↗ OpenReview ↗ NeurIPS Homepage ↗ Chat
TL;DR#
Analyzing calcium imaging data in real-time is crucial for closed-loop neuroscience experiments, but existing methods are often too slow. This paper introduces challenges in real-time processing of such data, highlighting the need for fast and robust algorithms. The limitations of current real-time approaches are discussed, emphasizing problems such as handling incomplete knowledge of cell identities and false-positive activity detections.
The paper’s main contribution is realSEUDO, an improved version of the SEUDO algorithm. It is optimized for speed and robustness, simultaneously identifying neurons and inferring their activity in real time. The algorithm’s performance is demonstrated through experiments, showing comparable results to offline methods and superior performance to existing online techniques with an average processing speed of 120 Hz. This enables the use of calcium imaging in closed-loop studies requiring high-speed feedback.
Key Takeaways#
Why does it matter?#
This paper is important because it presents realSEUDO, a novel real-time algorithm for analyzing calcium imaging data. This addresses a critical need in closed-loop neuroscience experiments, where quick analysis of neural activity is crucial for adapting experiments on the fly. Its speed and robustness open doors for more sophisticated closed-loop studies and advances our understanding of neural dynamics. The approach also has implications for other real-time image analysis problems.
Visual Insights#

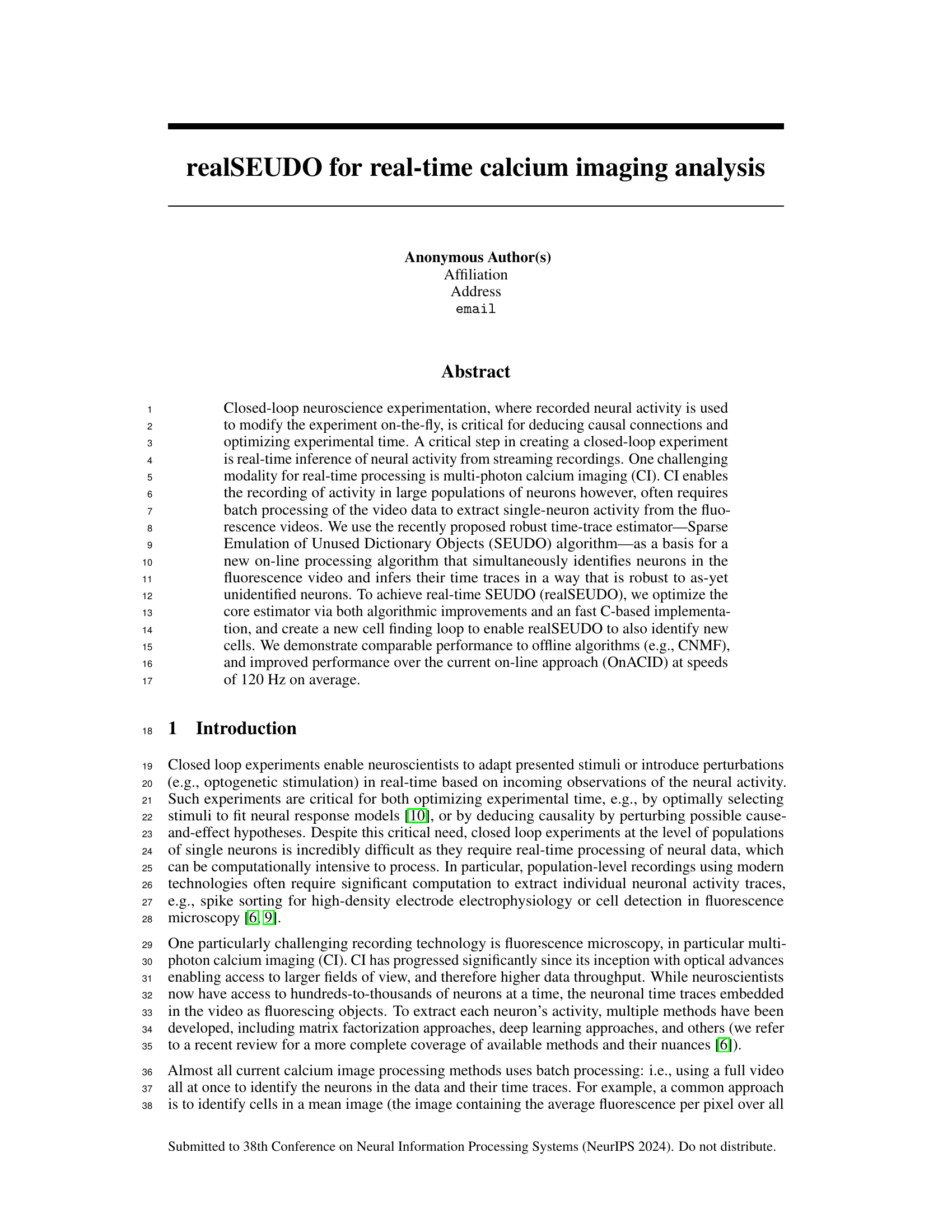
This figure describes the realSEUDO algorithm workflow. Panel A shows a schematic of a closed-loop experiment using calcium imaging, highlighting the need for real-time processing. Panel B illustrates how realSEUDO uses the SEUDO algorithm to handle missing or unknown cells, preventing false positives in activity detection. Panel C details the real-time cell identification process within realSEUDO, showing how the algorithm iteratively identifies and updates the list of known cells.

