framework, explicitly addresses these shortcomings, providing context-sensitive explanations that scale effectively to large bag sizes. By tracing the flow of relevance throughout the model, LRP reveals which instances most strongly support or refute the model’s decision, thus leading to more faithful and insightful explanations. This granular level of interpretability is crucial for building trust in MIL models, fostering knowledge discovery, and facilitating effective model debugging in applications with high stakes.
Histopathology Insights#
Histopathology insights derived from AI models are revolutionizing the field by enabling finer-grained analysis of tissue samples. Explainable AI (XAI) techniques are crucial for translating model predictions into actionable information for pathologists. Faithful explanation methods, such as layer-wise relevance propagation, are particularly effective at disentangling instance interactions within tissue slides and highlighting areas of positive and negative evidence, exceeding the capabilities of traditional attention mechanisms. This allows for more accurate identification of disease areas, uncovering novel associations between visual features and disease subtypes or biomarkers, facilitating knowledge discovery, and improving model debugging. The ability to pinpoint relevant tissue regions and distinguish positive and negative evidence within the context of the whole slide image greatly enhances diagnostic capabilities, leading to improved accuracy in diagnoses and predictions for a wide array of histopathological tasks.
Limitations of MIL#
The section on “Limitations of MIL” in this histopathology research paper would likely delve into the inherent challenges of applying Multiple Instance Learning (MIL) to this domain. Instance ambiguity, arising from the small size and inherent noise in tissue patches, would be a central theme. The paper likely points out that the limited information content of individual instances makes reliable classification difficult, often requiring the integration of evidence across multiple patches. Positive, negative, and class-wise evidence within a single bag present another difficulty, as some patches might support different classifications simultaneously. The model must effectively weigh these conflicting signals. Furthermore, a discussion on instance interactions is crucial because the impact of one patch often depends on the context of other patches within the same tissue sample. This means the model has to consider not just individual patches but their complex relationships within the bag to make accurate predictions. Finally, the authors would probably address the computational limitations of certain MIL explanation methods, especially for large bag sizes typical in histopathology, highlighting the challenges in achieving both accuracy and efficient interpretation.
Future Research#
Future research directions stemming from this work could explore more sophisticated methods for integrating instance interactions within the MIL framework, moving beyond pairwise relationships to capture higher-order dependencies. Investigating alternative aggregation functions beyond attention mechanisms, especially those better suited for handling diverse instance characteristics in histopathology, would be valuable. Further research could focus on developing more robust and efficient explanation methods for MIL that scale well to very large datasets, addressing computational limitations of current approaches. Finally, exploring the application of xMIL and xMIL-LRP to other weakly supervised learning domains beyond histopathology, such as time-series analysis and remote sensing, could reveal further insights and broaden the impact of this work.
