↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Current neural network interatomic potentials (NNIPs) struggle with scalability due to complex domain constraints designed to improve accuracy. These constraints hinder the model’s ability to learn effective representations and generalize well to larger datasets. This ultimately limits the potential applications of NNIPs in computationally intensive tasks like large-scale materials simulations.
This research introduces the Efficiently Scaled Attention Interatomic Potential (ESCAIP), a novel architecture specifically designed for scalability. By leveraging attention mechanisms within graph neural networks, ESCAIP efficiently scales model size, significantly improving computational efficiency and accuracy. It achieves state-of-the-art results on a broad range of datasets and demonstrates a 10x speedup in inference and 5x less memory usage compared to existing NNIP models. This work suggests that focusing on scalable architectures rather than complex domain-specific constraints is a more effective approach for enhancing both accuracy and efficiency in NNIP development.
Key Takeaways#
Why does it matter?#
This paper significantly advances the field of neural network interatomic potentials by introducing ESCAIP, a scalable architecture that achieves state-of-the-art performance on various datasets while dramatically improving efficiency. This opens new avenues for research, particularly in materials science and catalysis, where large-scale simulations are crucial.
Visual Insights#
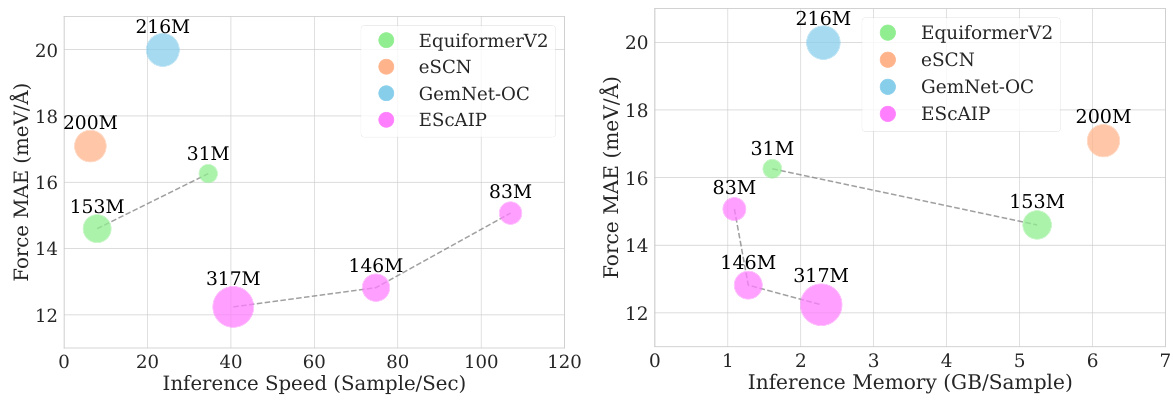
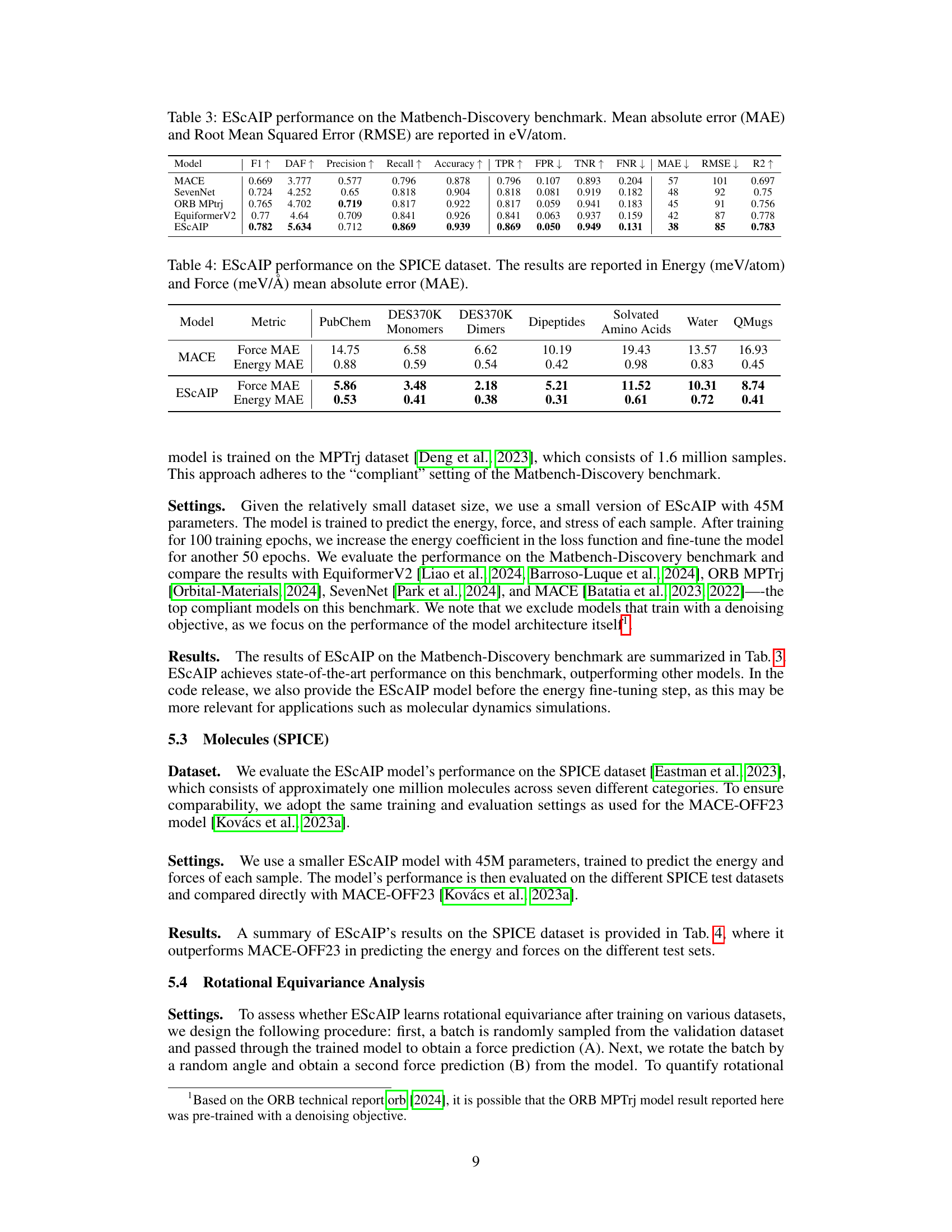
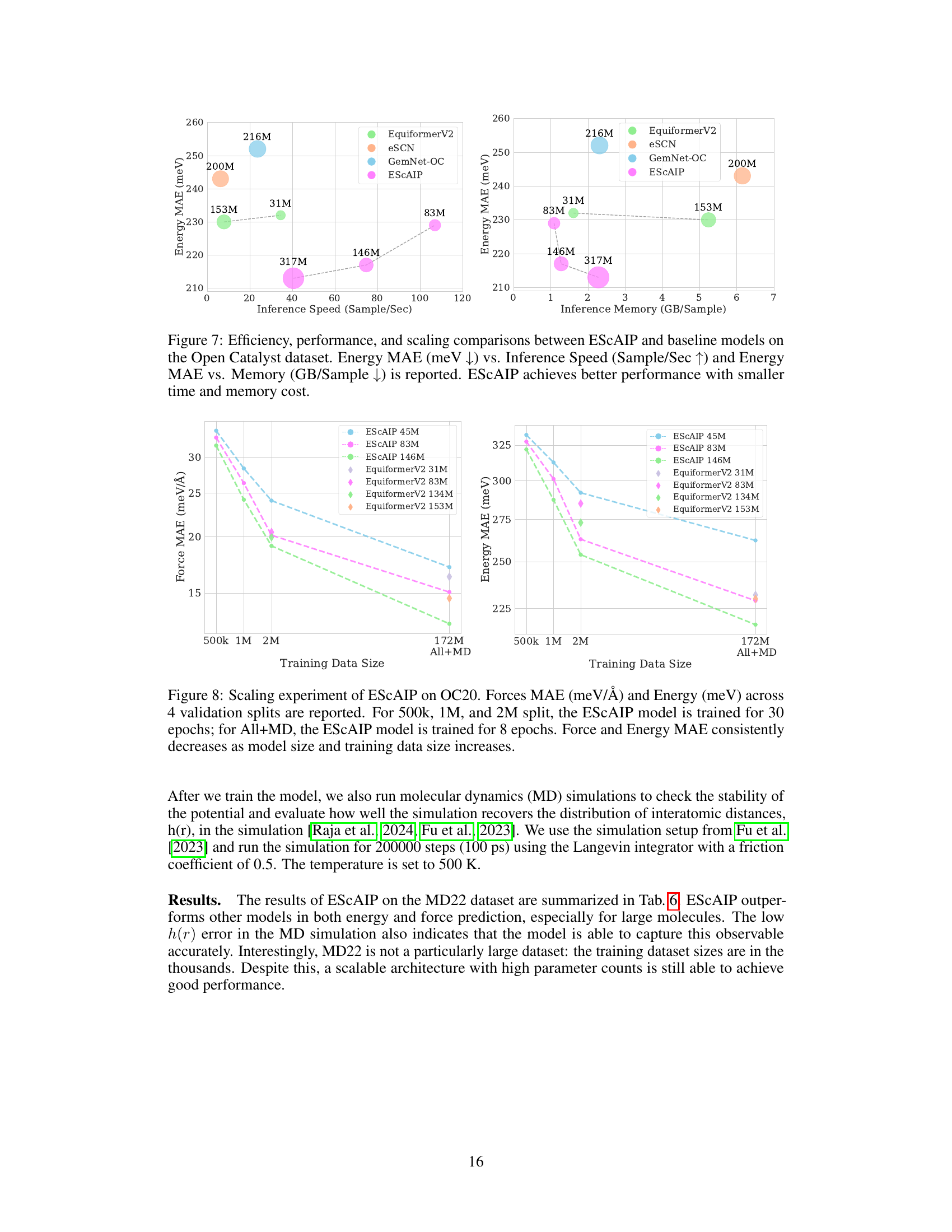
This figure compares the performance of ESCAIP with other neural network interatomic potential (NNIP) models on the Open Catalyst 20 dataset. Two plots are shown: one illustrating the relationship between inference speed and force mean absolute error (MAE), and another showing the relationship between memory usage and force MAE. Both plots demonstrate that ESCAIP achieves state-of-the-art performance with significantly improved efficiency (faster speed and lower memory usage) compared to other models.