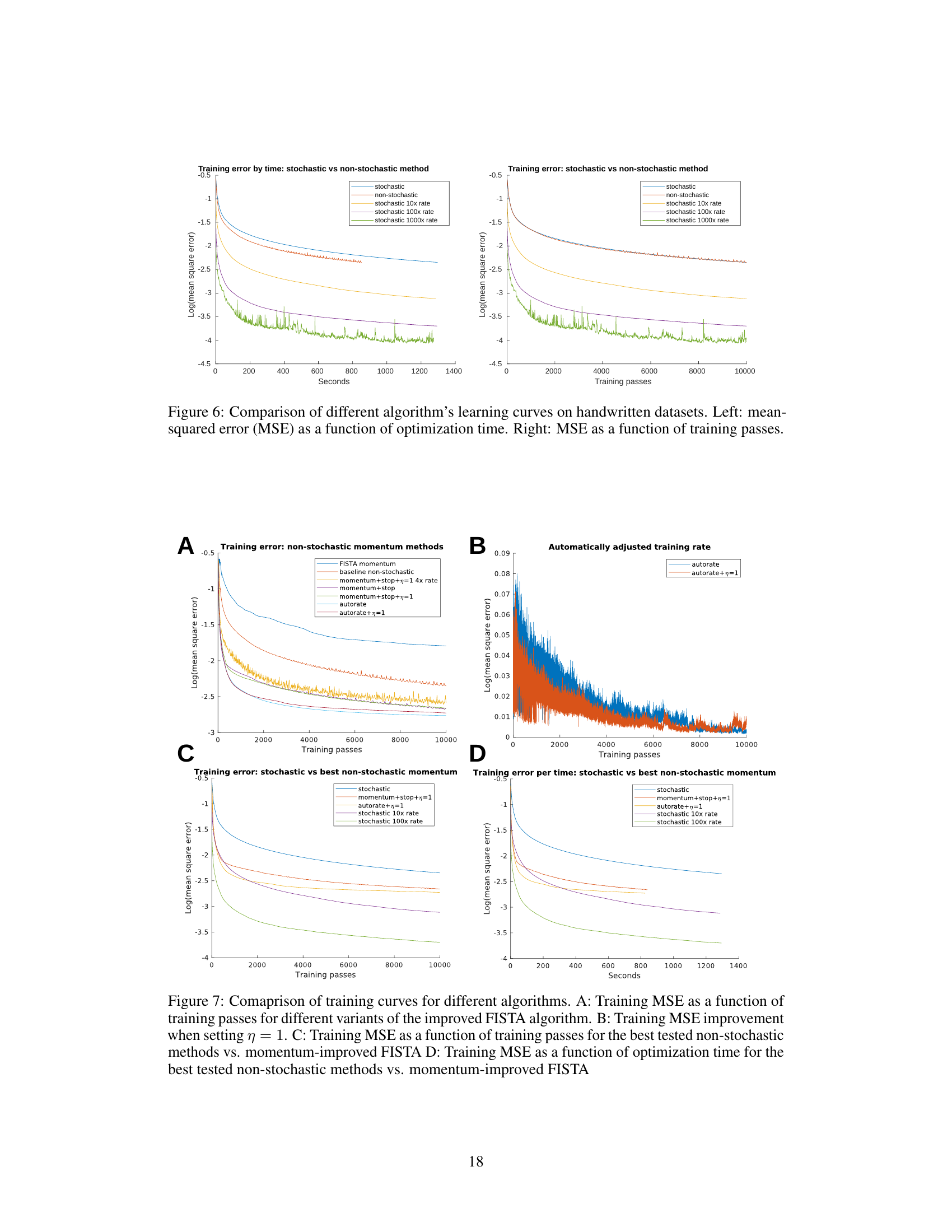
This table presents the performance comparison of different algorithms on a handwritten dataset. The algorithms compared include various versions of FISTA (Fast Iterative Shrinkage-Thresholding Algorithm), both stochastic and non-stochastic, with and without modifications like momentum stopping and automatic rate adjustments. The performance is measured using the mean squared error (MSE) and the logarithm of the MSE, showing error rate and squared mean error. The table highlights the impact of these modifications on the algorithms’ efficiency and accuracy in solving the optimization problem.