More visual insights#
More on figures

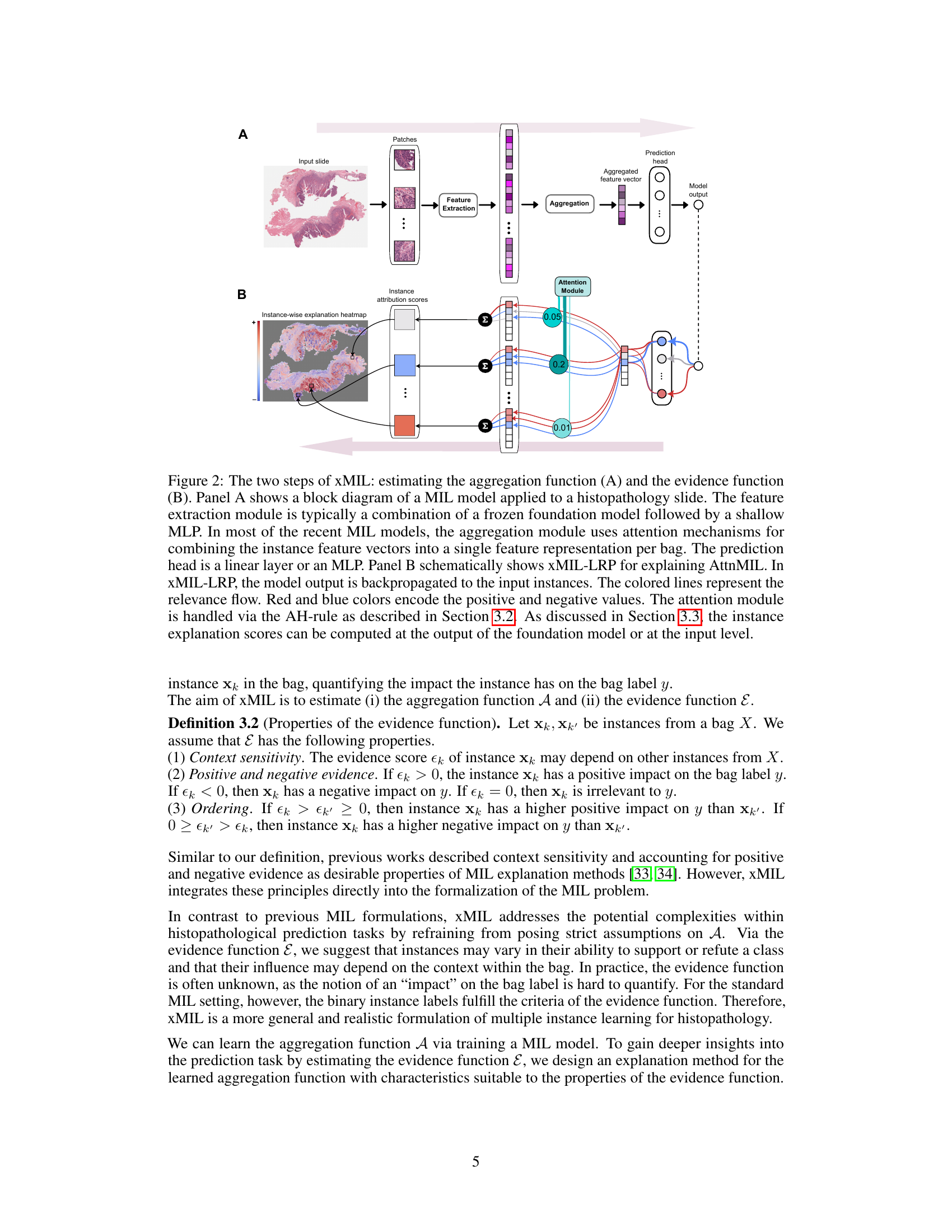
This figure illustrates the two main steps of the xMIL framework: aggregation function estimation and evidence function estimation. Panel A shows the standard MIL model architecture applied to a histopathology slide, while Panel B details the xMIL-LRP method for explaining AttnMIL, highlighting the backpropagation of model output to input instances and the use of color-coded relevance flow to represent positive and negative evidence.