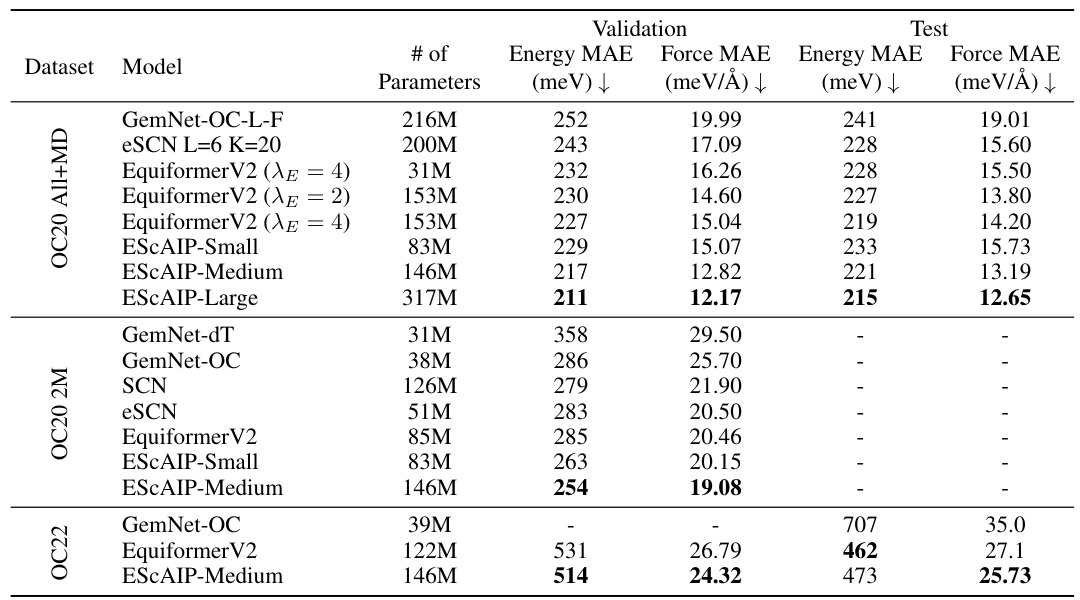
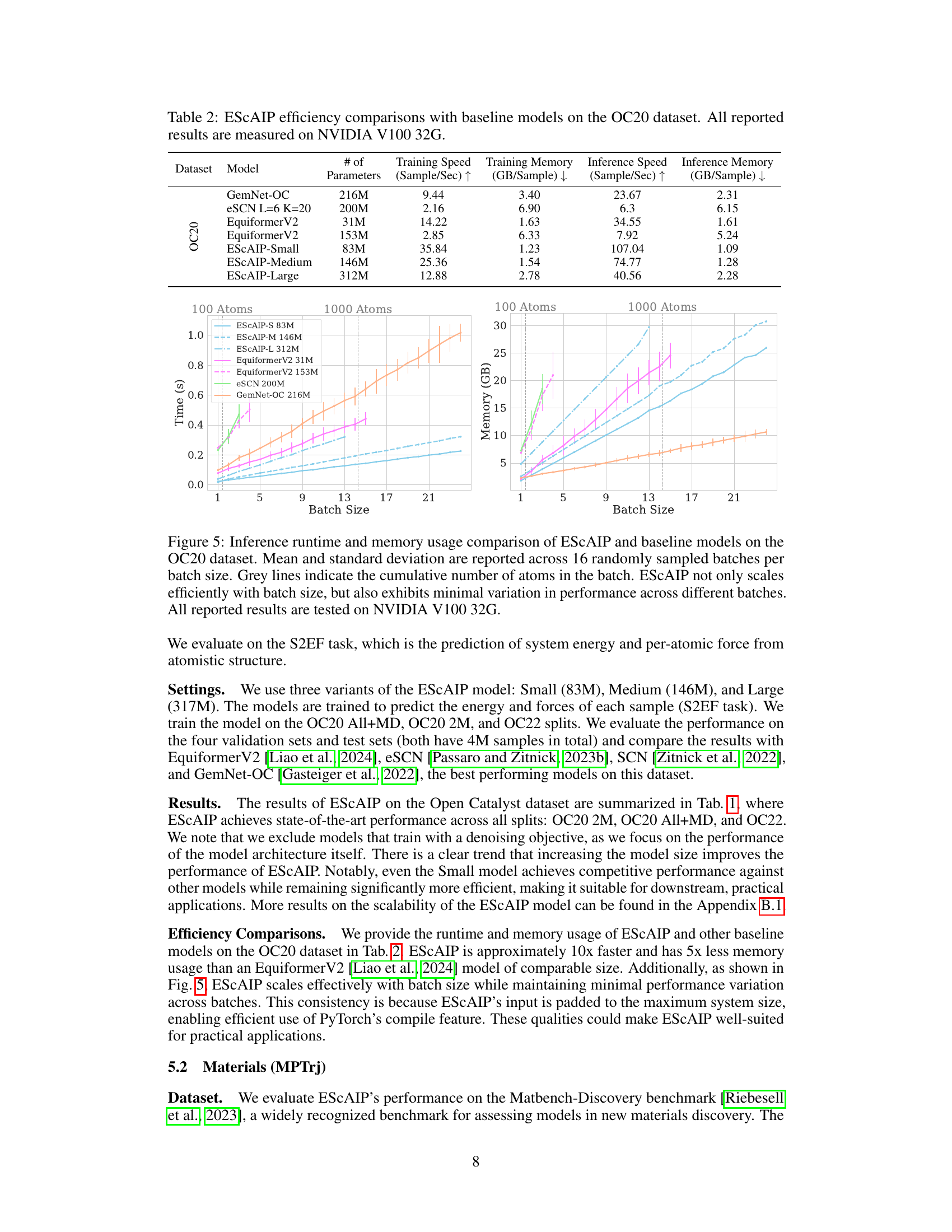
This table presents a comparison of the performance of the Efficiently Scaled Attention Interatomic Potential (ESCAIP) model against other state-of-the-art models on three different datasets: OC20 All+MD, OC20 2M, and OC22. The performance is measured in terms of mean absolute error (MAE) for both energy and force predictions. The table also shows the number of parameters for each model, highlighting ESCAIP’s efficiency and comparable or superior performance despite using fewer parameters.
In-depth insights#
Scalable NNIPs#
The concept of “Scalable NNIPs” revolves around the ability of neural network interatomic potentials to efficiently handle larger datasets and model sizes. Current NNIPs often struggle with scalability due to the incorporation of numerous physical constraints, such as symmetry constraints, which limit the model’s flexibility and computational efficiency. A key challenge lies in balancing the desire for accurate, physically-informed models with the need for efficient scaling. This necessitates a shift towards general-purpose architectures that prioritize efficient computation over strict adherence to pre-defined constraints. The development of attention mechanisms within NNIPs, particularly at the neighbor level, presents a promising approach, offering improvements in model expressivity and computational efficiency, thus enabling scalability. This approach allows for the development of general-purpose NNIPs that achieve better expressivity and efficiency while scaling with increased computational resources and training data.
Attention Mechanisms#
The effective use of attention mechanisms is a central theme in the paper, significantly boosting the efficiency and accuracy of neural network interatomic potentials (NNIPs). The authors highlight that scaling model parameters within attention mechanisms, rather than through other methods like increasing the rotation order (L) in equivariant models, proves to be a more effective strategy. This approach leads to substantial improvements in both model performance and computational efficiency. Optimized GPU kernels further enhance the speed of ESCAIP’s attention mechanisms, resulting in considerable speedups compared to existing NNIP models. The paper emphasizes that the application of attention is not merely about a specific architecture, but a philosophy for developing scalable, general-purpose NNIPs. This novel approach to NNIP development suggests a paradigm shift from incorporating complex, physics-based constraints to leveraging general-purpose architectures, and highlights the power of attention in achieving this goal.