In-depth insights#
Real-time SEUDO#
The heading ‘Real-time SEUDO’ suggests a significant advancement in calcium imaging analysis. It implies the adaptation of the existing SEUDO (Sparse Emulation of Unused Dictionary Objects) algorithm, originally designed for offline processing, to operate in real-time. This is crucial for closed-loop neuroscience experiments requiring immediate feedback. Real-time processing is achieved through algorithmic optimizations and efficient C-based implementation, allowing for processing speeds exceeding 30 Hz. The algorithm likely incorporates a novel cell identification loop, enabling it to simultaneously identify neurons and infer their activity traces from streaming video data. The core innovation lies in the real-time capability while maintaining robustness to the presence of unidentified neurons, a key challenge in online calcium imaging. By optimizing the core SEUDO estimator and using patch-based parallelization, the approach likely enhances both speed and scalability. Overall, ‘Real-time SEUDO’ represents a substantial step towards enabling real-time closed-loop experiments at the level of large neuronal populations. This likely paves the way for more sophisticated, dynamic studies of neural circuit function.
FISTA Optimization#
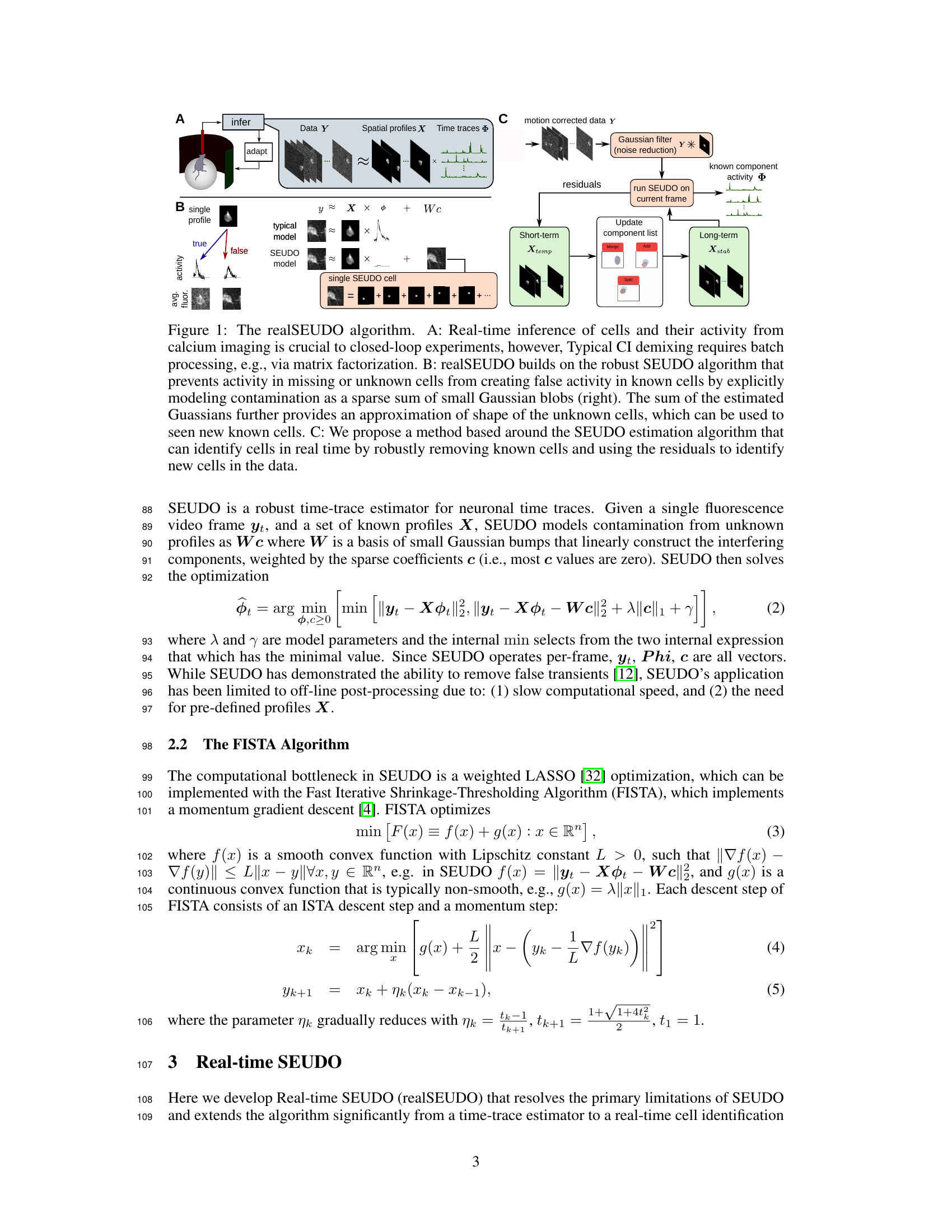
The core of the real-time SEUDO algorithm hinges on efficiently solving a weighted LASSO optimization problem. This is achieved through the Fast Iterative Shrinkage-Thresholding Algorithm (FISTA), a first-order optimization method known for its speed and effectiveness in handling such problems. FISTA’s iterative nature, involving gradient descent steps coupled with momentum updates, allows for a relatively quick convergence to the optimal solution, crucial for real-time applications. Significant algorithmic improvements were introduced to boost FISTA’s performance further. These included reducing the number of iterations required, exploiting inherent parallelism in computations, optimizing internal calculations (like smoothness parameter estimation), and simplifying the overall problem structure without sacrificing accuracy. These changes dramatically accelerated processing speed, paving the way for real-time calcium imaging data analysis.
