↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Inferring high-resolution fMRI features from readily available EEG data has been a challenge in neuroscience due to the complex relationship between neural activity and fMRI hemodynamic responses, and the spatial ambiguity of EEG signals. Existing EEG-fMRI translation methods are limited in the brain areas studied and the experimental conditions used. They often lack generalizability, hindering broader applicability.
This paper introduces NeuroBOLT, a multi-dimensional transformer-based framework for EEG-to-fMRI synthesis. NeuroBOLT overcomes previous limitations by learning representations from multiple domains (temporal, spatial, spectral) to translate raw EEG data into fMRI activity across various brain regions (primary sensory, high-level cognitive, deep subcortical). It achieves state-of-the-art accuracy and demonstrates remarkable generalizability across resting-state and task conditions, as well as across subjects and sites.
Key Takeaways#
Why does it matter?#
This paper is important because it presents NeuroBOLT, a novel and generalizable framework that successfully reconstructs resting-state fMRI signals from EEG data with state-of-the-art accuracy. This significantly advances the integration of EEG and fMRI, two commonly used but distinct neuroimaging modalities. The research opens new avenues for investigating brain function, particularly in resting-state conditions, and has potential clinical applications by enabling cost-effective, portable brain imaging.
Visual Insights#
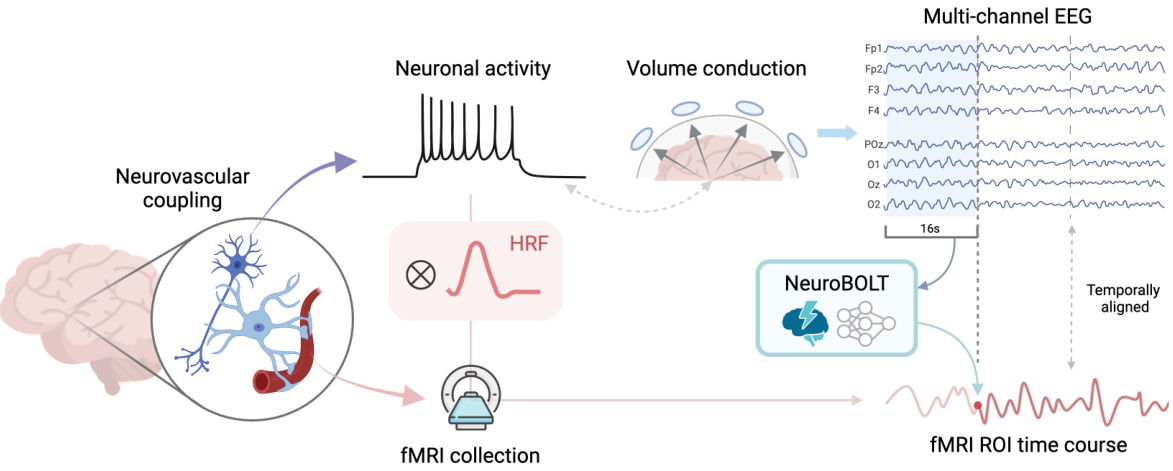
This figure illustrates the process of translating EEG signals into fMRI signals using the NeuroBOLT model. It starts with neuronal activity generating electrical potentials, which are then measured by EEG electrodes on the scalp. Due to volume conduction, the EEG signals do not directly reflect the location of neural activity. These EEG signals, after a 16-second time window, are processed by the NeuroBOLT model. The model outputs a temporally aligned fMRI ROI (region of interest) time course, which represents the predicted fMRI activity. The figure highlights the challenges of EEG-fMRI translation (volume conduction) and how NeuroBOLT attempts to overcome these to synthesize the high-resolution fMRI activity from EEG.