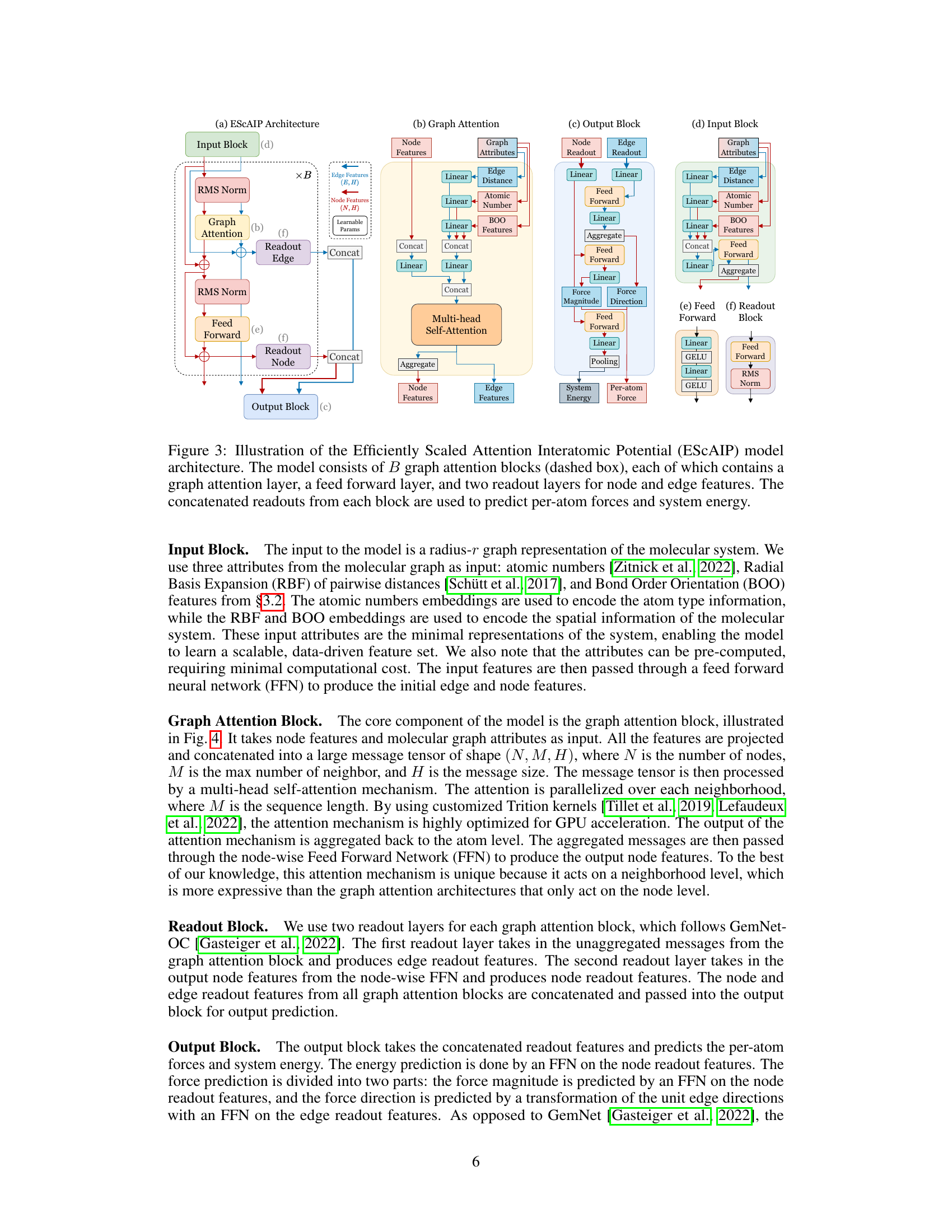
ESCAIP Architecture#
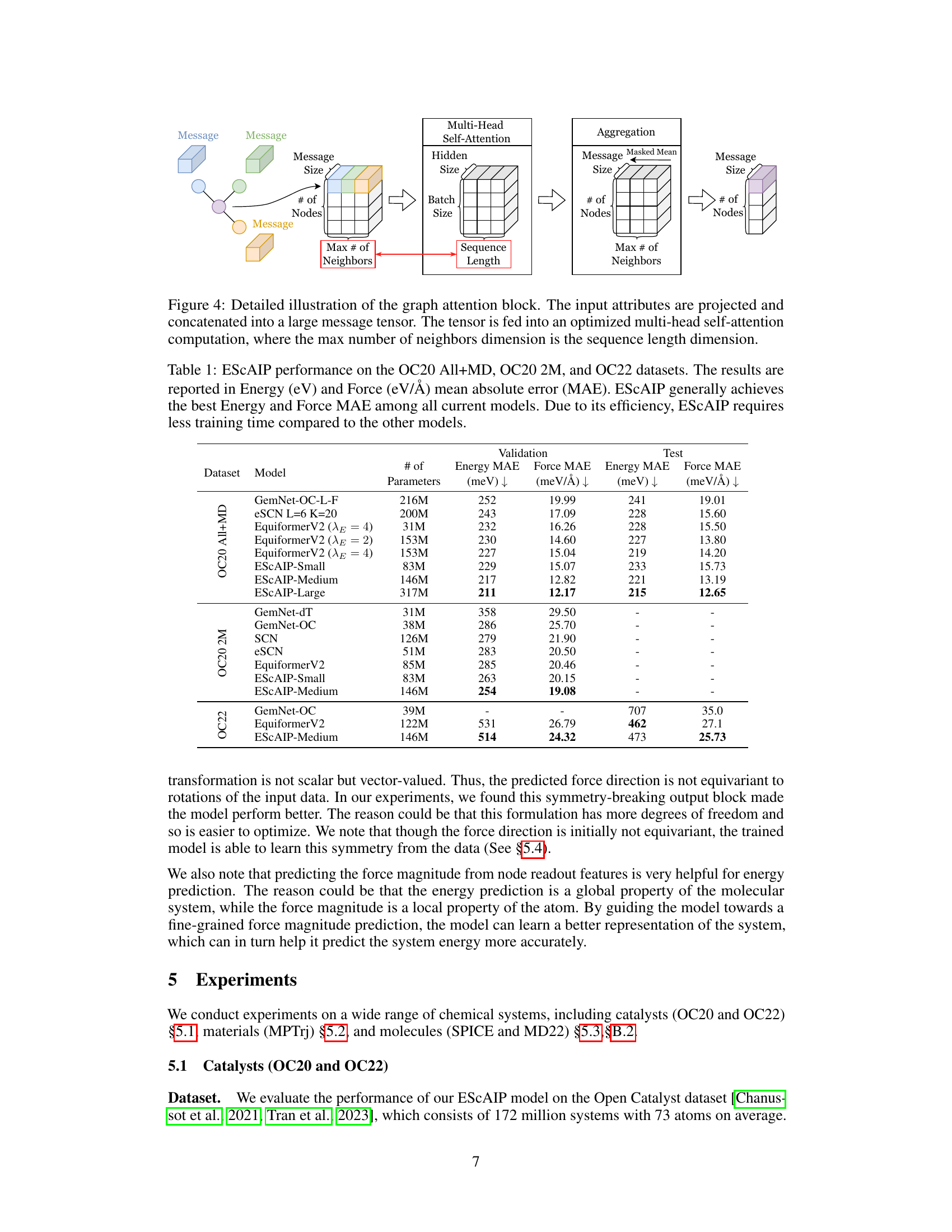
The Efficiently Scaled Attention Interatomic Potential (ESCAIP) architecture is a novel approach to neural network interatomic potentials (NNIPs), prioritizing scalability and efficiency. ESCAIP leverages a multi-head self-attention mechanism within graph neural networks, operating on neighbor-level representations rather than just node-level features. This design choice enhances model expressivity and allows for highly optimized GPU implementations. Unlike many existing NNIPs that incorporate numerous domain-specific constraints like rotational equivariance, ESCAIP opts for a more general-purpose architecture, leading to substantial performance gains and improved efficiency in inference time and memory usage. This design philosophy emphasizes the importance of achieving better expressivity through scaling, making ESCAIP a significant advancement in the field of NNIPs.
Benchmark Results#
A dedicated ‘Benchmark Results’ section would be crucial for a research paper on neural network interatomic potentials (NNIPs). It should present a rigorous comparison of the proposed NNIP model (e.g., ESCAIP) against existing state-of-the-art models across multiple datasets and metrics. This comparison would need to cover several aspects: Quantitative performance metrics: It should include mean absolute error (MAE), root mean squared error (RMSE), and other relevant metrics for both energy and forces predictions. Datasets should be carefully selected to represent a diversity of chemical systems, including molecules, materials, and catalysts. The results should clearly demonstrate ESCAIP’s advantages (e.g., improved accuracy, efficiency, or scalability) compared to baselines. The discussion should analyze the model’s performance across different scales, perhaps including an investigation of scaling behavior with respect to model size, data size, and computational resources. Finally, it should discuss any limitations or unexpected behavior observed during benchmarking and potential future directions for improvement.
Future Directions#