Cell Identification#
Accurate cell identification in calcium imaging data is crucial for meaningful analysis, yet it remains a significant challenge. Real-time approaches are particularly difficult, demanding efficient algorithms that balance speed with accuracy. The paper explores this challenge by adapting the SEUDO algorithm for real-time use, focusing on identifying cells amidst noise and potential interference from unknown cells. The approach cleverly tackles incomplete knowledge of the cell population, which is a common limitation in real-time processing, by integrating a novel feedback loop. This loop dynamically updates the cell model as new cells are detected, effectively integrating new cells into the analysis without requiring a complete re-processing. The paper emphasizes the robustness of the method, ensuring accurate identification even when dealing with noisy data and cells not initially identified. This cell identification process, integrated within a larger real-time processing framework, contributes to a major step towards reliable and efficient analysis of high-throughput calcium imaging data for closed-loop neuroscience experiments.
Algorithmic Advance#
The core algorithmic advance centers on optimizing the SEUDO algorithm for real-time performance. Key improvements include a faster C++ implementation replacing the original MATLAB version, algorithmic optimizations to reduce computational steps in the core FISTA algorithm, and a novel patch-based parallelization strategy to handle large datasets efficiently. The authors also introduce a novel feedback loop for automatic cell identification, enabling realSEUDO to continually update its neuron model by identifying newly appearing cells in the data stream. This combined approach results in a significant speed increase, enabling frame processing rates exceeding 30 Hz. This contrasts with previous methods that required batch processing and couldn’t achieve such speeds. The performance gains stem from both efficient implementation choices and refinements to the underlying optimization algorithms, significantly advancing real-time capabilities for calcium imaging analysis.