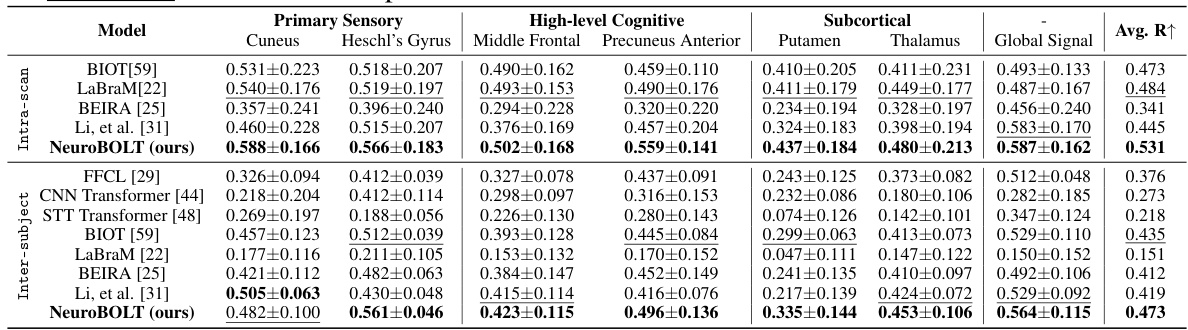
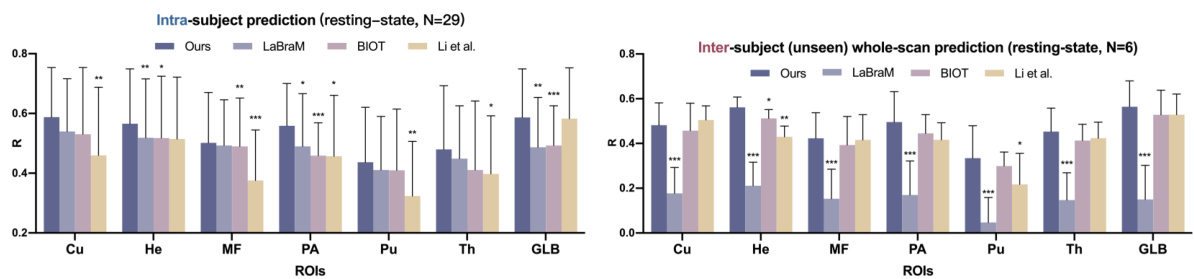
This table presents the Pearson correlation coefficient (R) values achieved by different models in both intra-subject and inter-subject experiments. The intra-subject results show the model performance when training and testing are performed on the same subject’s data, while inter-subject results indicate the model’s generalizability to unseen subjects. Higher R values indicate better performance. The best and second-best performing models for each ROI (region of interest) are highlighted.
In-depth insights#
EEG-fMRI Synthesis#
EEG-fMRI synthesis aims to bridge the gap between the high temporal resolution of EEG and the high spatial resolution of fMRI. Deep learning methods have emerged as promising tools, translating EEG data into fMRI-like brain activity maps. Challenges remain in accurately modeling the complex hemodynamic response and addressing spatial ambiguities inherent in EEG. Successful synthesis requires careful consideration of the different biophysical origins and spatial-temporal scales of the two modalities. Multi-dimensional feature mapping (incorporating temporal, spatial, and spectral information) has shown promising results, improving predictive power and generalizability across brain regions and experimental conditions. Generalizable frameworks that overcome subject-specific limitations and handle resting-state data are vital to broader adoption of EEG-fMRI synthesis in clinical and research settings. While still in development, this synthesis offers the potential for affordable, portable, and high-temporal resolution neuroimaging.
NeuroBOLT Model#
The NeuroBOLT model is a novel, multi-dimensional transformer-based framework for translating raw EEG data into corresponding fMRI activity signals across the entire brain. Its core innovation lies in leveraging multi-dimensional representations from temporal, spatial, and spectral domains, overcoming limitations of previous EEG-fMRI synthesis methods. NeuroBOLT effectively integrates spatiotemporal and spectral information, capturing the complexity of neural dynamics and the projection from neural to BOLD signals. This comprehensive approach enables the accurate reconstruction of fMRI signals from unseen resting-state data, particularly showing promise in primary sensory and subcortical brain regions. A key strength is its generalizability, demonstrated through experiments on both resting-state and task-based fMRI datasets, and its potential for cross-subject prediction. While impressive results are shown, future directions include scaling to high-resolution fMRI reconstruction and addressing potential biases from the limited dataset size.
Multi-Scale Fusion#
Multi-scale fusion, in the context of EEG-fMRI synthesis, is a crucial technique for effectively capturing the complex temporal dynamics of brain activity. EEG signals exhibit a wide range of frequencies reflecting different neural processes, while fMRI data has a much lower temporal resolution. A multi-scale approach addresses this mismatch by incorporating spectral features from multiple frequency bands, each with its own temporal resolution. This allows the model to capture both high-frequency, rapid changes and low-frequency, sustained activity patterns, providing a more comprehensive representation of neural activity for accurate fMRI signal prediction. The fusion of information across these multiple scales is critical, enabling the model to learn a robust mapping between the different modalities, overcoming limitations of relying on a single frequency or timescale. The success of this approach highlights the importance of considering the multifaceted nature of brain activity for accurate cross-modal synthesis. Careful selection and integration of spectral scales is essential for optimal performance, and further research should investigate the optimal multi-scale strategies for various brain regions and cognitive tasks.