Future research directions in neural network interatomic potentials (NNIPs) should prioritize scaling model size and data further, exploring strategies beyond simply increasing model parameters. This includes investigating the interplay of attention mechanisms and model expressivity, coupled with highly optimized GPU kernels to ensure efficient training and inference. General-purpose NNIP architectures that transcend specific domain constraints are key, enabling better generalization across chemical domains and minimizing the need for handcrafted features. Furthermore, a significant focus on generating and utilizing larger, more comprehensive datasets is crucial for improving model performance and accuracy. Addressing this data gap through collaborative efforts will enable the development of truly general-purpose NNIPs capable of efficiently handling larger-scale simulations. Finally, exploring the use of model distillation and pre-training techniques for smaller datasets and specific applications may enhance the efficiency and accessibility of NNIP technology.
More visual insights#
More on figures

This figure presents the results of an ablation study conducted on the EquiformerV2 model to investigate how different strategies for scaling model parameters affect the model’s performance in predicting energy and forces. The study systematically varies the number of parameters in different parts of the model while keeping the total number of parameters roughly constant across different configurations. The results demonstrate that increasing the attention mechanism parameters is particularly effective in improving performance, while other scaling strategies may not be as effective.
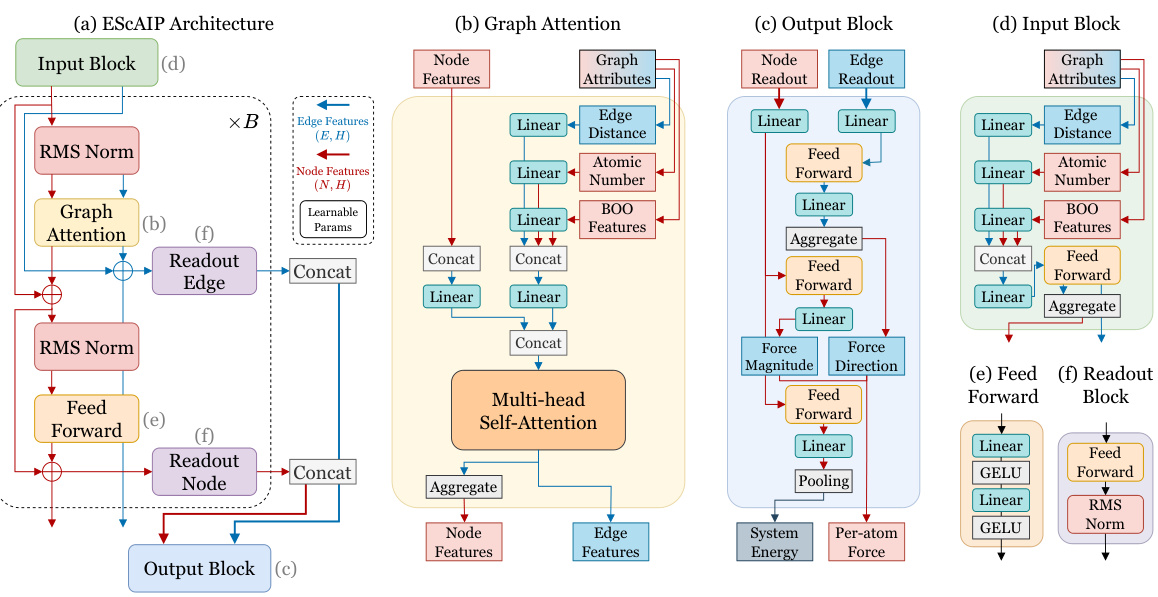
This figure illustrates the architecture of the Efficiently Scaled Attention Interatomic Potential (ESCAIP) model. The model comprises multiple graph attention blocks, each block sequentially processing node and edge features. Each block includes a graph attention layer, a feed-forward network, and two readout layers which aggregate information to predict both the per-atom forces and total system energy. The input features are atomic numbers, radial basis expansion of pairwise distances, and bond orientational order (BOO).