Future Directions#
The research paper’s ‘Future Directions’ section would ideally explore several promising avenues. Improving real-time performance remains a crucial goal, perhaps through more sophisticated hardware acceleration or algorithmic optimizations beyond FISTA. Expanding to other imaging modalities like two-photon microscopy with different fluorophores or voltage imaging would significantly broaden the impact. Addressing incomplete cell identification is also key; robust methods for detecting and incorporating new cells as they emerge, while mitigating false positives, are needed. The method could also benefit from more rigorous testing, encompassing a wider range of experimental conditions and cell densities, to validate its generalizability. Finally, exploring the integration of closed-loop applications and investigating feedback mechanisms to dynamically adapt stimulus selection based on real-time neural activity could reveal powerful new insights into neural circuits.
More visual insights#
More on figures

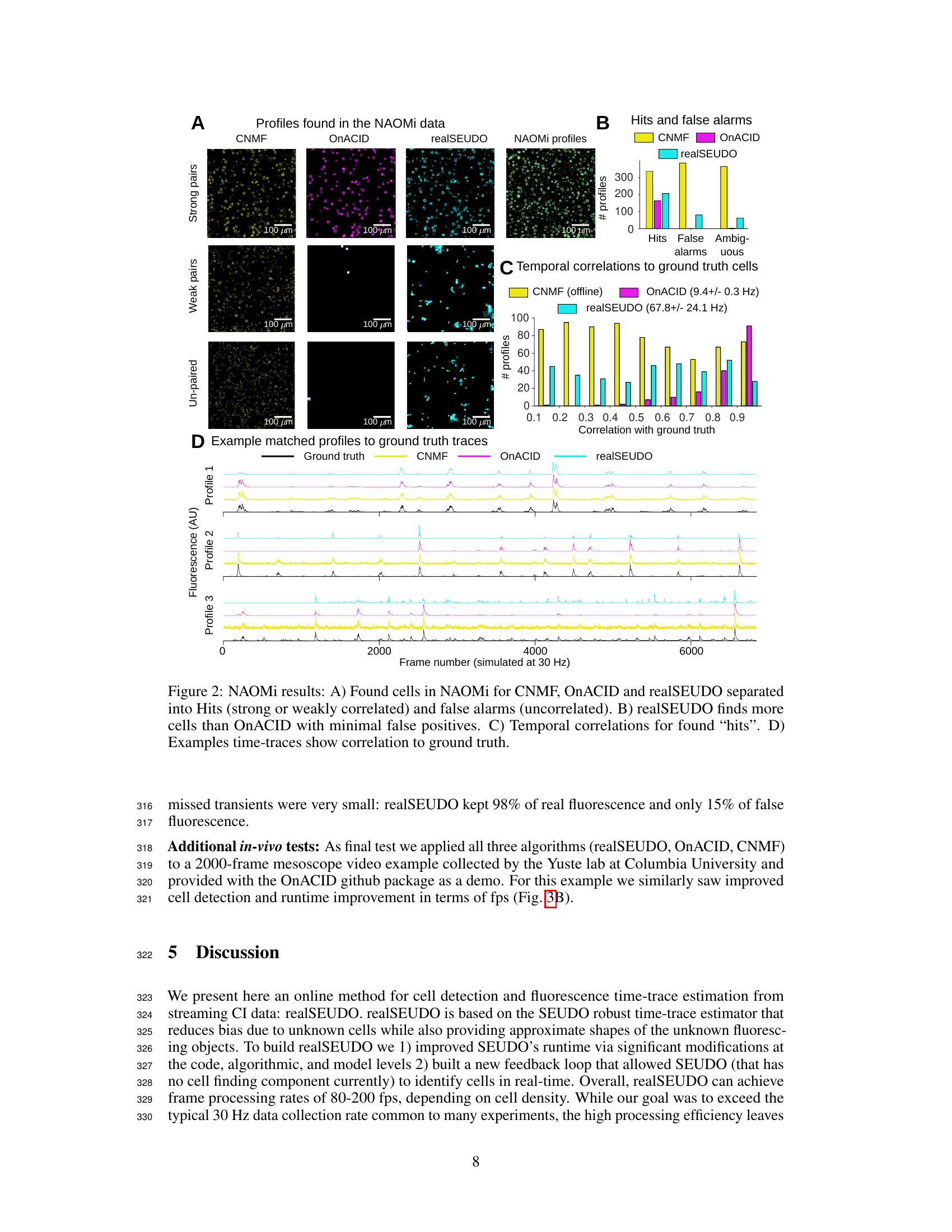
This figure demonstrates the performance of three different algorithms, CNMF, OnACID, and realSEUDO, on a simulated calcium imaging dataset generated by NAOMi. Panel A shows the spatial distribution of cells identified by each algorithm, with strong, weak, and unpaired matches indicated. Panel B compares the number of hits (true positives), false alarms (false positives), and ambiguous cases for each algorithm. Panel C displays the temporal correlation between identified cells and ground truth cells, with realSEUDO exhibiting higher correlation. Finally, Panel D shows example time traces for selected cells, illustrating the agreement between the algorithms and ground truth.