Generalization Tests#
Generalization tests in a research paper are crucial for establishing the robustness and reliability of the model or method presented. They assess the model’s ability to perform well on data that it was not specifically trained on. This involves evaluating the model’s performance on unseen datasets, different conditions, or even on different populations. A successful generalization test would demonstrate that the model’s effectiveness isn’t limited to the specific training data and can be applied more broadly. This is crucial for demonstrating practical applicability. Key aspects of generalization tests to consider include the diversity of test data, clear metrics to evaluate performance, and a thoughtful discussion on the reasons behind any successes or failures in generalization. Proper analysis of the limitations and boundary conditions for generalization provides valuable insights for future research and development. The results could highlight the need for further refinement of the model or the collection of more diverse data. Ultimately, the strength of the claims made within the paper is directly tied to the success of these generalization tests.
Future Directions#
Future research could explore enhancing NeuroBOLT’s generalizability by incorporating domain adaptation techniques to better handle variations in EEG acquisition and fMRI protocols across different labs or populations. Investigating the model’s performance with larger and more diverse datasets, including those with various pathologies, would also strengthen its robustness and clinical utility. A key area for improvement is scaling up NeuroBOLT to handle higher-resolution fMRI data, enabling more precise spatial localization of brain activity. Further research should also focus on improving the model’s interpretability, potentially through techniques like attention visualization, to enhance understanding of its underlying decision-making processes. Finally, exploring the potential of NeuroBOLT for real-time applications such as brain-computer interfaces or neurofeedback could unlock new avenues for clinical interventions and personalized treatment.
More visual insights#
More on figures
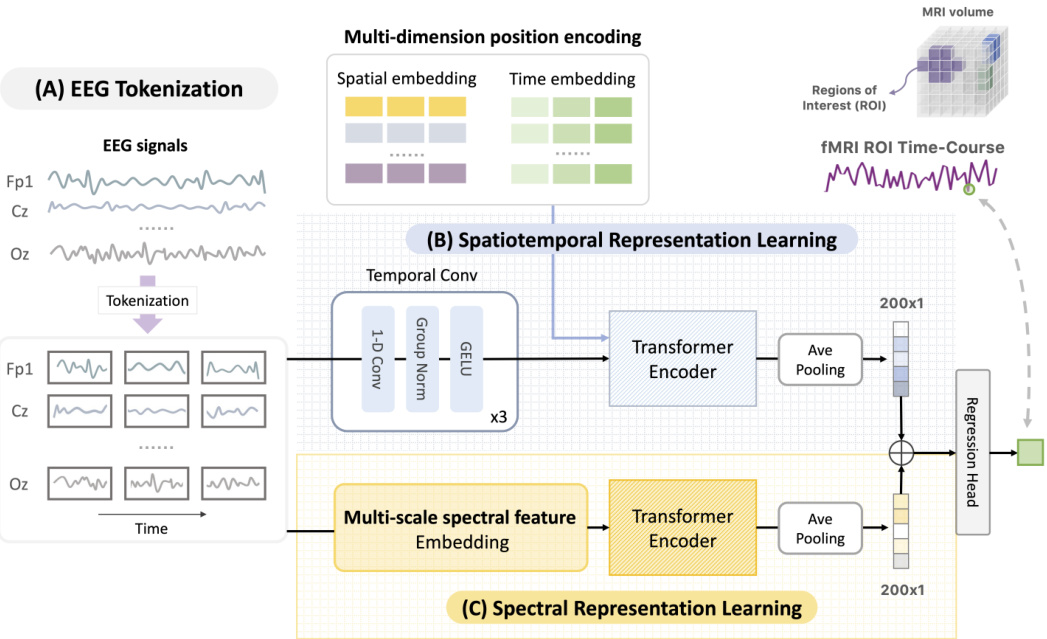
This figure illustrates the architecture of NeuroBOLT, a deep learning model for translating EEG signals into fMRI signals. The model takes an EEG window as input and processes it through two parallel modules: a spatiotemporal module and a spectral module. The spatiotemporal module extracts temporal and spatial features from the EEG, while the spectral module extracts frequency information. Both modules generate embeddings, which are then combined and fed into a regression head that outputs the predicted fMRI time series.