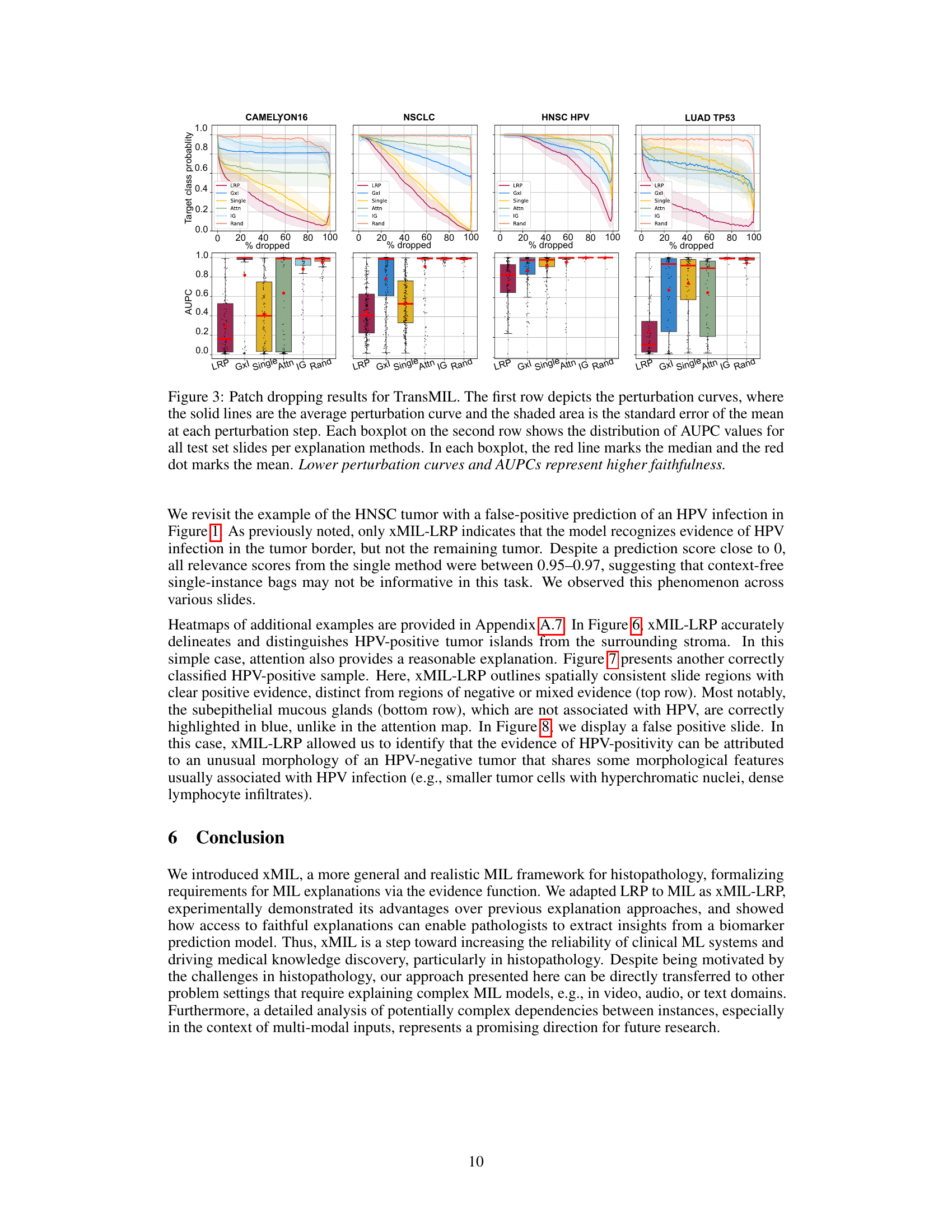
This figure displays the results of a faithfulness evaluation for TransMIL, a method for multiple instance learning (MIL). The top row shows perturbation curves, which illustrate how the model’s prediction changes as progressively more of the most relevant patches (as determined by different explanation methods) are removed from the input. The bottom row shows box plots summarizing the area under the perturbation curve (AUPC) for each explanation method across all slides. Lower AUPC values indicate higher faithfulness, meaning the explanation method’s rankings of patch importance more accurately reflect the model’s predictions.

This figure displays the results of a faithfulness evaluation for TransMIL model using a patch-dropping method. The top row shows perturbation curves illustrating how the model’s prediction changes as progressively more of the most relevant patches are removed. The bottom row presents boxplots summarizing the area under the perturbation curves (AUPC) for each explanation method across all slides. Lower AUPC values indicate higher faithfulness, meaning the explanation method better reflects the model’s predictions.

This figure compares heatmaps from three different MIL explanation methods: xMIL-LRP, Attention, and Single. It uses a head and neck tumor slide as an example, showing how each method highlights different areas of the slide as important for predicting HPV status. xMIL-LRP offers a more detailed and nuanced explanation, distinguishing between positive and negative evidence, while the other methods provide less insightful results.