This figure shows the detailed steps of the graph attention block in the ESCAIP model. It begins by projecting and concatenating input attributes into a large message tensor. This tensor is then processed by a multi-head self-attention mechanism, optimized for GPU acceleration using custom Triton kernels. The attention mechanism is parallelized over each neighborhood, with the maximum number of neighbors determining the sequence length. Finally, the resulting messages are aggregated back to the atom level.

This figure shows the results of an ablation study on the EquiformerV2 model, investigating the effect of scaling parameters through attention mechanisms and spherical channels on model performance with varying training dataset sizes. It demonstrates that scaling parameters in attention mechanisms leads to faster performance improvements with larger datasets compared to scaling spherical channels or using the original model.
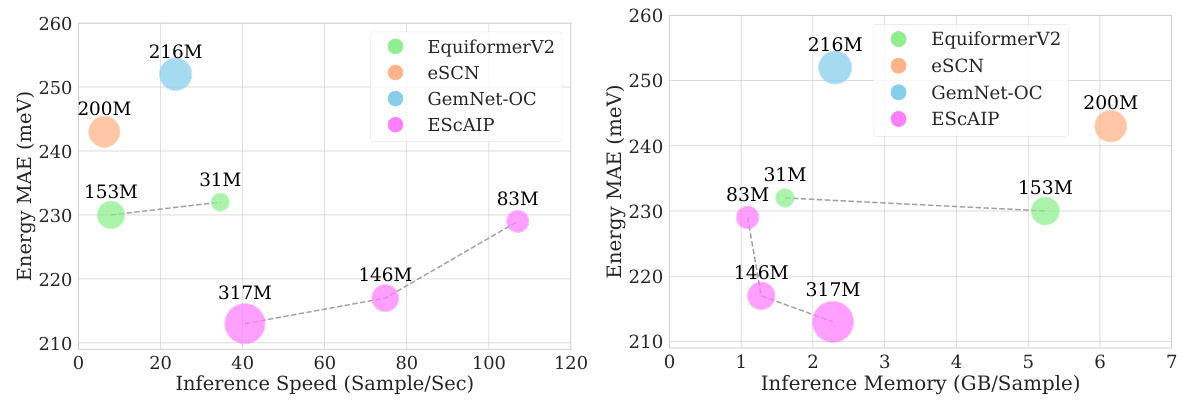
This figure compares the efficiency, performance, and scalability of the proposed Efficiently Scaled Attention Interatomic Potential (ESCAIP) model against three other baseline models (EquiformerV2, eSCN, and GemNet-OC) on the Open Catalyst 20 dataset. The plots show the relationship between force mean absolute error (MAE), inference speed, and memory usage for each model. Lower MAE values are better, indicating higher accuracy; higher inference speed is better, indicating faster computation; and lower memory usage is better, indicating less resource consumption. ESCAIP consistently outperforms the baseline models across all three metrics.
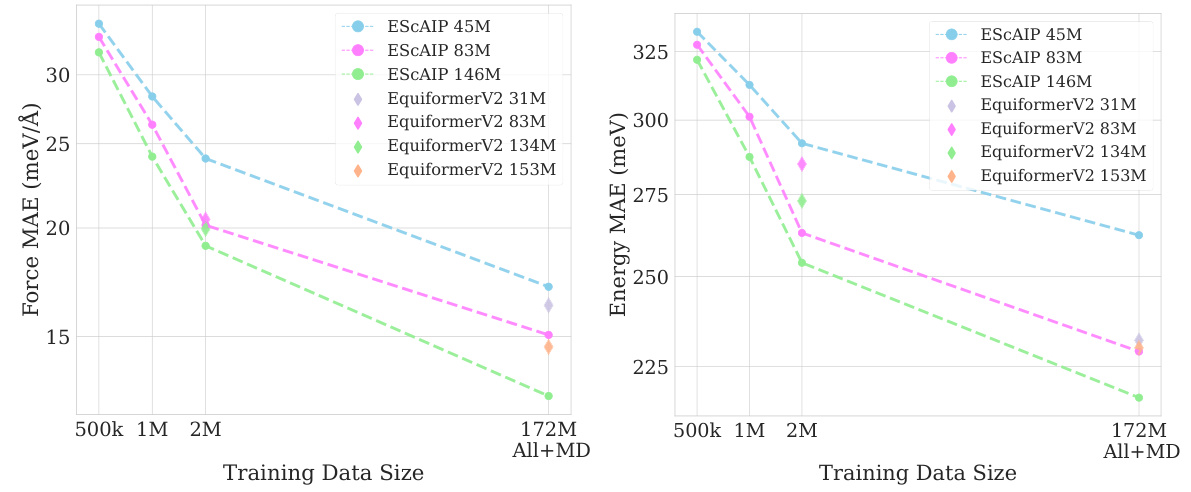
This figure shows the results of scaling experiments performed on the Open Catalyst 20 dataset (OC20) using the Efficiently Scaled Attention Interatomic Potential (ESCAIP) model. It demonstrates how both force MAE (Mean Absolute Error) and energy MAE decrease as the model size and the amount of training data increase. The different data sizes used for training are indicated: 500k, 1M, 2M, and the full All+MD dataset. The number of training epochs also varies depending on the dataset size, with fewer epochs used for the larger datasets. The consistent downward trend in MAE signifies that ESCAIP scales well with increased data and model size.