This figure illustrates the multi-scale spectral feature embedding method used in NeuroBOLT. It shows how EEG signals are processed to capture spectral features at multiple scales (Scale 1 to Scale L), using Short-Time Fourier Transforms (FFT) with varying window sizes. The resulting multi-scale spectral representations are then integrated using trainable frequency embeddings to provide a comprehensive representation of EEG spectral dynamics.
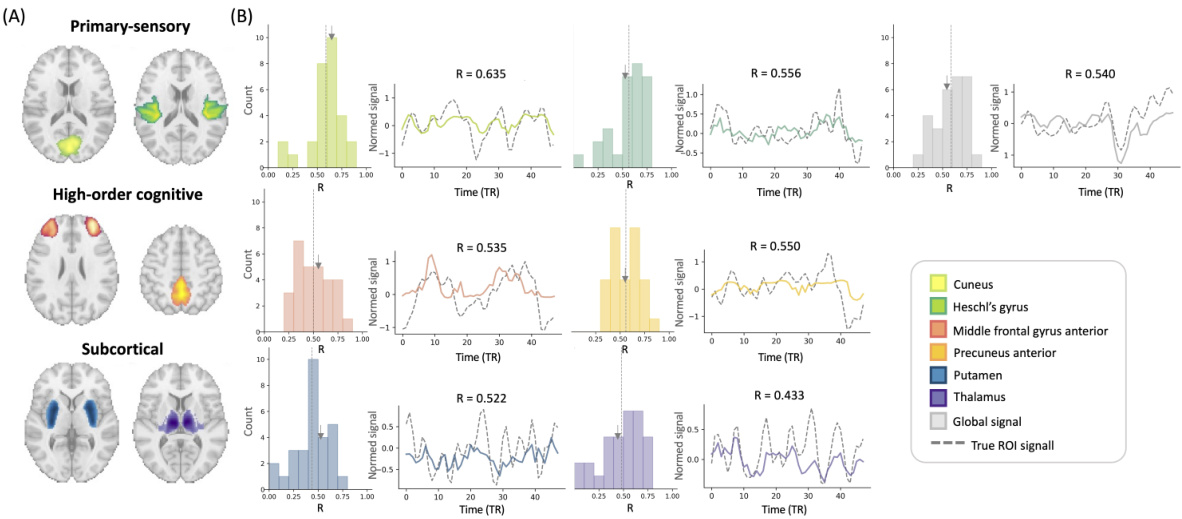
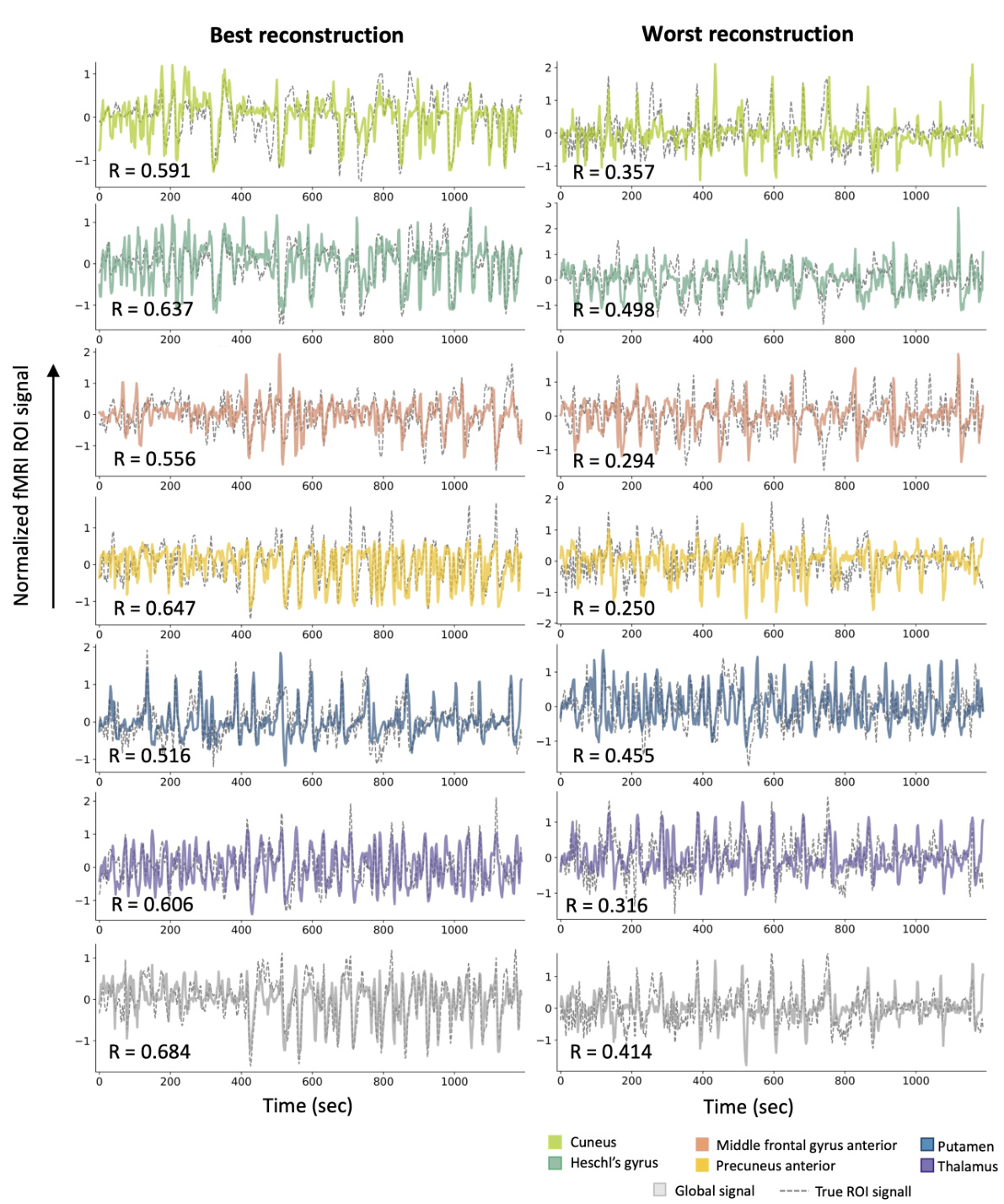
This figure visualizes the intra-subject prediction results. Panel (A) shows the brain regions (ROIs) used in the experiment. Panel (B) presents the distribution of prediction performances (correlation coefficients) and example time-series reconstructions for