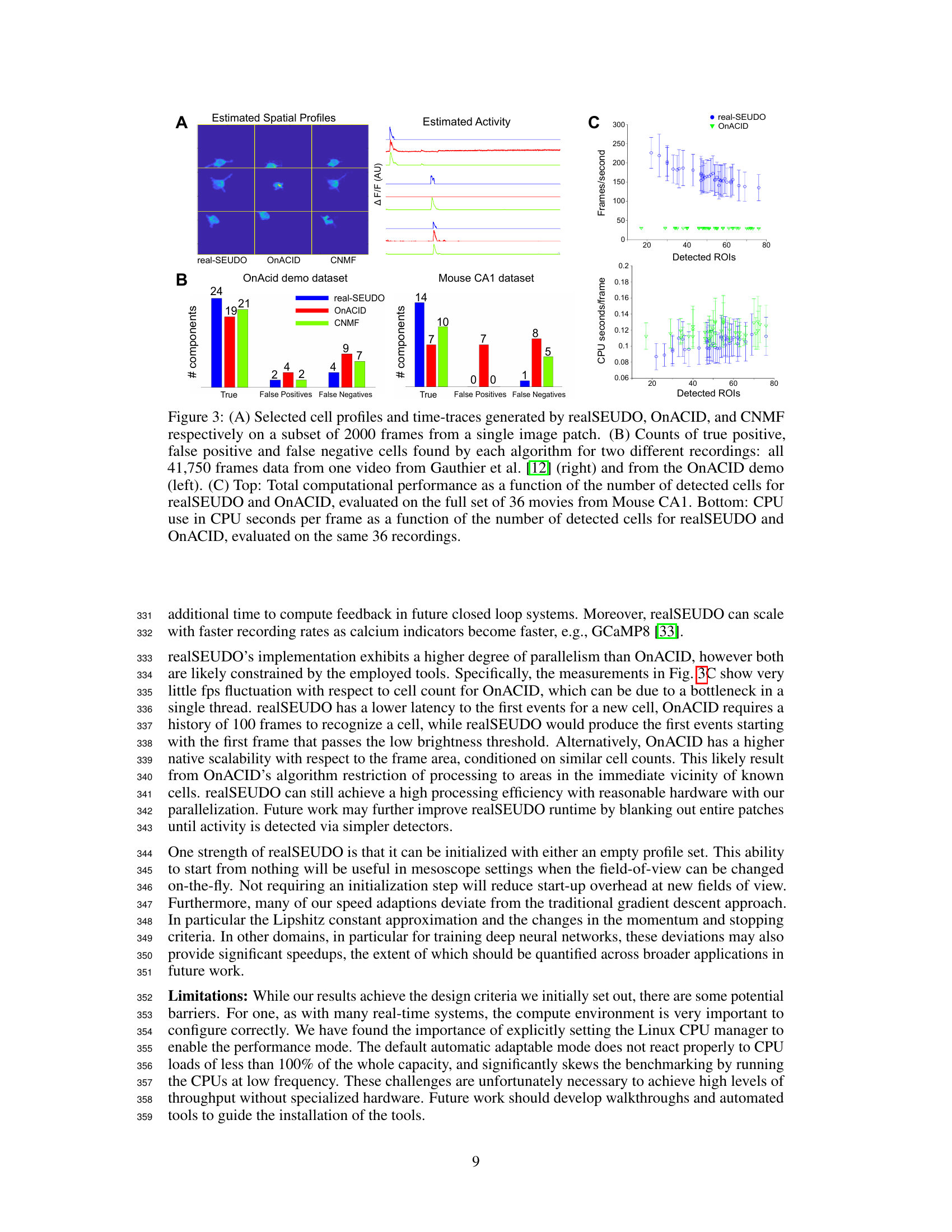
This figure presents a comparison of realSEUDO, OnACID, and CNMF performance on calcium imaging data. Panel A shows example spatial profiles and corresponding temporal traces for a subset of cells detected by the three methods. Panel B provides a quantitative comparison of the number of true positives, false positives, and false negatives for each method on two different datasets. Panel C shows a comparison of the computational performance (frames per second and CPU seconds per frame) of realSEUDO and OnACID as a function of the number of detected ROIs, demonstrating the superior efficiency of realSEUDO.