More on tables
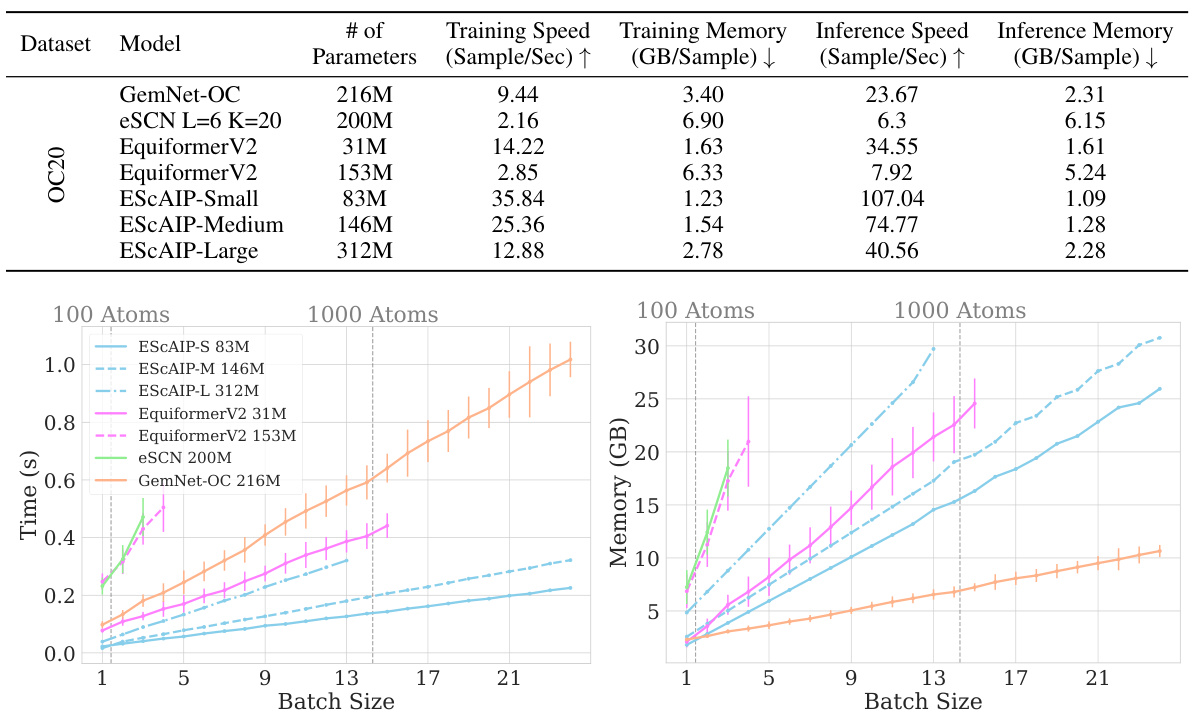
This table compares the training speed, training memory, inference speed, and inference memory of ESCAIP with baseline models (GemNet-OC, eSCN, EquiformerV2) on the Open Catalyst 20 dataset. It shows that ESCAIP is significantly more efficient than the other models, achieving substantial speedups and memory savings, especially as the model size increases.
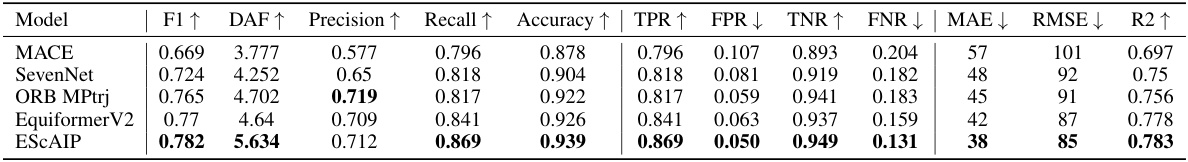
This table presents the performance of the ESCAIP model, along with several other models, on the Matbench-Discovery benchmark. The benchmark assesses the ability of models to predict material properties. The table shows various metrics such as F1 score, mean absolute error (MAE), root mean squared error (RMSE), and R-squared (R2), providing a comprehensive evaluation of the model’s accuracy and predictive power. Higher values for F1, Precision, Recall, Accuracy, TPR, and R2 and lower values for MAE, RMSE, FPR, and FNR indicate better performance.
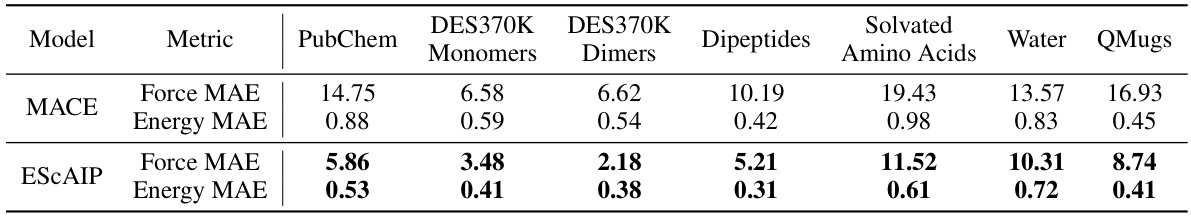
This table presents the results of the Efficiently Scaled Attention Interatomic Potential (ESCAIP) model on the SPICE dataset. It compares ESCAIP’s performance against the MACE model, showing mean absolute errors (MAE) for both energy (meV/atom) and force (meV/Å) across various categories of molecules in the dataset: PubChem, Monomers, Dimers, Dipeptides, Amino Acids, Water, and QMugs. Lower MAE values indicate better performance.
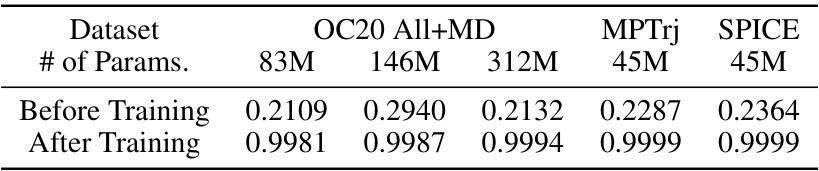
This table presents the results of a rotational equivariance analysis performed on the ESCAIP model after training on various datasets (OC20 All+MD, MPTrj, and SPICE). The analysis involved predicting forces on atomistic systems, rotating those systems, predicting forces again, and then calculating the cosine similarity between the two sets of force predictions. A high cosine similarity (consistently above 0.99) indicates that ESCAIP accurately predicts rotations.