each ROI. The dashed lines in histograms represent the average correlation values, while the grey arrows in the histograms indicate the mean correlation values. This figure demonstrates the model’s ability to predict fMRI signals from EEG data within a single subject.
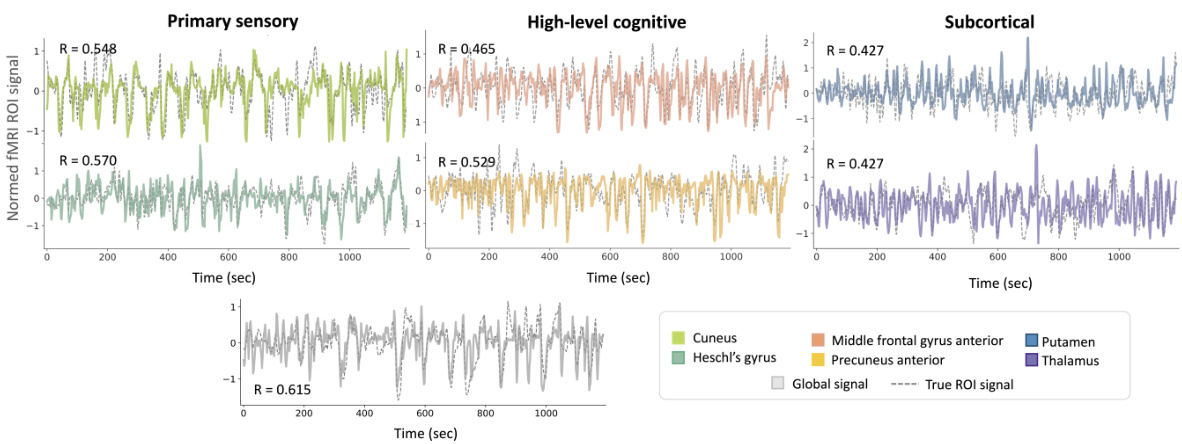
This figure presents the results of intra-subject prediction experiments. Panel (A) shows the brain regions (ROIs) that were selected for analysis. Panel (B) shows histograms and example time series plots of the correlation between predicted and true fMRI signals for each ROI. The histograms show the distribution of correlation coefficients across all subjects, and the time series plots show examples of predictions for ROIs with correlation coefficients close to the average.