This figure compares the results of three different calcium imaging analysis methods: realSEUDO, OnACID, and CNMF. It shows the detected cells for each method, color-coded for easy identification. The figure also includes selected time traces for matching cells across the three methods. This allows for a visual comparison of the cell detection accuracy and the similarity of the resulting time traces across the different algorithms. The visual comparison highlights the strengths and weaknesses of each method in terms of cell detection and time trace estimation.

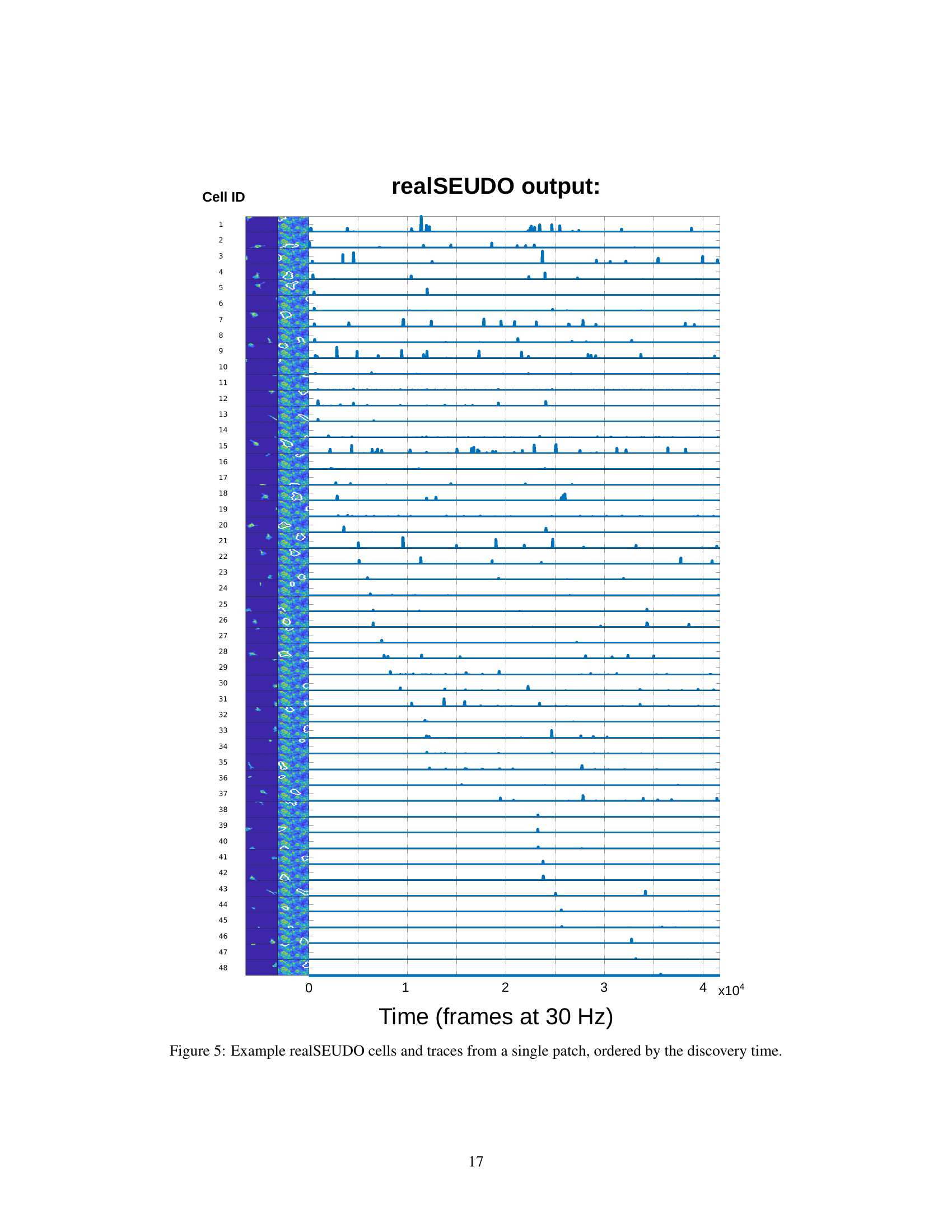
This figure displays the output of the realSEUDO algorithm for a single patch. It shows the detected cells (on the left) and their corresponding activity traces (on the right). The cells are ordered by the time they were discovered during the real-time processing. Each row represents a single cell, with the left side showing the spatial location and shape of the cell, and the right side showing its activity trace over time. The y-axis represents the cell ID, and the x-axis represents the time in frames at 30Hz. The color intensity in the activity trace indicates the level of activity. The figure demonstrates realSEUDO’s ability to identify and track the activity of individual neurons in real time.