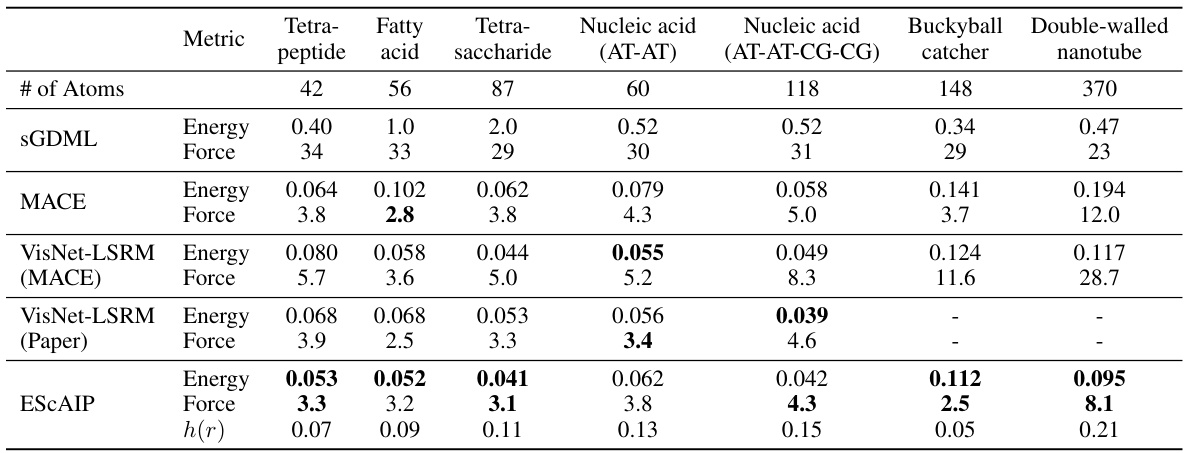
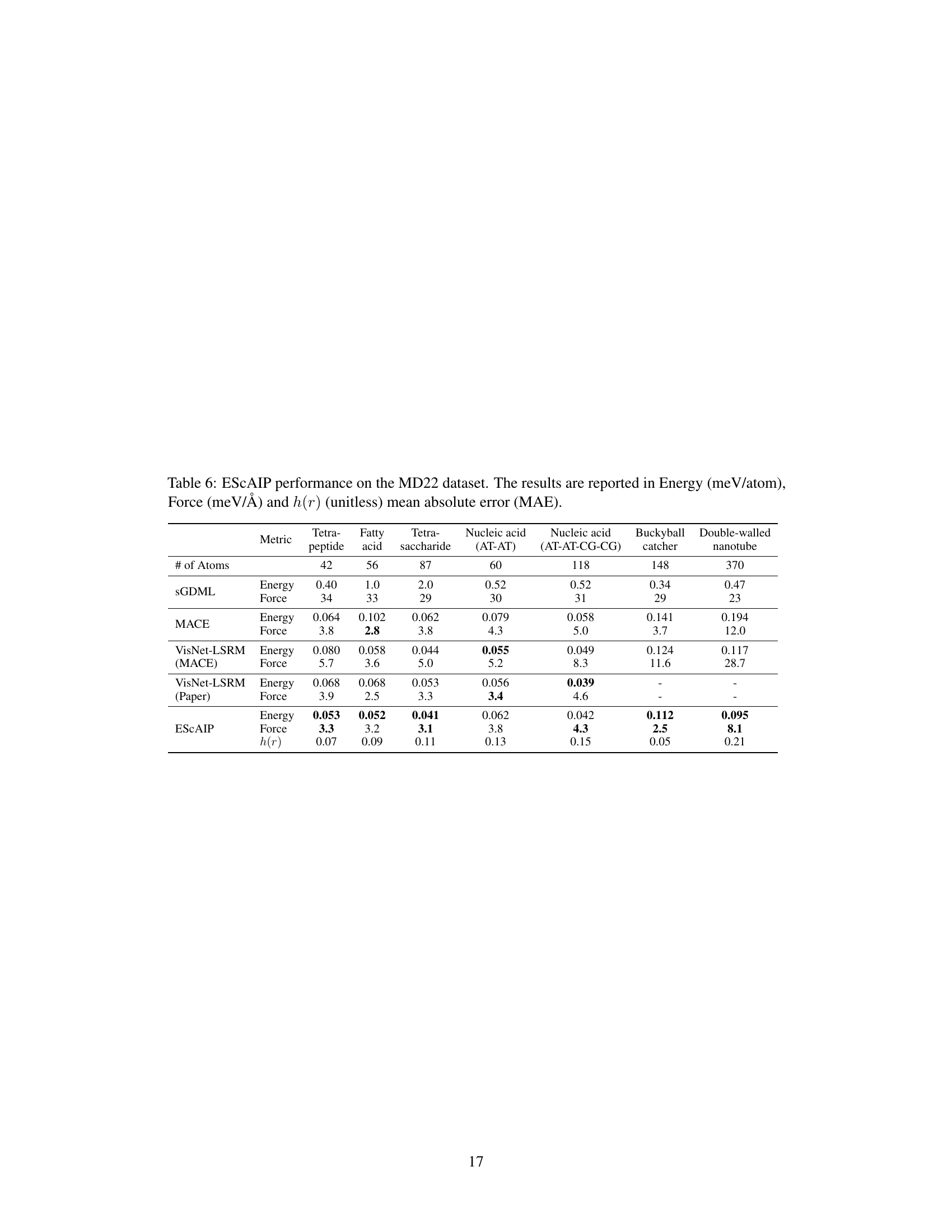
This table presents a comparison of the performance of various models, including ESCAIP, on the MD22 dataset. The performance is measured by the mean absolute error (MAE) for energy (meV/atom) and force (meV/Å), and also includes the MAE of the radial distribution function h(r) which is a measure of the structural accuracy of the simulation. The models are evaluated on several different molecules of varying sizes and complexities.
Full paper#