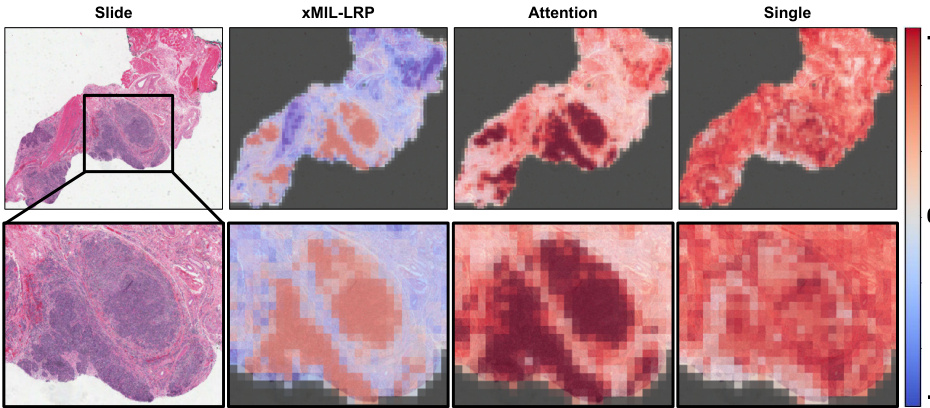
The figure shows heatmaps generated by three different MIL explanation methods (xMIL-LRP, Attention, Single) applied to a head and neck tumor slide. The heatmaps highlight the areas of the slide that are most important for the model’s prediction of HPV status. xMIL-LRP provides a more detailed and accurate explanation than the other two methods, showing both positive and negative evidence for the prediction. The other methods fail to provide such detailed insights.
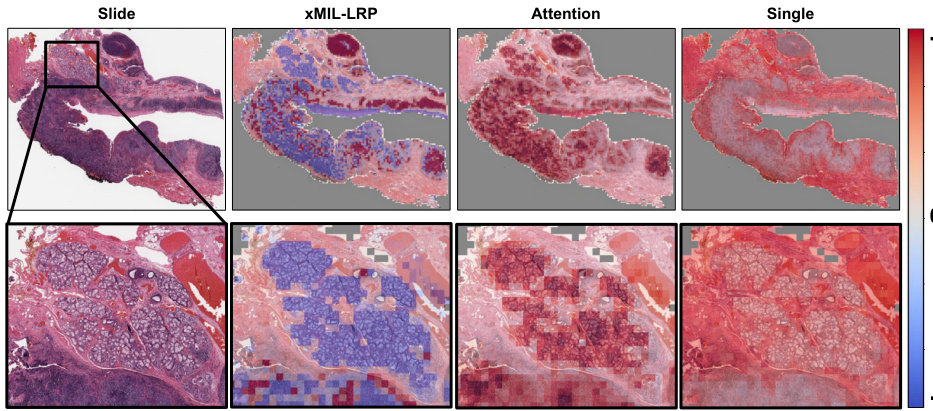
This figure shows heatmaps generated by three different MIL explanation methods: xMIL-LRP, Attention, and Single. The heatmaps highlight regions of a tissue slide that are most important for a model’s prediction of HPV status. xMIL-LRP provides a more detailed explanation, differentiating between positive (red) and negative (blue) evidence, which allows pathologists to better understand the model’s reasoning. The other two methods provide less detailed and less informative heatmaps.

This figure compares heatmaps from three different MIL explanation methods (xMIL-LRP, Attention, and Single) applied to a head and neck tumor slide. The goal is to predict HPV status. xMIL-LRP provides a more detailed and accurate heatmap highlighting areas of positive and negative evidence, allowing pathologists to understand the model’s decision-making process. In contrast, the other two methods provide less informative heatmaps.
Full paper#