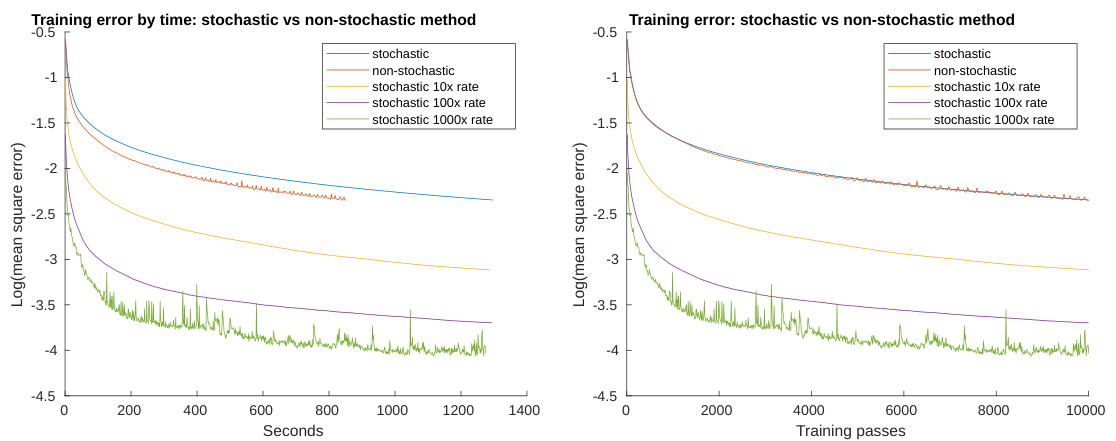
This figure compares the performance of different optimization algorithms on a handwritten digit recognition task. The left panel shows the mean squared error (MSE) as a function of the time taken for optimization. The right panel displays the MSE as a function of the number of training passes. The algorithms being compared include a stochastic gradient descent method and several variations at different learning rates (10x, 100x, 1000x), and a non-stochastic momentum method. The plot illustrates the trade-offs between speed and accuracy in these different approaches to optimization.

This figure displays the complete sets of strongly correlated activity traces obtained using three different calcium imaging analysis methods: CNMF (an offline method), OnACID (an online method), and the proposed realSEUDO algorithm. Each column represents the results from one of these methods. The figure showcases the temporal dynamics of neuronal activity, allowing for a visual comparison of the accuracy and performance across the three algorithms.

This figure compares the results of three different calcium imaging analysis algorithms: CNMF, OnACID, and realSEUDO. For each algorithm, it shows a heatmap representation of the activity of multiple neurons over time. The heatmaps visualize the fluorescence intensity of each neuron across a series of frames (simulated at 30 Hz). The x-axis represents the frame number, and the y-axis represents the neuron ID. Each color in the heatmap corresponds to a range of fluorescence intensities. The figure also displays the spatial profiles of neurons detected by each algorithm. This visual comparison allows for a direct assessment of the performance of each algorithm in terms of its accuracy, efficiency, and robustness in identifying neurons and extracting their temporal activity patterns from the raw fluorescence data.
Full paper#