This figure illustrates the architecture of NeuroBOLT, a multi-dimensional transformer-based framework for EEG-to-fMRI synthesis. The framework consists of three main stages: (A) EEG Tokenization, where raw EEG signals are divided into uniform patches; (B) Spatiotemporal Representation Learning and (C) Spectral Representation Learning, where two parallel modules extract spatiotemporal and spectral features from the EEG patches respectively; and finally, a regression head that combines the features from both modules to predict the fMRI signals. The figure shows a clear overview of the data flow and processing steps within the NeuroBOLT architecture.
This figure presents the results of intra-subject prediction on resting-state fMRI data. Panel (A) shows the brain regions (ROIs) used in the study. Panel (B) displays the distribution of Pearson correlation coefficients between the predicted and actual fMRI signals for each ROI. Example time-series reconstructions are included to illustrate the performance of the model for ROIs near the average correlation value.

This figure illustrates the architecture of NeuroBOLT, a multi-dimensional transformer-based EEG encoding framework. It shows how the model processes raw EEG data. First, EEG data is tokenized into patches. Then, two parallel modules (spatiotemporal and spectral representation learning) process these patches separately, generating embeddings. Finally, these embeddings are combined and fed into a regression head to produce the fMRI prediction.
More on tables
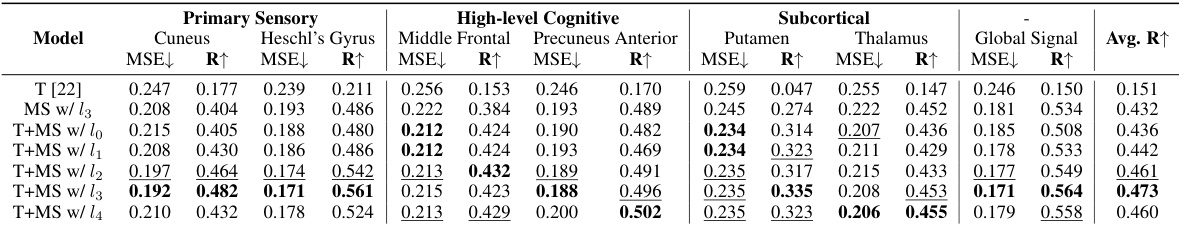
This table presents the Mean Squared Error (MSE) values for both intra-subject and inter-subject experiments conducted on resting-state data. The MSE is a measure of the average squared differences between predicted and actual fMRI signals. Lower MSE indicates better prediction accuracy. The table shows MSE values for seven regions of interest (ROIs): Cuneus, Heschl’s gyrus, Middle Frontal, Precuneus Anterior, Putamen, Thalamus, and Global Signal, for several models including NeuroBOLT and baselines. This allows for a comparison of the prediction performance of different models across different brain regions.

This table presents the Pearson correlation coefficient (R) values for the model’s performance in predicting fMRI signals from EEG data. It shows the results for both intra-subject (using the same subject’s data for training and testing) and inter-subject (using different subjects’ data for training and testing) experiments. The table breaks down the results by brain region (primary sensory, high-level cognitive, subcortical) and includes multiple models for comparison, allowing for easy evaluation of NeuroBOLT’s performance relative to other methods.
This table presents the Mean Squared Error (MSE) values for both intra-subject and inter-subject experiments conducted on resting-state fMRI data. It compares the MSE values of NeuroBOLT with several other baseline models across different regions of interest (ROIs). Lower MSE values indicate better performance in predicting fMRI signals from EEG data.
This table shows the mean squared error (MSE) values achieved by NeuroBOLT and other baseline models in both intra-subject and inter-subject prediction experiments using resting-state data. The MSE is a measure of the average squared difference between the predicted and actual fMRI signals across all time points. Lower MSE values indicate better performance. The results are broken down for each of the seven brain regions of interest (ROIs).

This table presents the Mean Squared Error (MSE) values for both intra-subject and inter-subject experiments conducted on resting-state fMRI data. It provides a quantitative comparison of the prediction accuracy of NeuroBOLT and several baseline models across seven different regions of interest (ROIs) in the brain: Cuneus, Heschl’s Gyrus, Middle Frontal, Precuneus Anterior, Putamen, Thalamus, and Global Signal. Lower MSE values indicate better prediction accuracy.
Full paper#